Abstract
Pathogens secrete effector proteins to suppress host immunity, mediate nutrient uptake and subsequently enable parasitism. However, on non-adapted hosts, effectors can be detected as non-self by host immune receptors and activate non-host immunity. Nevertheless, the molecular mechanisms of effector triggered non-host resistance remain unknown. Here, we report that a small cysteine-rich protein PstSCR1 from the wheat rust pathogen Puccinia striiformis f. sp. tritici (Pst) activates immunity in the non-host solanaceous model plant Nicotiana benthamiana. PstSCR1 homologs were found to be conserved in Pst, and in its closest relatives, Puccinia graminis f. sp. tritici and Puccinia triticina. When PstSCR1 was expressed in N. benthamiana with its signal peptide, it provoked the plant immune system, whereas no stimulation was observed when it was expressed without its signal peptide. PstSCR1 expression in N. benthamiana significantly reduced infection capacity of the oomycete pathogens. Moreover, apoplast-targeted PstSCR1 triggered plant cell death in a dose dependent manner. However, in Brassinosteroid insensitive 1-Associated Kinase 1 (SERK3/BAK1) silenced N. benthamiana, cell death was remarkably decreased. Finally, purified PstSCR1 protein activated defence related gene expression in N. benthamiana. Our results show that a Pst-secreted protein, PstSCR1 can activate surface mediated immunity in non-adapted hosts and contribute to non-host resistance.
Introduction
Surface localized pattern recognition receptors (PRRs) mediate pathogen associated molecular pattern (PAMP)-triggered immunity (PTI) against a variety of plant pathogens1–3. PTI initiates the generation of reactive oxygen species (ROS), synthesis of salicylic acid, plant defence gene expression, stomatal closure and callose accumulation4. In some cases, surface immune receptors stimulate localized programmed cell-death, known as the hypersensitive response (HR)5, although what determines the decision for cellular suicide remains unclear. Nevertheless, adapted pathogens can either evade or suppress PTI by secreting a wide range of effector proteins into the apoplast, cytoplasm, and other host subcellular compartments6–8. Apoplastic effectors can interact with surface proteins and other extracellular molecules8 such as defence-related enzymes to perturb their functions, enabling parasitism9–13. Some known apoplastic effectors include cell wall-degrading enzymes, toxins, ethylene inducing peptides, and small cysteine-rich (SCR) proteins. Since SCR effectors can form disulfide bonds, they are thought to be more stable in the harsh conditions of apoplast8, 14. Some effectors are known to stimulate cell death through surface localized immune receptors8, 15–18. Despite major progress made in previous decades, the biochemical functions and the host interactors of apoplastic effectors are largely unknown. Particularly in pathogenic fungi, it has been difficult to determine whether an effector is apoplastic, or host-translocated, due to the absence of canonical amino acid sequence motifs as seen in oomycete RXLR effectors19, 20. Discovering how effectors operate in different subcellular compartments is critical for understanding the mechanisms of host-pathogen interactions that will eventually lead to new ways of engineering plant disease resistance.
Wheat yellow rust disease, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the major threats to global wheat production21. Although genomics studies helped identification of numerous candidate Pst effectors22–24, their modes of action are yet to be discovered. Dissecting the functions of effectors has been difficult due to absence of effective tools for functional gene analysis in crops25, 26. As an alternative, N. benthamiana serves as a model plant to study molecular plant-microbe interactions27. Recently, Petre et al. have reported that N. benthamiana is a feasible experimental tool to functionally analyse candidate effectors from Pst, a fungal pathogen of wheat28.
In this work, using N. benthamiana as a model system, we studied subcellular localization, function and response to pathogen infections of PstSCR1, which was previously predicted as a candidate effector23. Our data allowed us to conclude that PstSCR1 is an apoplastic effector of Pst, which is recognized in PAMP-triggered immunity. In non-adapted hosts, effectors can assist to explore the components of non-host resistance and discover novel participants of plant immunity.
Results
PstSCR1 is a Puccinia specific effector induced during infection
Fifteen candidate Pst effector gene expressed sequence tags (ESTs) were reported previously23. Among these, six have been further examined for developmental stage-specific gene expression23. The EST “GH737102” sequence that encodes PstSCR1 appeared as a full-length cDNA possessing a putative signal peptide (SP) and is expressed nearly 120 times more in infected leaves of wheat than in urediniospores23. Blastp showed that the candidate effector has 14 hypothetical homologues in Pst, Puccinia graminis f. sp. tritici and Puccinia triticina. We noted that PstSCR1 (also known as Pstha2a523) protein sequence has three conserved (Y/F/W)x(C) motifs (Supplementary Fig. S1), one of which is located at the N-terminus as described in many wheat rust and other fungal effector candidates29–32. In order to test whether PstSCR1 is expressed during Pst infection of wheat, we employed qPCR (Supplementary Fig. S2) using infected samples collected at different time points (24-h, 72-h, 8-d and 10-d). The PstSCR1 was expressed highly between 72-h post-infection (hpi) to 8-d post-infection (dpi), but its expression was reduced at 10-dpi (Supplementary Fig. S3). Sequencing of the isolated PCR product at 8-dpi showed a perfect match with the reported EST sequence of PstSCR1. These results show that PstSCR1 family is exclusively conserved within three closely related Puccinia species.
