Abstract
Mycothiol (MSH) is the major low molecular weight (LMW) thiol in Actinomycetes. Here, we used shotgun proteomics, OxICAT and RNA-seq transcriptomics to analyse protein S-mycothiolation, reversible thiol-oxidations and their impact on gene expression in Mycobacterium smegmatis under hypochlorite stress. In total, 58 S-mycothiolated proteins were identified under NaOCl stress that are involved in energy metabolism, fatty acid and mycolic acid biosynthesis, protein translation, redox regulation and detoxification. Protein S-mycothiolation was accompanied by MSH depletion in the thiol-metabolome. Quantification of the redox state of 1098 Cys residues using OxICAT revealed that 381 Cys residues (33.6%) showed >10% increased oxidations under NaOCl stress, which overlapped with 40 S-mycothiolated Cys-peptides. The absence of MSH resulted in a higher basal oxidation level of 338 Cys residues (41.1%). The RseA and RshA anti-sigma factors and the Zur and NrdR repressors were identified as NaOCl-sensitive proteins and their oxidation resulted in an up-regulation of the SigH, SigE, Zur and NrdR regulons in the RNA-seq transcriptome. In conclusion, we show here that NaOCl stress causes widespread thiol-oxidation including protein S-mycothiolation resulting in induction of antioxidant defense mechanisms in M. smegmatis. Our results further reveal that MSH is important to maintain the reduced state of protein thiols.
Introduction
Eukaryotes and Gram-negative bacteria utilize glutathione (GSH) as their major low molecular weight (LMW) thiol. However, most Gram-positive bacteria do not produce GSH1. Instead, the Actinomycetes, including streptomycetes, mycobacteria and corynebacteria, synthesize mycothiol (AcCys-GlcN-Ins, MSH) as GSH-surrogate and major LMW thiol1–3. MSH functions in detoxification of ROS, alkylating agents, toxins, antibiotics, heavy metal stress and aromatic compounds2, 4–7. Apart from MSH, mycobacteria produce the histidine-derived alternative LMW thiol ergothioneine (EGT) and both, MSH and EGT are important to maintain redox and bioenergetics homeostasis and are required for virulence in Mycobacterium tuberculosis 8.
In previous studies, we have shown in Corynebacterium glutamicum that MSH is engaged in redox regulation and thiol-protection of proteins under hypochlorite stress by the formation of mixed disulfides, termed as S-mycothiolations9. The identified S-mycothiolated proteins function in many metabolic pathways, such as the central carbon metabolism, the biosynthesis of amino acids, nucleotides, thiamine and myo-inositol-1-phosphate as well in protein translation. Some S-mycothiolated proteins are conserved and abundant targets for S-thiolations across Gram-positive bacteria, including ribosomal proteins, GuaB, SerA and MetE9, 10. S-mycothiolated proteins include also antioxidant enzymes, such as peroxiredoxins (Tpx, Mpx) and methionine sulfoxide reductases (MsrA). The reduction of S-mycothiolated proteins is catalyzed by the mycoredoxin-1 (Mrx1)/MSH/mycothiol disulfide reductase (Mtr) pathway as well as by the thioredoxin (Trx)/thioredoxin reductase pathways that control the activity of Tpx, Mpx and MsrA in vitro 11. Mpx and MsrA form intramolecular disulfides and S-mycothiolations under H2O2 treatment in vitro and require both the Trx and Mrx1 pathways for regeneration12, 13. The Mrx1/Mtr/MSH pathway is also involved in reduction of the peroxiredoxin AhpE in M. tuberculosis 14.
Mycobacteria produce high levels of 20 mM MSH, but the impact of MSH to maintain the reduced state of protein thiols and its role in protein S-mycothiolation under oxidative stress are unknown. This knowledge about the role of MSH in redox modifications is particularly important since MSH plays an important role for virulence in M. tuberculosis 15. Thus, conserved S-mycothiolated proteins and major redox-switches in mycobacteria could be future drug targets to treat live-threatening tuberculosis disease. Here, we combined shotgun-proteomics, OxICAT and “Voronoi redox treemaps” to monitor protein S-mycothiolation and reversible thiol-oxidations and to analyze the role of MSH for the redox balance in the model bacterium Mycobacterium smegmatis under hypochlorite stress. Using RNA-seq transcriptomics, the regulatory impact of thiol-oxidation of NaOCl-sensitive transcription factors on the changes in gene expression was analyzed.
Results
Mycobacterium smegmatis tolerates high NaOCl concentrations resulting in strongly increased protein S-mycothiolation
We were interested to study the overall extent of protein S-mycothiolation under NaOCl stress in M. smegmatis since NaOCl was previously shown to cause strongly increased protein S-thiolations in several bacteria1. First, we determined the physiological NaOCl-concentration that reduced the growth-rate, but still was sub-lethal in M. smegmatis allowing recovery of growth. M. smegmatis wild type was grown in Hartman’s-de Bont minimal medium (HdB) with glycerol as carbon source and exposed to different concentrations of NaOCl stress during the exponential growth16. The sub-lethal NaOCl-concentration that reduced the growth rate was determined as 1 mM, while 500 µM did not affect the growth (Fig. 1A). Thus, M. smegmatis is able to survive and recover in growth after treatment with high doses of 1 mM NaOCl. To analyse the effect of MSH in this high NaOCl resistance, we compared the growth of the wild type with that of the mshC mutant. In contrast to the wild type, the mshC mutant was unable to grow with 1 mM NaOCl (Fig. 1B), which provides bona fide evidence for the role of MSH in the protection against NaOCl stress.