Apoplast targeted PstSCR1 enhances plant immunity against oomycete pathogens in N. benthamiana
To determine the extent to which PstSCR1 alters plant immunity, we expressed it in N. benthamiana leaves with its SP (PstSCR1) or without its SP (ΔSP-PstSCR1) and performed infection assays using the hemibiotroph Phytophthora infestans and an obligate biotroph Peronospora hyoscyami f. sp. tabacina. Expression of PstSCR1 targeted to the apoplast significantly reduced infection of both P. infestans (Fig. 1a and b) and P. tabacina (Fig. 1c) whereas expression of cytoplasmic ΔSP-PstSCR1 had no effect (Fig. 1). We validated the subcellular localizations of PstSCR1 and ΔSP-PstSCR1 using established subcellular markers in N. benthamiana. We showed that both RFP- and GFP-labeled PstSCR1 were secreted to the apoplast (Fig. 2), whereas ΔSP-PstSCR1 remained cytoplasmic (Fig. 2c).
Figure 1.
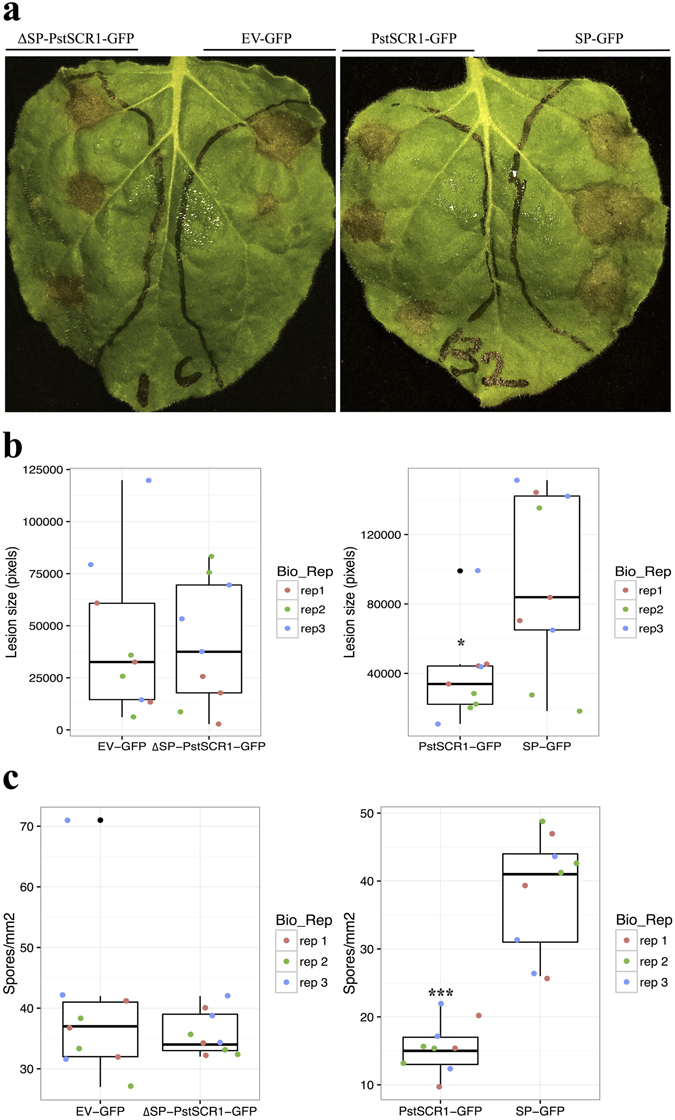
Expression of PstSCR1 reduces infection capacity of oomycete pathogens. (a) The infection of N. benthamiana leaf with P. infestans after expressing ΔSP-PstSCR1-GFP and PstSCR1-GFP constructs. Photographs were taken after 8-dpi. (b) P. Infestans lesion sizes were reduced in leaves expressing PstSCR1-GFP in a SP-dependent manner. Leaf patches expressing PstSCR1-GFP showed significantly smaller lesions compared to leaves expressing SP-GFP, whereas patches expressing ΔSP-PstSCR1-GFP showed similar lesion sizes to those expressing EV-GFP, as measured in pixels (by ImageJ tool). Asterisk indicates significant differences by ttest (*P ≤ 0.05). (c) Peronospora hyoscyami f. sp. tabacina spore count was reduced in leaves expressing PstSCR1-GFP in a SP-dependant manner. N. benthamiana leaves were infected with P. tabacina and spores were counted 8-dpi. Leaf patches expressing PstSCR1-GFP showed significantly less spores than patches expressing SP-GFP, whereas patches expressing ΔSP-PstSCR1-GFP showed similar spores to those expressing EV-GFP. Asterisks indicate significant differences by ttest (***P ≤ 0.001).
Figure 2.
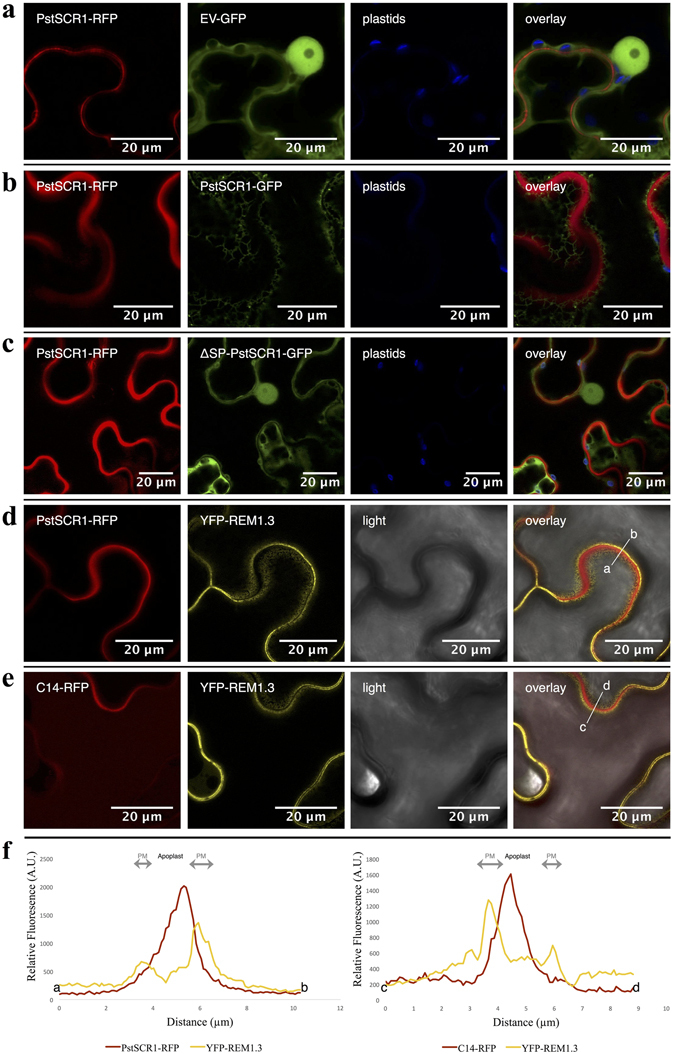
PstSCR1 accumulates in the plant apoplast. N. benthamiana plants were co-expressed by agro-infiltration using the following constructs: (a) pGWB454/PstSCR1-RFP and pK7FWG2/EV-GFP, (EV: empty vector), as nucleo-cytoplasmic marker; (b) pGWB454/PstSCR1-RFP and pK7FWG2/PstSCR1-GFP; (c) pGWB454/PstSCR1-RFP and pK7FWG2/ΔSP-PstSCR1-GFP. (d) pGWB454/PstSCR1-RFP and pK7/YFP-REM1.3. (e) pGWB554/C14 (Apoplast and vacuole marker47) and pK7WGY2/REM1.3 (Plasma membrane marker58–60). (d) and (e) indicate PstSCR1 expressed with its SP accumulates in apoplastic space but not at the plasma membrane. (f) The intensity plots illustrate relative RFP and YFP fluorescence signals along the line connecting the points; a-b and c-d in overlayed images of (d) and (e), respectively.
Our immunoblot analysis of immunoprecipitates obtained from total protein extracts expressing PstSCR1 fusion constructs revealed expected sized fragments (Supplementary Fig. S4). Nevertheless, apoplastic PstSCR1 was more stable than cytoplasmic ΔSP-PstSCR1 possibly due to inefficient folding in the cytosol (Supplementary Fig. S4). These results suggest that PstSCR1 is an apoplastic effector whose expression in N. benthamiana results in enhanced disease resistance either due to its activation of surface immune receptors or adverse effects of its virulence function in a non-host plant.
Overexpression of PstSCR1 induces cell death in N. benthamiana
Next, to further determine the effect of PstSCR1 on plant immunity, we transiently overexpressed PstSCR1 in N. benthamiana with and without a FLAG-tag. Four days post agro-infiltration (dpai) we observed cell death, whereas overexpression of either ΔSP-PstSCR1 with a FLAG-tag, or GFP with a SP (SP-GFP), did not result in cell death (Fig. 3). Therefore the PstSCR1 secretion signal is not only indispensible for apoplastic targeting (Fig. 2) but also for its cell death–inducing activity (Fig. 3)15, 33, 34. We found that cell death was only observable with PstSCR1-FLAG when expressed in the strong pTRBO vector. Expressing PstSCR1 in pK7FWG2 vector did not generate any observable cell death (Supplementary Fig. S5).
Figure 3.
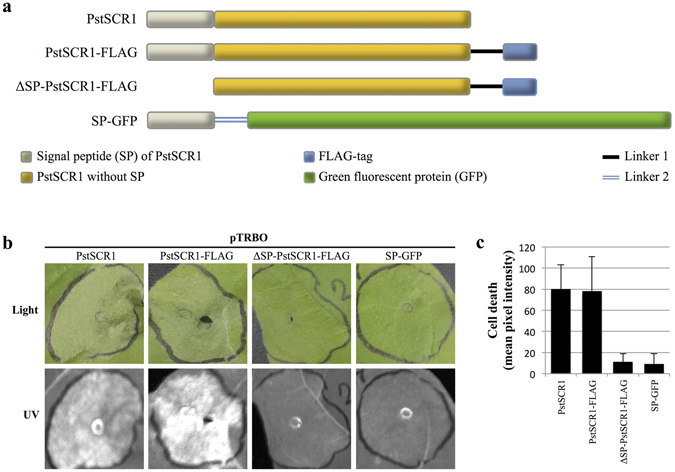
Secretion of PstSCR1 is required to induce cell death in N. benthamiana. (a) The schematic view of constructs used in the experiment. (b) PstSCR1 overexpressed the pTRBO vector with a secretion signal causes HR. Shown are representative N. benthamiana leaf patches expressing pTRBO/PstSCR1, pTRBO/PstSCR1-FLAG, pTRBO/ΔSP-PstSCR1-FLAG and pTRBO/SP-GFP, 4-dpai. (c) Cell death quantification of infiltrated leaf regions. Pixel intensities were normalized by subtracting background in non-infiltrated zone. Error bars represent standard deviations.
To determine the extent to which apoplastic PstSCR1 stimulates cell death, we performed cell death assays using apoplastic extracts in the presence or absence of PstSCR1 (Fig. 4). We observed cell death in a dose dependent manner in leaves infiltrated with apoplastic fluid with PstSCR1, but not with the secreted GFP control (Fig. 4). We analysed apoplastic fluid and protein extract of remnant leaves that were used for apoplastic fluid isolation on immunoblots (Supplementary Fig. S6). The theoretical size of the PstSCR1-FLAG is smaller than the size observed on blots, which implies post-translational modification(s). In the control samples, Anti-GFP western blotting revealed that GFP is present both in apoplast and total protein extract. However, Anti-FLAG antibody did not detect SP-GFP-FLAG in the apoplastic extract, suggesting FLAG-tag was cleaved and SP-GFP-FLAG is prone to proteolysis in the apoplast (Supplementary Fig. S6). These results demonstrate PstSCR1 is stable in the apoplast, thus providing the possibility that it is functional.