Figure 1.
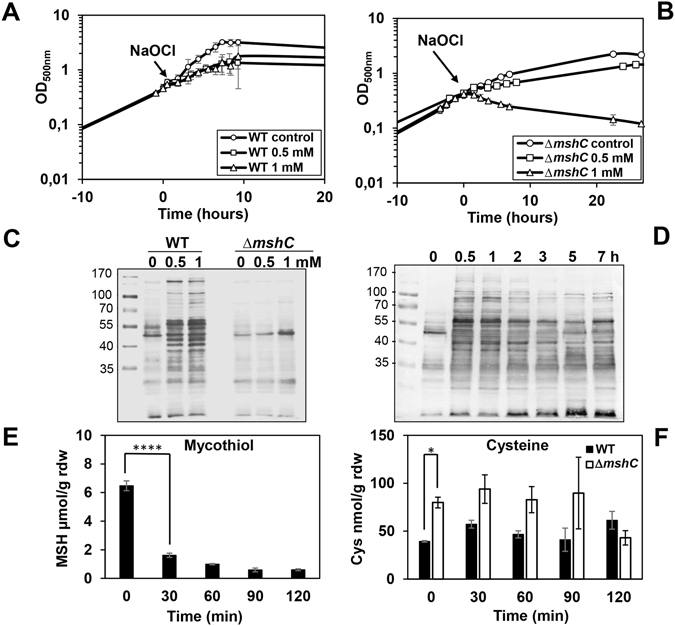
M. smegmatis tolerates high doses of 1 mM NaOCl leading to strongly increased protein S-mycothiolation and depletion of MSH in the thiol-metabolome. (A,B) The M. smegmatis wild-type and the ΔmshC mutant strains were cultivated in HdB minimal medium and exposed to sub-lethal concentrations of 0.5–1 mM NaOCl at an OD500 of 0.4. In contrast to the wild type, the ∆mshC mutant was unable to grow with 1 mM NaOCl. (C,D) Protein S-mycothiolation was increased in the wild type after exposure to 0.5–1 mM NaOCl stress as shown using non-reducing MSH-specific immunoblot analysis. (E) Thiol-metabolomics revealed the strong depletion of MSH in the wild type in response to 1 mM NaOCl stress indicating that MSH is used for protein S-mycothiolation. The MSH level decreased from 6.5 to 1.6 ± 0.25 µmol/g rdw after 30 min of NaOCl stress (One-way ANOVA, n = 15, P < 0.0001 for Co/NaOCl). (F) The Cys-levels in the control were calculated as 39.4 ± 1.33 nmol/g rdw in the wild type and 79.9 ± 9.75 nmol/g rdw in the ∆mshC mutant (Unpaired t-test, n = 6, p = 0.0173 for WT/∆mshC at t = 0 min). No significant changes in the Cys levels were measured after NaOCl stress in both strains (One-Way ANOVA, n = 15, P > 0.05 for WT and the mshC mutant Co/NaOCl).
Using MSH-specific Western blot analyses, the extent of protein S-mycothiolation was analysed in a time-dependent manner after exposure to 1 mM NaOCl (Fig. 1C,D). NaOCl stress resulted in strongly increased levels of S-mycothiolated proteins in the M. smegmatis wild type already 30 min after NaOCl stress. The level of S-mycothiolated proteins was decreased again after seven hours of NaOCl exposure correlating with the resumed growth of cells. Thus, this strongly increased protein S-mycothiolation in M. smegmatis is consistent with the high MSH level as determined previously17.
Protein S-mycothiolation should result in a depletion of MSH in the metabolome and was analyzed using thiol-metabolomics (Fig. 1E). The intracellular MSH concentration was determined as 6 µmol/g raw dry weight (rdw) in untreated wild-type cells and decreased 5-fold after 30 min of NaOCl exposure. The MSH depletion in the thiol-metabolome confirms that MSH is used for protein S-mycothiolation under NaOCl stress. In contrast to MSH, the cysteine levels were low with 40–60 nmol/g rdw in the wild type and 80–90 nmol/g rdw in the mshC mutant (Fig. 1F). Thus, cysteine cannot compensate for the absence of MSH as LMW thiol in mycobacteria.
Shotgun LC-MS/MS analysis identifies 58 S-mycothiolated proteins under NaOCl-stress conditions
Next, we used shotgun LTQ-Orbitrap LC-MS/MS analysis, to analyze S-mycothiolated Cys-peptides in the NEM-alkylated protein extracts of M. smegmatis under NaOCl stress based on their mass increase of +484 Da for MSH. In total, 58 proteins with S-mycothiolated Cys-peptides were identified (Table 1, Table S1A–C) which are visualized in Voronoi treemaps in relation to the total protein abundance of all proteins present in the proteome (Fig. 2). Among the identified S-mycothiolated proteins are known peroxiredoxins, such as Tpx, AhpC and OsmC that are S-mycothiolated at their active and/or resolving Cys residues (Table S1A,B). The inhibition of Tpx activity by S-mycothiolation and reactivation by the Mrx1/MSH/Mtr electron pathway was previously shown for Tpx of C. glutamicum 9. AhpC has been shown to function as peroxidase and peroxinitrite reductase together with AhpD, the dihydrolipoamide succinyl transferase (SucD) and the NADH-dependent dihydrolipoamide dehydrogenase (Lpd), thus linking the antioxidant response to regeneration of important enzymes of the intermediary metabolism18.
Table 1.
Selected S-mycothiolated proteins of Mycobacterium smegmatis wild type and quantification of their % oxidation by OxICAT.
| MSMEG-ID | Protein | Function | CysSSM peptide | % Ox NaOCl/co |
|---|---|---|---|---|
| Antioxidant enzymes | ||||
| MSMEG_4891 | AhpC | AhpC peroxiredoxin | (K)DFTFVC61(+484)PTEIAAFGK(L) | 6,70 |
| MSMEG_2421 | OsmC | OsmC family protein | (R)AVDQVC116(+484)TVGR(T) | 10,49 |
| MSMEG_3479 | Tpx | Thiol peroxidase | (K)SVLLNIFPSVDTPVC60(+484)ATSVR(T) | 11,57 |
| (K)AASSGATVLC80(+484)VSK(D) | −9,09 | |||
| (R)FC93(+484)GAEGIENVTTASAFR(S) | 6,91 | |||
| Protein biosynthesis and quality control | ||||
| MSMEG_1436 | RplC | 50 S ribosomal protein L3 | (R)RPGSIGGC154(+484)ATPGR(V) | 7,32 |
| MSMEG_1521 | RpsM | 30 S ribosomal protein S13 | (R)KIEIGC86(+484)YQGLR(H) | 21,77 |
| MSMEG_6895 | RpsR2 | 30 S ribosomal protein S18 | (R)VTGNC57(+484)VQHQR(D) | 10,66 |
| MSMEG_0839 | Lon1 | ATP-dependent protease Lon | (R)IIDC72(+484)QNLGANR(Y) | 25,70 |
| MSMEG_0832 | Def | Peptide deformylase | (R)LFVYDC68(+484)APTR(G) | 5,76 |
| Transcriptional regulation | ||||
| MSMEG_2750 | IdeR | Iron-dependent repressor IdeR | (R)LLVDVIGLPWEDVHAEAC102(+484)R(W) | — |
| MSMEG_4953 | TetR2 | TetR-family transcriptional regulator | (R)LIDAAETC21(+484)LR(A) | — |
| MSMEG_0227 | TetR1 | TetR-family transcriptional regulator | (R)LTAILLGPEPGTAC143(+484)R(V) | — |
| Biosynthesis of cofactors | ||||
| MSMEG_0913 | UmaA | Methoxy mycolic acid synthase 1 | (K)LDLKPGMTLLDVGC76(484)GWGGALER(A) | 10,46 |
| MSMEG_6904 | Ino1 | Inositol-3-phosphate synthase | (R)VAIVGVGNC18(+484)ASSLVQGVQYYR(N) | 6,31 |
| MSMEG_0793 | ThiG | Thiazole synthase | (R)LGIAALPNTAGC75(+484)R(G) | 10,25 |
| Energy metabolism | ||||
| MSMEG_6242 | Adh2 | Putative glycerol dehydrogenase | (R)AISEHIQDDWC398(+484)TPGNPR(E) | 4,94 |
| MSMEG_6759 | GlpK3 | Glycerol kinase | (K)NGLLTTVC294(+484)YR(L) | 10,73 |
| (R)ATLESIC389(+484)YQSR(D) | 5,67 | |||
| MSMEG_3086 | TpiA | Triosephosphate isomerase | (R)VAGAADAQEVC192(+484)K(A) | 2,04 |
| MSMEG_0911 | AceA | Isocitrate lyase | (K)NGLEPC268(+484)IAR(A) | 11,36 |
| MSMEG_5676 | CitA | Citrate synthase | (R)TIDEC143(+484)PTVTAR(F) | 14,23 |
| MSMEG_5049 | Kgd | 2-oxoglutarate metabolism enzyme | (R)SSEYC695(+484)TDVAK(M) | 4,60 |
| Metabolism of fatty acids and phospholipids | ||||
| MSMEG_5639 | EchA6 | Enoyl-CoA hydratase | (R)NALNC26(+484)ELVDSLR(E) | 4,73 |
| MSMEG_0531 | MSMEG_0531 | Acyl-CoA dehydrogenase | (R)AAYEYALDYAC285(+484)QR(E) | 4,50 |
| MSMEG_6208 | MSMEG_6208 | Acyl-CoA thioesterase | (R)DGDVFC21(+484)IREPEPNTIER(L) | — |
| MSMEG_1813 | AccD5 | Propionyl-CoA carboxylase beta chain | (R)VEGRPVGIVANQPTQFAGC356(+484)LDINASEK(A) | 11,49 |
| MSMEG_4329 | AccD6 | Acetyl/propionyl-CoA carboxylase | (R)LGGC294(+484)LNSESAEK(S) | 12,96 |
| Metabolism of nucleotides | ||||
| MSMEG_2299 | NrdE2 | Ribonucleoside-diphosphate reductase | (K)ITHSNLC380(+484)SEILQVSTPSEFNDDLSYAK(V) | — |
| MSMEG_1602 | GuaB | Inosine-5′-monophosphate dehydrogenase | (K)VGVGPGSIC325(+484)TTR(V) | 19,34 |
| MSMEG_3634 | GuaB2 | Inosine-5′-monophosphate dehydrogenase | (K)VGVGPGAmC302(+484)TTR(M) | 33,37 |
| MSMEG_2656 | Pnp | Polyribonucleotide nucleotidyltransferase | (K)ALC248(+484)AAQQELADR(A) | 5,67 |
The M. smegmatis wild type was exposed to 1 mM NaOCl for 30 min and 58 S-mycothiolated proteins were identified using shotgun LC-MS/MS analysis using the Scaffold proteome software based on the mass increase of 484 Da (+MSH) at Cys peptides. The table lists for 29 selected mycothiolated proteins the MSMEG-ID, the protein name, function and the S-thiolated Cys peptide sequence. The OxICAT data were extracted from Tables S3 and S4 for the S-mycothiolated Cys peptides. The full table of the 58 S-mycothiolated proteins is presented in Table S1A,B.