Figure 4.
Apoplastic fluid containing the secreted PstSCR1 triggers cell death in N. benthamiana in a dose-dependent manner. The infiltration of N. benthamiana was conducted with apoplastic fluid containing processed PstSCR1. (a) Shown is a representative leaf of N. benthamiana infiltrated with apoplastic fluid (with no dilution (600–700 μg/μL), 1:3 and 1:10 dilutions in ddH2O) from N. benthamiana expressing PstSCR1-FLAG and SP-GFP-FLAG. Four days after apoplastic fluid infiltration, the leaves were examined under normal light and UV exposure. (b) Cell death quantification of infiltrated leaf regions. Pixel intensities were normalized by subtracting background in non-infiltrated zone. Error bars represent standard deviations.
NbBAK1 silencing leads to reduced cell death triggered by PstSCR1
The plant receptor-like kinase SERK3/BAK1 is involved in response to PAMP molecules and is a key participant of the PTI response35–37. To check whether SERK3/BAK1 is involved in cell death by PstSCR1, we tested SERK3/BAK1-silenced N. benthamiana plants with apoplastic fluid from tissue expressing secreted PstSCR1. In SERK3/BAK1 silenced N. benthamiana leaves (Supplementary Fig. S7) cell death was decreased (Fig. 5). On the other hand, in the GFP-silenced samples no change was observed (Fig. 5). Therefore, this indicates that PsrSCR1 triggered cell death is a BAK1-dependent process, which presumably requires a surface immune receptor yet to be characterized.
Figure 5.
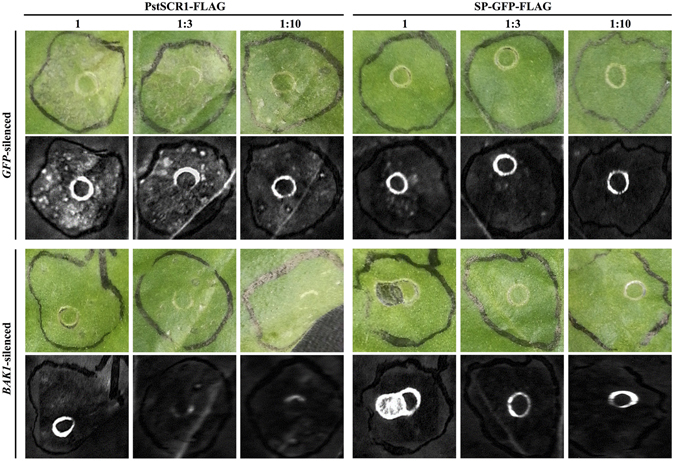
NbBAK1 is required for PstSCR1 triggered cell death. Apoplastic fluid from plant samples expressing PstSCR1-FLAG and SP-GFP-FLAG were infiltrated into NbBAK1 and GFP silenced N. benthamiana with varying amounts; 1 ((no dilution), 1:3 and 1:10) dilutions. Following 4-d after apoplastic fluid infiltration, the leaves were examined under normal light and UV exposure.
PTI marker genes are induced with PstSCR1 injection
To further illustrate that PstSCR1 triggers PTI-like responses, we decided to analyse its effect on defence-related gene up-regulation. We chose two N. benthamiana genes namely NbCYP71D20 (a putative cytochrome P450) and NbACRE31 (a putative calcium-binding protein), which are induced upon PAMP treatment38, 39. We purified PstSCR1 from the apoplastic fluid and infiltrated it into N. benthamiana leaves. The defence gene, NbACRE31 was activated early, 2-d after purified PstSCR1 infiltration and remained stable34 at 4-d but the activation of NbCYP71D20 took place later (4-d) (Supplementary Fig. S8). The expression of these defence genes was not detected in control leaves that were treated with SP-GFP-FLAG immunoprecipitated by Anti-FLAG from apoplastic fluid of N. benthamiana leaves.
Discussion
The apoplast is a hostile environment and critical barrier for plant pathogens to overcome. Although remarkable progress has been made in identifying defence-related pathways targeted by pathogen effectors, the information on how pathogens pass the early barrier of plant immunity is very limited40. Here we report that a small, cysteine-rich effector-like protein PstSCR1 secreted by Pst triggers PTI responses at the cell surface. Consistent with this, heterologous expression of the PstSCR1 targeted to the extracellular space in N. benthamiana enhanced disease resistance against the oomycete pathogens P. infestans and P. tabacina. Thus, PstSCR1 secreted by the yellow rust fungus carries the characteristics of a proteinaceous PAMP that activates immunity in a non-host solanaceous plant N. benthamiana.
During infection of wheat with the Pst-78 strain, PstSCR1 gene expression was highly induced at 3-dpi, with peak expression observed at 8-dpi, which is consistent with previous gene expression studies of PstSCR1 23. The up-regulation of PstSCR1 coincides with development of the fungal mycelium (2- to 8-dpi), in which generation of haustoria and haustorial mother cells, and formation of the pustule bed occurs for development of the uredinium and sporulation41. While we failed to detect PstSCR1 gene expression at 24 hpi on the host plant wheat, we cannot exclude the possibility that it is expressed at early time points of infection on a non-host such as N. benthamiana. It is likely that multiple factors contribute to non-host resistance, and indeed other Pst effectors or secreted proteins may be involved. According to the phylogenetic tree we generated (Fig. S1B), the clade of PstSCR1 exclusively includes homologs from Pst. Moreover, from our database search, all PstSCR1 homologs found are conserved within closely related Puccinia species. Thus, PstSCR1 homologs may have evolved rapidly within Puccinia species.