Figure 2.
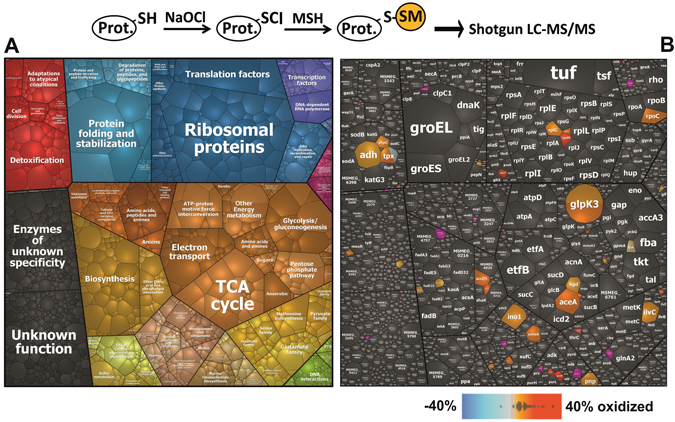
Voronoi treemaps show protein abundance of S-mycothiolated proteins identified in M. smegmatis under NaOCl stress using Orbitrap LC-MS/MS analysis. (A) The treemap legend shows the classification of the M. smegmatis proteome according to TIGRfam annotations. (B) The total spectral counts determine the cell size of each protein identified in the proteome dataset and classified according to TIGRfam. The identified 58 S-mycothiolated proteins are color-coded using an orange-red color gradient based on their Cys oxidation level as quantified by the OxICAT data (Tables 2, S3 and S4). Non-modified proteins are colored in grey and S-mycothiolated proteins that were not identified using the OxICAT approach are shown in pink.
Interesting S-mycothiolated proteins include further transcriptional regulators that could be redox-controlled by protein S-mycothiolation, such as global regulator for iron uptake of the DtxR-family (IdeR) that was S-mycothiolated at Cys102, its primary iron-binding site (Fig. S1A). In Corynebacterium diphtheriae, the Cys-Asp mutant in the related DtxR-repressor was incompatible in DNA-binding to its target operators19. Apart from DtxR, two unknown TetR-family regulators (MSMEG_4953 and MSMEG_0227) were identified as S-mycothiolated transcription factors. Future studies should elucidate whether these transcriptional regulators function as redox-switches under NaOCl stress to control their specific target genes.
The largest group of S-mycothiolated proteins functions in energy metabolism that include abundant enzymes of the different routes of glycerol catabolism, glycolysis, the glyoxylate shunt and gluconeogenesis (MSMEG_6759 or GlpK3, MSMEG_6242 or Adh2, TpiA, CitA, AceA, Kgd) (Fig. 3, Tables 1, S1 and S2). Since M. smegmatis was grown with glycerol as sole carbon and energy source, the glycerol kinase GlpK3 and the glycerol dehydrogenase MSMEG_6242 (Adh2) are abundant S-mycothiolated proteins and specify two branches of the glycerol catabolic pathways (Fig. 3)20. GlpK3 uses ATP for phosphorylation of glycerol to glycerol-3-phosphate which is converted to dihydroxyacetone phosphate (DHAP) by the glycerol-3-phosphate dehydrogenase MSMEG_6761 (GlpD2) (Table S1D). The Adh2 enzyme (MSMEG_6242) reduces glycerol to DHA which is then phosphorylated by the DHA kinase (DhaKLM) to DHAP. S-mycothiolation of GlpK3 and Adh2 could prevent glycerol degradation under NaOCl stress to save the source of carbon and energy.
Figure 3.
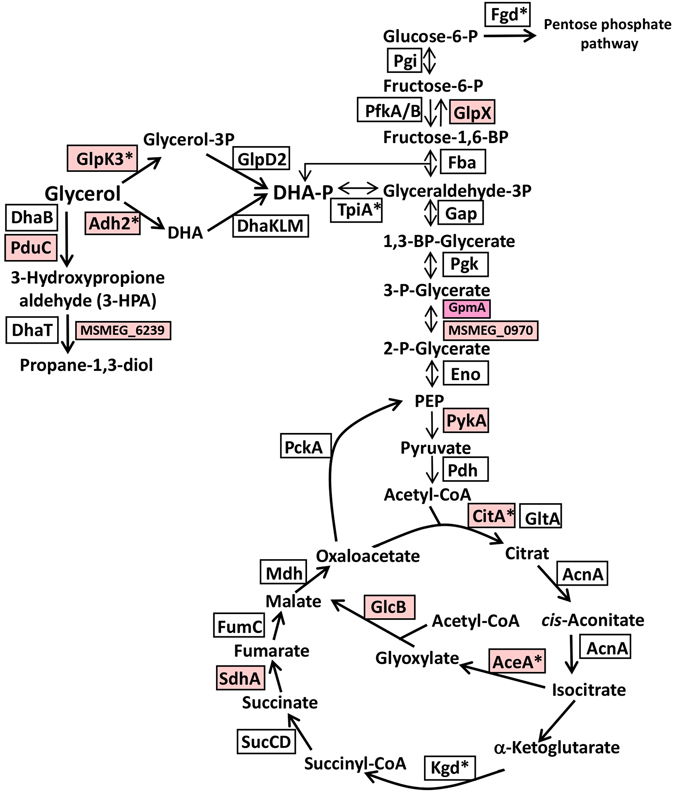
Schematics of the glycerol catabolism, glycolysis, TCA cycle, glyoxylate cycle and gluconeogenesis in M. smegmatis highlighting NaOCl-sensitive thiol-switches. The reversibly oxidized NaOCl-sensitive enzymes are color-coded in light and dark pink indicating 10% and 20% thiol-oxidation increase under NaOCl stress, respectively. The S-mycothiolated proteins are labelled with an asterisk. The pathways of the glycerol catabolism include the aerobic oxidation of glycerol to dihydroxyacetone-phosphate (DHA-P) and the propane-diol-pathway that are catalyzed by (1) the glycerol kinase (GlpK3 or MSMEG_6759) and glyceraldehyde dehydrogenase (GlpD2), (2) the glycerol dehydrogenase (Adh2 or MSMEG_6242) and dihydroxyacetone kinase complex (DhaKLM) and (3) the B12-dependent glycerol dehydratase (DhaB or MSMEG_1546-49) and propane-1,3-diol-dehydrogenase (MSMEG_6239). DHAP enters the glycolysis, TCA and glyoxylate shunt and gluconeogenesis for energy and biomass production. The gluconeogenesis enzymes include GlpX (fructose-1,6-Bis-phosphatase) and PckA (PEP-carboxykinase), while Pgi (glucose-6-phosphate isomerase), Fba (fructose-bisphosphate aldolase), Gap (glyceraldehyde-3-phosphate dehydrogenase), Pgk (phosphoglycerate kinase), GpmA and MSMEG_0970 (phosphoglycerate mutase) and Eno (enolase) are involved in both glycolysis and gluconeogenesis. The glyoxylate shunt includes the isocitrate lyase (AceA) and malate synthase (GlcB).
We further identified the isocitrate lyase AceA and the myo-inositol-1-phosphate synthase Ino1 as abundant S-mycothiolated proteins in M. smegmatis. AceA is the key enzyme of the glyoxylate bypass of the TCA cycle (Fig. 3) and required for growth on fatty acids in M. tuberculosis to enable the use of carbon for biomass production via gluconeogenesis21. AceA inhibition and protection by S-mycothiolation under oxidative stress could be important for survival of mycobacteria. Ino1 functions in MSH and phosphatidylinositol biosynthesis and was S-mycothiolated also in C. glutamicum (Fig. S1B).
Furthermore, enzymes involved in the biosynthesis of fatty acids as precursors for mycolic acids are essential in M. tuberculosis and abundant targets for S-mycothiolation in M. smegmatis (Fig. 2)22, 23. Among the S-mycothiolated proteins are acetyl-CoA carboxylases involved in the carboxylation of acetyl-CoA to malonyl-CoA as first step of the fatty acid biosynthesis (AccD5 and AccD6), the enoyl-CoA hydratase (EchA6), the methoxy mycolic acid synthase (UmaA), the acyl-CoA dehydrogenase (MSMEG_0531) and the acyl-CoA-thioesterase (MSMEG_6208). UmaA is S-mycothiolated at the conserved Cys76 that is required for S-adenosylmethionine binding.
S-mycothiolated enzymes participate in nucleotide biosynthesis, such as PurC, Pnp, NrdE2 and the conserved inosine-5′-monophosphate (IMP) dehydrogenases GuaB and GuaB2. Both GuaB homologs are S-mycothiolated at their conserved active sites, Cys325 and Cys302, respectively, that form the thioimidate intermediate (Fig. S1C). Other targets for S-mycothiolation are biosynthesis enzymes for thiamine (ThiG, MSMEG_4827), cobalamin (CobN), iron sulfur-cluster assembly (YfhF2) and translation proteins, such as ribosomal proteins (RplC, RpsM, RpsR2) and amino acyl tRNA synthetases (GatC, PheT). RplC and RpsM are S-mycothiolated at their conserved Cys154 and Cys86, respectively. The conservation of ribosomal proteins as targets for S-thiolation suggests an inhibition of protein biosynthesis under oxidative stress.
In previous studies, we have identified S-cysteinylated proteins in the absence of bacillithiol and mycothiol9, 10. Here, we detected only 11 S-cysteinylated proteins in the mshC mutant (Table S1A), including Tpx, AccD5, AccD6, CitA, RplC, RpsM and RpsR2 that were S-mycothiolated in the wild type. Thus, S-cysteinlyation cannot compensate for the loss of MSH in protein protection and redox-regulation. Mycobacteria also produce ergothioneine (EGT) as histidine-derived LMW thiol that is important to maintain redox and bioenergetics homeostasis in M. tuberculosis 8, 17, 24. However, we did not found EGT-mixed disulfides in the mshC mutant since these are probably instable modifications and escape the identification using mass spectrometry.