Like apoplastic effectors, fluorescent fusion proteins of PstSCR1 show accumulation in the apoplast in a signal peptide dependent manner (Fig. 2)15, 33, 42. Expression of apoplast-targeted PstSCR1 in N. benthamiana increased resistance to oomycete pathogens, whilst cytoplasmic PstSCR1 did not (Fig. 1). The enhanced disease resistance against the oomycete pathogens triggered by PstSCR1 could be due to activation of PTI by PstSCR1 and/or direct competition between PstSCR1 and oomycete effectors for the host susceptibility factors. In line with this, purified PstSCR1 triggered defence related gene expression, which is one of the hallmarks of the PTI responses. Thus, our results are consistent with the view that PstSCR1 is a secreted rust protein that triggers PTI responses.
When highly expressed by the tobacco mosaic virus (TMV) based pTRBO vector43, full length PstSCR1 triggered cell death but not in the absence of its signal peptide. The cell death did not occur when PstSCR1 was expressed by the weaker 35S promoter (Fig. S5), however it was sufficient to limit oomycete infections (Fig. 1). The cell death triggered by higher expression of PstSCR1 can be explained by two possibilities: Firstly, over production of PstSCR1 could hyper activate the PTI machinery resulting in HR like cell death. Secondly, PstSCR1 might show a toxic effect due to its overrepresentation in the apoplast perhaps by non-specifically perturbing the extracellular environment. However, our results favour the former, as silencing of NbBAK1, one of the main components of the PTI signalling pathways, compromised cell death induced by PstSCR1 (Fig. 5). Thus, the cell death phenotype observed with higher gene expression of PstSCR1 is most likely due to over activation of the PTI signalling pathway.
Our results suggest that PstSCR1 is recognized in non-host plant N. benthamiana by an undetermined surface immune receptor that requires NbBAK1 for signalling. The finding that the oomycetes were incapable of fully suppressing this immune response indicates that this could be a divergent PTI pathway that cannot be suppressed by the oomycete effectors effectively. This would explain the increased resistance against oomycete pathogens stimulated by PstSCR1 when expressed at low levels that does not activate HR.
The molecular mechanisms that mediate non-host resistance are poorly characterized. It is likely that multiple factors affect this phenomenon. Our results demonstrate that pathogen secreted proteins such as PstSCR1 might fail at immune evasion in non-host plants and contribute to non-host resistance rather than serving as virulence cues. In line with this, Kettles et al., showed that effector candidates from the wheat pathogen Zymoseptoria tritici are recognized in N. benthamiana 34. Previous studies showed that many PRRs strongly associate with NbBAK1 upon ligand binding. Recently, Saur et al., demonstrated that BAK1 can be used as a bait to identify PRRs upon ligand activation44. Thus, elicitors like PstSCR1 can be exploited as tools to discover novel immune receptors that mediate non-host resistance. Interfamily transfer of plant PRRs were proven to be effective to engineer enhanced disease resistance45. Therefore, once identified, the PRR that responds to PstSCR1 in Nicotiana can be transferred to wheat plants to improve disease resistance.
Materials and Methods
Bioinformatics tools
The cDNA sequence of the candidate effector was obtained from National Center of Biotechnology Information (NCBI) with EST accession number of GH737102. The open reading frame (ORF) of the EST was predicted by ORF-Finder of NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The signal peptide (SP) sequence was determined by SignalP 4.1 online tool (http://www.cbs.dtu.dk/services/SignalP/)46. The amino acid sequence was analysed via blastp at the NCBI to detect the closely related sequences.
Cloning
The primers used in the study are listed in Table S1, presented in supplementary information. The vectors and the constructs, pTRV1, pTRBO, pTRBO/GFP and pTRBO/FLAG-RFP, were gifts from Kamoun Lab, Sainsbury laboratory, Norwich, UK. The vectors (pGWB554/C14 and pK7WFY2/REM1.3) expressing C14:RFP and YFP:REM1.3 were as in Bozkurt et al. and Raffaele et al., respectively47, 48. The PstSCR1 gene had made synthesized with SP, and PacI and NotI restriction site extensions corresponding to N-terminus and C-terminus, respectively and the construct was obtained as pBSK/PstSCR1 (GeneScript). For the subcellular localization of PstSCR1 experiments, the gene was re-amplified with and without SP from pBSK/PstSCR1 with CACC-SP or CACC-ATG-SCR1 (without SP) as forward primer and SCR1-noSTP as reverse primer and cloned into pENTR/D-TOPO vector (Invitrogen) and recombined with the two different destination vectors; pK7FWG249 and pGWB45450 through LR clonase reaction (Invitrogen). For the overexpression, the effector with (PstSCR1) and without SP (ΔSP-PstSCR1) and FLAG-Tag on the C-terminus was constructed by PCR using forward primers; PacI-SP-fw or PacI-noSP-SCR1-fw and reverse primers; SCR1C-FLAGRev2 and SCR1C-FLAGRev1. The final PCR product was obtained with PacI restriction site on the 5′-end and FLAG-Tag and NotI restriction site on the 3′-end. The PCR products were cloned into pTRBO (pJL48) vector43 and labelled as pTRBO/Pst SCR1-FLAG and pTRBO/ΔSP-PstSCR1-FLAG. To express secreted GFP (SP-GFP), SP was amplified with CACC-SP and SP-noSTP primers using pBSK-PstSCR1 as a template; then, cloned into pENTR/D-TOPO, and followed by LR recombination (Invitrogen) into pK7FWG249 and labelled as pK7FWG2/SP-GFP. For the cloning of SP-GFP-FLAG, SP-GFP was first amplified with PacI-SP-fw and GFP-FLAG-Rev primers using pK7FWG2/SP-GFP as a template. SP-GFP-FLAG was generated using the amplified product as a template with the primers PacI-SP-fw and SCR1-C-FLAGRev1 as forward and reverse primers, respectively. The SP-GFP-FLAG was cloned into pTRBO (pJL48)43 and denoted as pTRBO/SP-GFP-FLAG. For the cloning and the amplification of the plasmids, the constructs were maintained in E. coli Top10 strain. In Agro-infiltration experiments, the constructs were introduced into Agrobacterium tumefaciens GV3101 strain by electroporation.
Agro-infiltration assays
Nicotiana benthamiana plants were grown at 20–24 °C with 16-light/8-dark cycle in a growth room. Four to six week-old plant middle leaves were used for agro-infiltration assays. Agrobacterium-mediated gene transfer was conducted with minor modifications as described elsewhere51. A. tumefaciens (GV3101, pMP90) culture was pelleted by 5 min, at 4000 rpm in room temperature (RT) and the cell pellet was washed with distilled water, washing was repeated two more times. The cells were suspended in Agro-induction medium (10 mM MES pH 5.6, 10 mM MgCl2). The concentration of the suspension was adjusted to 0.2 A600 for infiltration. The infiltrated leaves were collected after 2–4 days post-agroinfiltration (dpai) depending on the expression level for microscopic imaging, apoplastic fluid isolation and total protein extraction.
Apoplastic fluid isolation
The Agrobacterium (with pTRBO/PstSCR1-FLAG or pTRBO/SP-GFP-FLAG) infiltrated N. benthamiana leaves of 2–3 dpai were used to obtain apoplastic fluid as in the method previously described52. Briefly, the leaves were detached, rinsed with distilled water, carefully folded, placed in a 60-mL syringe filled with distilled water, and vacuum was applied for 5–15 seconds repeatedly until the leaves appeared as dark translucent. The leaves were wiped with clean tissue or filter paper, and sandwiched in parafilm sheets, rolled and placed in a 20-mL syringe, centrifuged in 50 mL falcon tube for 10 min at 1,000 g, at 4 °C. The collected apoplastic fluid (400–500 μL/1.0–1.5 g leaf sample) was centrifuged at 15,000 g for 5 min. The supernatant, having 600–700 μg/μL total protein concentration, was transferred to a fresh tube on ice and stored at −80 °C later use or utilized on the same day.
Apoplastic fluid infiltration
Apoplastic fluid samples obtained from various constructs containing Agrobacterium-infiltrated N. benthamiana were infiltrated into fresh N. benthamiana leaves until the injected area reaches to the size of a penny. The samples and the controls were infiltrated side by side on the same leaf with no or various dilutions (1 (no dilution), 1:3 and 1:10) in ddH2O. The presence of or the level of hypersensitive response (HR) was examined 4–5 days after apoplastic fluid infiltration.
PstSCR1 purification
Anti-FLAG M2 affinity gel (Sigma, A2220) was mixed gently by pipetting with a cut tip. After resin completely suspended, 50 (or 250 for large scale) μL of it was placed into a 50 mL falcon tube containing 250 μL (25 mL for large scale) apoplastic fluid or protein extract diluted (1:1) with IP buffer ((10% glycerol, 25 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA) freshly mixed, 0.1% (v/v) Tween 20). The tube was mixed turning end-over-end at 4 °C for 3-hour. After the incubation, resin was pelleted at 800 g for 30 sec, and supernatant was removed; then pellet was re-suspended in 1 mL IP buffer and transferred into fresh microcentrifuge tube. The wash was repeated four times. After last wash, the remnant liquid was removed very carefully by using syringe with needle not to remove the beads. Immobilized proteins were eluted from the beads in re-suspension solution of 500 μL IP buffer containing 150 ng/μL FLAG tag peptide and the tube was gently shaken in horizontal position for 2-hour, at 4 °C. Eluted proteins (supernatants) were transferred into fresh tube.
Infiltration of purified PstSCR1 into N. benthamiana
Apoplastic-purified PstSCR1 samples were diluted (1:10) with ddH2O and injected into fresh N. benthamiana leaves until the whole leaf infiltrated. As a control, immunoprecipitation sample obtained from apoplastic fluid of N. benthamiana expressing SP-GFP-FLAG was used. Two and four days after purified PstSCR1 protein infiltration, the expression levels of NbCYP71D20 and NbACRE31 were determined by qPCR.
Virus induced gene silencing of NbBAK1
NbBAK1 was silenced using the agrobacterium-mediated co-infiltration of the clones containing Tobacco Rattle Virus as pTRV2/BAK1 and pTRV1 with A600 ratio of 2:1 (0.4:0.2, respectively). As a viral control, Agro-pTRV2/GFP:pTRV1 (2:1) were co-infiltrated53. The newly emerged leaves of three weeks post-silenced tobacco were used for the injection of apoplastic fluid obtained from PstSCR1 expressing leaves.
Infection assay
The effect of PstSCR1 expression during pathogen growth was assayed by transiently expressing pK7FWG2/PstSCR1-GFP on one half and pK7FWG2/SP-GFP on the other half of N. benthamiana leaves (4–5 weeks old). Following 4–5 hour post-infiltration (hpi), leaves were detached and either Phytophthora infestans 88069 or Peronospora hyoscyami f. sp. tabacina were inoculated as 3 spots of 10 μL of cultures of each, into each half of the infiltrated area of the leaves54, 55. The lesion diameter of Phytophthora infestans growth and the number of spores of Peronospora hyoscyami f. sp. tabacina were recorded at 8-dpi. The infection assay experiments were repeated on at least three independent N. benthamiana leaves.