Quantification of the redox status of 1098 Cys residues by OxICAT and visualization of the thiol-oxidation levels using “Voronoi redox treemaps”
The shotgun proteomics method identified only 58 S-mycothiolated proteins. However, this method is not quantitative due to the unstable nature of the MSH modification. Thus, we used the quantitative thiol-redox proteomics approach OxICAT to determine more comprehensively the thiol-oxidation state of all Cys residues including those that are S-mycothiolated in the M. smegmatis wild type. In addition, the mshC mutant was included in the OxICAT analysis25, 26 to reveal the role of MSH for the thiol- redox state of proteins in M. smegmatis.
The principle of the OxICAT method relies on the differential labelling of reduced and oxidized thiol-residues using isotope-coded affinity tags (ICAT)25, 26. Reduced Cys residues are labelled with light 12C-ICAT, followed by reduction of reversible thiol-oxidations (e.g. protein disulfides and S-thiolations) and subsequent labelling of previously oxidized thiols with heavy 13C-ICAT reagent. Light and heavy ICAT-labeled peptide pairs show a mass difference of 9 Da after separation using mass spectrometry. The percentage of thiol-oxidation for each Cys-peptide is calculated based on the intensity of the heavy ICAT-labeled Cys-peptide in relation to the total intensity of the light and heavy-ICAT-labelled Cys-peptides25, 26.
Using OxICAT, the percentages of thiol-oxidation levels were quantified for 1098 Cys residues under control and NaOCl stress in M. smegmatis wild-type cells (Tables 2, S2–S4; Fig. 4). The Cys-peptides were color-coded according to their percentages of thiol-oxidations and visualized in “Voronoi redox treemaps” that are based on the TIGRfam classification (Fig. 5A–D). In the wild-type control, 857 Cys residues (78.1%) showed an oxidation state of <25%, that included 444 Cys residues (40.4%) with <10% oxidation. This indicates that the majority of all detected thiols are reduced under non-stress conditions in M. smegmatis (Fig. 5A). A minor part of 241 Cys residues (21.8%) had higher oxidation levels of >25% in the control. These basal level oxidized proteins belong to membrane proteins and ABC transporter components involved in transport functions, protein secretion, folding and quality control.
Table 2.
Selected NaOCl-sensitive proteins with >10% increased thiol-oxidations under NaOCl stress in M. smegmatis as revealed using the OxICAT method.
| Locus tag | Gene name | Protein function | Cys (a, b) | Buried/Exposed (d) | OxICAT Wild type | OxICAT ΔmshC | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| % Diff NaOCl/Co (e) | % ox Co (f) | % ox NaOCl (f) | % Diff NaOCl/Co (e) | % ox Co (f) | % ox NaOCl (f) | |||||
| Detoxification and adaptation to atypical environments | ||||||||||
| MSMEG_0127 | adhE1 | Alcohol DH, zinc-containing | 48* | B | 32,0 | 13,5 | 45,5 | 13,8 | 19,5 | 33,3 |
| MSMEG_0217 | adhB | Alcohol DH, zinc-containing | 105* | B | 39,1 | 20,1 | 64,8 | |||
| MSMEG_5866 | adhB2 | Alcohol DH, zinc-containing | 106* | B | 35,7 | 20,9 | 55,2 | 16,9 | 28,8 | 45,7 |
| MSMEG_4340 | adhE2 | Alcohol DH, zinc-containing | 107* | B | 33,1 | 23,8 | 54,2 | 15,3 | 34,8 | 50,0 |
| 145 | B | 34,6 | 18,0 | 43,8 | 18,5 | 28,7 | 47,2 | |||
| MSMEG_1138 | MSMEG_1138 | Alcohol DH, zinc-containing | 113* | B | 26,0 | 16,5 | 43,8 | |||
| MSMEG_4400 | MSMEG_4400 | Alcohol DH, zinc-containing | 65 | B | 27,8 | 6,4 | 34,1 | |||