Immunoblotting
N. benthamiana leaves were grinded in liquid nitrogen, and 1 g of leaf powder was dissolved in a 2-mL extraction buffer GTEN (10% glycerol, 25 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA) freshly mixed with 2% (w/v), polyvinylpolypyrrolidone, 1X protease inhibitor, (Thermo #88666) and 10 mM dithiothreitol, 0.1% (v/v) Tween 20. The cell debris was pelleted by centrifugation and supernatant was collected in a fresh tube (first with 3,000 g for 10 min, and again twice with 21,000 g for 10 min in microcentrifuge at 4 °C). For western blot analysis, anti-FLAG (Thermo MA1-91878) and anti-GFP (Thermo MA5-15256) antibodies were used as the primary, and anti-mouse Alkaline Phosphatase conjugated antibody (Chemicon International #AP308A) was used as the secondary antibody.
Confocal microscopy
N. benthamiana leaves after 2–3 dpai were cut in small pieces, immersed in distilled water and imaged on Leica 385 TCS SP5 confocal microscope (Leica Microsystems, Germany). The excitation of GFP, YFP and RFP probes were performed by 488, 514 and 561 nm laser diodes, respectively, and fluorescent emissions were detected at 495–550 (GFP and YFP) and 570–620 nm (RFP). For chloroplast autofluorescence, far infrared (>800 nm) excitation and emission were used.
qPCR
The total RNA of 100 mg leaf sample was extracted using RNeasy Mini Kit (Qiagen). The first strand cDNA was synthesized in 20 μL reaction volume from 800 ng total RNA using Transcriptor First strand cDNA synthesis Kit (Roche) according to the instructions suggested by the manufacturer. qPCR was conducted using AccuPower GreenStarTM qPCR Premix (BIONEER) with 10 μL of 1:20 diluted cDNA. To quantitative analysis of NbSERK3/BAK1, NbCYP71D20 and NbACRE31, combinations of NbSerk3-qRT-F and NbSerk3-qRT-R, NbCYP71D20-F and NbCYP71D20-R, and NbACRE31-F and NbACRE31-R primers were used, respectively56. EF1α-qRT-F and EF1α-qRT-R were used amplification of Elongation factor 1 alpha gene (EF1α), used as constitutively expressed reference gene15, 57. PstSCR1-SP-5-UTR-F and PstSCR1-Rev primers used to amplify the PstSCR1 transcripts. The primer sequences used in qPCR amplifications are illustrated in Supplementary Table S1.
Electronic supplementary material
Acknowledgements
This study was funded by TUBITAK with projects KBAG-110T445 and COST-113Z350 (call FA1208), and METU research funds, PI: MSA. BD was supported by TUBITAK (BIDEB-2215) and granted by COST-STSM-FA1208. ACO and AA were supported by TUBITAK (BIDEB-2211/C). We acknowledge Dr. Xianming Chen for supplying yellow rust Pst-78 spores. We thank Kamoun Lab, Sainsbury Laboratory, Norwich, UK for providing some of the vectors, and Nakagawa Lab, Shimane University, Matsue, Japan for pGWB454 clone. We are grateful to Prof. Shan-Ho Chou of National Chung Hsing University, Taiwan for expressing PstSCR1 in E. coli in attempts to crystallize, and Assist. Prof. Salih Ozcubukcu of Chemistry Department of METU for synthesis of FLAG peptide.
Author Contributions
All of the experiments were conducted and optimized by the leading author, B.D., as part of the PhD thesis study under supervision of PI, MSA; except the qPCR and wheat infection with Pst-78, which were conducted and performed by A.C.O. and A.A., respectively. M.S.A. supervised the research; designed and evaluated the data with B.D., T.O.B. supervised B.D. during COST-STSM FA1208. C.D. repeated the experiment presented in Supplementary Fig. S5. C.D. also performed intensity plot in Figure 2 and quantification of cell death in Figures 3 and 4.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01100-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 3.Win J, et al. Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 2012;77:235–247. doi: 10.1101/sqb.2012.77.015933. [DOI] [PubMed] [Google Scholar]
- 4.Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009;150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomma BPHJ, Nürnberger T, Joosten MHAJ. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell. 2011;23:4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Current Opinion in Plant Biology. 2012;15:483–492. doi: 10.1016/j.pbi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 8.Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 9.De Jonge R, Bolton MD, Thomma BPHJ. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Current Opinion in Plant Biology. 2011;14:400–406. doi: 10.1016/j.pbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Oliva R, et al. Recent developments in effector biology of filamentous plant pathogens. Cellular Microbiology. 2010;12:705–715. doi: 10.1111/j.1462-5822.2010.01471.x. [DOI] [PubMed] [Google Scholar]
- 11.Rooney HC, et al. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science (80-.) 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- 12.Shabab M, et al. Fungal Effector Protein AVR2 Targets Diversifying Defense-Related Cys Proteases of Tomato. Plant Cell. 2008;20:1169–1183. doi: 10.1105/tpc.107.056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian M, et al. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007;143:364–77. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders DGO, et al. Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS One. 2012;7:e29847. doi: 10.1371/journal.pone.0029847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Z, et al. A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell. 2015;27:2057–2072. doi: 10.1105/tpc.15.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postma J, et al. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 2016;210:627–42. doi: 10.1111/nph.13802. [DOI] [PubMed] [Google Scholar]
- 17.Rivas S, Thomas CM. Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 2005;43:395–436. doi: 10.1146/annurev.phyto.43.040204.140224. [DOI] [PubMed] [Google Scholar]
- 18.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–6. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, X. et al. Production of small cysteine-rich effector proteins in Escherichia coli for structural and functional studies. Molecular Plant Pathology, doi:10.1111/mpp.12385 (2016). [DOI] [PMC free article] [PubMed]
- 20.Zhang S, Xu JR. Effectors and Effector Delivery in Magnaporthe oryzae. PLoS Pathog. 2014;10:e1003826. doi: 10.1371/journal.ppat.1003826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen XM. Review/Synthèse Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Bot. 2005;27:314–337. [Google Scholar]
- 22.Cantu D, et al. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics. 2013;14:270. doi: 10.1186/1471-2164-14-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin C, et al. Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. Tritici. BMC Genomics. 2009;10:626. doi: 10.1186/1471-2164-10-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng W, et al. High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 2013;4:2673. doi: 10.1038/ncomms3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petre B, Joly DL, Duplessis S. Effector proteins of rust fungi. Front. Plant Sci. 2014;5:416. doi: 10.3389/fpls.2014.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upadhyaya NM, et al. A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol. Plant-Microbe Interact. 2014;27:255–264. doi: 10.1094/MPMI-07-13-0187-FI. [DOI] [PubMed] [Google Scholar]
- 27.Bombarely A, et al. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant-Microbe Interact. 2012;25:120809142432005. doi: 10.1094/MPMI-06-12-0148-TA. [DOI] [PubMed] [Google Scholar]
- 28.Petre B, et al. Heterologous expression screens in nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PLoS One. 2016;11:e0149035. doi: 10.1371/journal.pone.0149035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantu D, et al. Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One. 2011;6:e24230. doi: 10.1371/journal.pone.0024230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duplessis S, et al. From the Cover: Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA. 2011;108:9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godfrey D, et al. Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics. 2010;11:317. doi: 10.1186/1471-2164-11-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morais do Amaral A, Antoniw J, Rudd JJ, Hammond-Kosack KE. Defining the Predicted Protein Secretome of the Fungal Wheat Leaf Pathogen Mycosphaerella graminicola. PLoS One. 2012;7:e49904. doi: 10.1371/journal.pone.0049904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang A, et al. Identification and Characterization of Plant Cell Death-Inducing Secreted Proteins From Ustilaginoidea virens. Mol. Plant-Microbe Interact. 2016;29:405–416. doi: 10.1094/MPMI-09-15-0200-R. [DOI] [PubMed] [Google Scholar]
- 34.Kettles GJ, Bayon C, Canning G, Rudd JJ, Kanyuka K. Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana. New Phytol. 2017;213:338–350. doi: 10.1111/nph.14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan L, et al. Bacterial Effectors Target the Common Signaling Partner BAK1 to Disrupt Multiple MAMP Receptor-Signaling Complexes and Impede Plant Immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zipfel C. Pattern-recognition receptors in plant innate immunity. Current Opinion in Immunology. 2008;20:10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Xin DW, et al. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 2012;8:e1002707. doi: 10.1371/journal.ppat.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaparro-Garcia A, et al. Phytophthora infestans RXLR-WY effector AVR3a associates with dynamin-related protein 2 required for endocytosis of the plant pattern recognition receptor FLS2. PLoS One. 2015;10:e0137071. doi: 10.1371/journal.pone.0137071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doehlemann G, Hemetsberger C. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytologist. 2013;198:1001–1016. doi: 10.1111/nph.12277. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Wellings C, Chen X, Kang Z, Liu T. Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2014;15:433–446. doi: 10.1111/mpp.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali, S. et al. Analysis of putative apoplastic effectors from the nematode, Globodera rostochiensis, and identification of an expansin-like protein that can induce and suppress host defenses. PLoS One10 (2015). [DOI] [PMC free article] [PubMed]
- 43.Lindbo Ja. TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 2007;145:1232–40. doi: 10.1104/pp.107.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saur IML, et al. NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA. 2016;113:1511847113. doi: 10.1073/pnas.1511847113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lacombe S, et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010;28:365–369. doi: 10.1038/nbt.1613. [DOI] [PubMed] [Google Scholar]
- 46.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 47.Bozkurt TO, et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raffaele S, et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karimi M, Inzé D, Depicker A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa T, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 51.Win J, Kamoun S, Jones AME. Purification of effector-target protein complexes via transient expression in Nicotiana benthamiana. Methods Mol. Biol. 2011;712:181–194. doi: 10.1007/978-1-61737-998-7_15. [DOI] [PubMed] [Google Scholar]
- 52.O’Leary, B. M., Rico, A., McCraw, S., Fones, H. N. & Preston, G. M. The infiltration-centrifugation technique for extraction of apoplastic fluid from plant leaves using Phaseolus vulgaris as an example. J. Vis. Exp. e52113, doi:10.3791/52113 (2014). [DOI] [PMC free article] [PubMed]
- 53.Chaparro-Garcia A, et al. The receptor-like kinase serk3/bak1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS One. 2011;6:e16608. doi: 10.1371/journal.pone.0016608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giannakopoulou A, et al. Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. 2015;28:1316–1329. doi: 10.1094/MPMI-07-15-0147-R. [DOI] [PubMed] [Google Scholar]
- 55.Lee HA, et al. Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol. 2014;203:926–938. doi: 10.1111/nph.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segonzac C, et al. Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 2011;156:687–99. doi: 10.1104/pp.110.171249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu D, et al. Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana benthamiana Using Quantitative Real-Time PCR. PLoS One. 2012;7:e46451. doi: 10.1371/journal.pone.0046451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bozkurt TO, et al. The Plant Membrane-Associated REMORIN1.3 Accumulates in Discrete Perihaustorial Domains and Enhances Susceptibility to Phytophthora infestans. Plant Physiol. 2014;165:1005–1018. doi: 10.1104/pp.114.235804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 60.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