| MSMEG_1977 | MSMEG_1977 | Alcohol DH, zinc-containing | 39* | B | 22,8 | 14,5 | 30,0 | 11,9 | 12,6 | 27,5 |
| MSMEG_0595 | MSMEG_0595 | Fe-S oxidoreductase | 142* | B | 13,9 | 12,4 | 23,4 | 7,6 | 30,2 | 37,8 |
| MSMEG_0690 | MSMEG_0690 | Fe-S oxidoreductase | 637* | B | 11,5 | 22,7 | 38,8 | 8,2 | 38,5 | 45,2 |
| MSMEG_0768 | MSMEG_0768 | Rhodanese domain protein | 83* | B | 42,6 | 17,2 | 55,5 | 23,0 | 30,5 | 53,8 |
| MSMEG_6425 | MSMEG_6425 | Rhodanese-domain protein | 66* | B | 14,0 | 7,1 | 20,9 | 7,8 | 13,1 | 22,7 |
| MSMEG_1416 | MSMEG_1416 | Pyridine nucleotide-disulfide oxidoreductase | 159 | B | 11,4 | 12,0 | 20,3 | 12,5 | 14,3 | 26,7 |
| MSMEG_1566 | MSMEG_1566 | Oxidoreductase | 122 | B | 14,2 | 15,1 | 24,9 | |||
| MSMEG_2263 | hybC | Cytochrome-c3 hydrogenase | 58 | B | 29,3 | 21,7 | 49,7 | 3,8 | 30,7 | 37,9 |
| MSMEG_2297 | nrdH | Glutaredoxin | 11* | B | 14,2 | 45,7 | 56,1 | 15,0 | 57,7 | 69,2 |
| MSMEG_2421 | osmC | OsmC family protein | 48* | B | 26,2 | 13,1 | 38,4 | 4,9 | 23,2 | 29,2 |
| 116* (MSH) | B | 10,5 | 12,7 | 22,8 | 13,0 | 18,3 | 32,3 | |||
| MSMEG_2784 | msrB2 | Methionine sulfoxide reductase | 51* | B | 14,2 | 27,0 | 37,7 | 2,2 | 36,3 | 38,5 |
| MSMEG_3479 | tpx | Thiol peroxidase | 60* (MSH;Cys) | B | 11,6 | 29,1 | 39,9 | 8,9 | 37,8 | 48,0 |
| MSMEG_4085 | MSMEG_4085 | Nitrilotriacetate monooxygenase | 336 | B | 31,2 | 14,3 | 32,2 | |||
| MSMEG_4309 | ptpA | LMW protein-tyrosine-phosphatase | 10* | B | 22,2 | 11,2 | 30,6 | |||
| 58 | E | 41,1 | 47,7 | 79,9 | 10,2 | 17,3 | 31,5 | |||
| Transcription and Transcriptional regulators | ||||||||||
| MSMEG_0219 | MSMEG_0219 | RNA polymerase sigma factor | 271 | B | 17,5 | 10,3 | 28,9 | 5,7 | 16,8 | 26,8 |
| MSMEG_1367 | rpoB | RNA polymerase beta SU | 674 | B | 20,3 | 25,3 | 45,6 | 15,3 | 44,3 | 59,6 |
| MSMEG_1368 | rpoC | RNA polymerase beta’ SU | 48* | B | 19,6 | 14,3 | 29,5 | |||
| MSMEG_1515 | MSMEG_1515 | Two-component sensor histidine kinase | 5 | E | 35,6 | 13,1 | 48,5 | 13,2 | 39,2 | 58,3 |
| MSMEG_1831 | whiB2 | Transcriptional regulator WhiB2 | 67* | B | 12,6 | 20,0 | 31,7 | |||
| 99* | B | 10,5 | 33,6 | 44,9 | ||||||
| MSMEG_1874 | mtrA | Two-component response regulator MtrA | 68* | B | 10,1 | 2,1 | 11,1 | |||
| MSMEG_1915 | rshA | Anti-sigma-factor for SigmaH (RshA) | 76* | B | 38,2 | 15,8 | 53,2 | |||
| MSMEG_5071 | rseA | Anti-sigma-factor for SigmaE (RseA) | 67* | B | 37,5 | 41,8 | 63,4 | −3,4 | 49,1 | 45,7 |
| MSMEG_2743 | nrdR | Transcriptional repressor NrdR | 71* | B | 24,9 | 7,0 | 30,9 | |||
| MSMEG_4471 | MSMEG_4471 | MarR-family transcriptional regulator | 58 | B | 42,3 | 12,3 | 54,0 | 34,6 | 25,8 | 61,1 |
| MSMEG_4487 | furB | Ferric uptake regulator FurB | 124* | B | 17,4 | 30,2 | 42,6 | 10,9 | 35,4 | 47,2 |
| MSMEG_5768 | MSMEG_5768 | TetR family transcriptional regulator | 61 | E | 22,7 | 12,5 | 22,7 | |||
| Protein biosynthesis and quality control | ||||||||||
| MSMEG_1339 | rpmG | 50 S ribosomal protein L33-1 | 15* | B | 23,9 | 30,5 | 53,4 | |||
| MSMEG_1468 | rpsN | 30 S ribosomal protein S14 type Z | 27* | B | 19,6 | 41,5 | 60,1 | 13,0 | 32,5 | 45,5 |
| MSMEG_1520 | rpmJ | 50 S ribosomal protein L36 | 27* | B | 33,5 | 21,7 | 43,6 | 13,5 | 22,4 | 35,9 |
| MSMEG_1521 | rpsM | 30 S ribosomal protein S13 | 86* (MSH;Cys) | B | 21,8 | 10,4 | 32,9 | 20,2 | 14,7 | 34,4 |
| MSMEG_1579 | rimI | Alanine acetyltransferase | 55 | B | 38,4 | 9,7 | 51,0 | |||
| MSMEG_1878 | MSMEG_1878 | 30 S ribosomal protein S30 | 83 | E | 40,4 | 46,7 | 84,6 | 22,7 | 59,7 | 82,6 |
| MSMEG_2400 | rpmB | 50 S ribosomal protein L28 | 5* | B | 35,7 | 40,3 | 74,5 | 26,5 | 41,8 | 70,4 |
| 52 | B | 36,8 | 42,7 | 76,9 | 24,8 | 48,3 | 74,2 | |||
| MSMEG_4951 | rpmE | 50 S ribosomal protein L31 | 16* | B | 20,2 | 22,2 | 39,6 | 14,5 | 22,4 | 32,9 |
| MSMEG_6895 | rpsR2 | 30 S ribosomal protein S18-2 | 20* | B | 24,6 | 73,6 | 84,8 | 8,9 | 76,0 | 84,9 |
| 57* (MSH;Cys) | B | 10,7 | 11,4 | 21,2 | 7,3 | 15,2 | 23,4 | |||
| MSMEG_0839 | lon1 | ATP-dependent protease | 72 (MSH) | B | 25,7 | 11,1 | 38,5 | |||
| Glycolysis/Gluconeogenesis and TCA cycle | ||||||||||
| MSMEG_0935 | gpmA | 2,3-bisphosphoglycerate-mutase | 149 | E | 20,0 | 7,3 | 27,9 | 9,9 | 13,6 | 22,0 |
| MSMEG_0970 | MSMEG_0970 | Phosphoglycerate mutase | 146 | B | 10,6 | 13,4 | 21,5 | |||
| MSMEG_1547 | pduC | Glycerol dehydratase large SU | 156 | B | 12,2 | 20,6 | 31,0 | 8,3 | 37,5 | 46,5 |
| 168 | B | 11,1 | 19,3 | 31,6 | 4,9 | 32,6 | 37,6 | |||
| 342* | B | 15,4 | 7,9 | 20,6 | 12,4 | 21,4 | 33,1 | |||
| Selected NaOCl-sensitive proteins with >10% increased thiol-oxidations under NaOCl stress in M. smegmatis | ||||||||||
| MSMEG_3227 | pyk2 | Pyruvate kinase | 9* | B | 10,6 | 10,1 | 20,6 | 8,7 | 21,2 | 30,4 |
| MSMEG_5239 | glpX | Fructose-1,6-bisphosphatase | 205 | B | 12,5 | 13,7 | 23,9 | |||
| MSMEG_6759 | glpK3 | Glycerol kinase | 294 (MSH) | B | 10,7 | 8,9 | 16,8 | 9,9 | 19,9 | 29,8 |
| MSMEG_0911 | aceA | Isocitrate lyase | 191 | B | 11,3 | 8,5 | 20,8 | 11,0 | 22,3 | 33,7 |
| 268 (MSH) | B | 11,4 | 12,7 | 20,3 | 12,8 | 24,8 | 37,6 | |||
| MSMEG_1670 | sdhA2 | Succinate DH | 385 | B | 15,5 | 31,0 | 35,1 | |||
| MSMEG_3640 | glcB | Malate synthase G | 612* | B | 12,8 | 9,7 | 20,9 | |||
| MSMEG_4645 | orB | a-OG ferredoxin oxidoreductase, beta SU | 59* | B | 10,1 | 9,4 | 20,9 | |||
| MSMEG_5676 | citA | Citrate (Si) synthase | 143* (MSH;Cys) | E | 14,2 | 4,8 | 19,0 | 7,5 | 13,0 | 20,5 |
| Metabolism of Fatty acid and phospholipids | ||||||||||
| MSMEG_0913 | umaA | Methoxy mycolic acid synthase 1 | 76* (MSH) | B | 10,5 | 9,1 | 17,6 | |||
| MSMEG_1340 | MSMEG_1340 | (3 R)-hydroxyacyl-ACP dehydratase SU HadA | 105 | B | 15,5 | 4,7 | 19,7 | 16,9 | 12,7 | 29,6 |
| MSMEG_1342 | MSMEG_1342 | (3 R)-hydroxyacyl-ACP dehydratase SU HadC | 127 | B | 14,0 | 8,7 | 22,0 | 2,8 | 24,5 | 39,7 |
| MSMEG_1553 | eutB | Ethanolamine ammonia-lyase | 36 | B | 10,6 | 6,1 | 16,0 | 6,5 | 15,0 | 20,1 |
| MSMEG_1554 | eutC | Ethanolamine ammonia-lyase light chain | 204* | B | 11,7 | 15,7 | 26,4 | |||
| MSMEG_1807 | accA3 | Acetyl-/propionyl-CoA carboxylase alpha chain | 236* | B | 13,4 | 12,6 | 26,0 | 22,8 | 25,7 | 45,7 |
| MSMEG_1813 | accD5 | Methylmalonyl-CoA carboxyltransferase | 356 (MSH;Cys) | B | 11,5 | 17,2 | 26,4 | 10,7 | 30,9 | 41,6 |
| MSMEG_2207 | MSMEG_2207 | Beta-ketothiolase | 9 | B | 12,9 | 11,0 | 26,2 | −2,9 | 39,4 | 36,9 |
| MSMEG_4116 | MSMEG_4116 | 3-hydroxyacyl-CoA DH | 148 | B | 18,8 | 12,9 | 34,4 | |||
| MSMEG_4327 | kasA | 3-oxoacyl-(Acyl-carrier-protein) synthase 1 | 171* | B | 28,0 | 15,4 | 50,0 | |||
| MSMEG_4328 | kasB2 | 3-oxoacyl-(Acyl-carrier-protein) synthase 1 | 227 | B | 26,0 | 11,7 | 36,0 | 12,2 | 21,6 | 34,9 |
| MSMEG_4329 | accD6 | Acetyl/propionyl-CoA carboxylase (Beta SU) | 191 | B | 19,5 | 14,2 | 32,7 | 14,6 | 28,2 | 38,4 |
| 213 | E | 15,7 | 13,8 | 27,1 | ||||||
| 294 (MSH) | B | 13,0 | 9,6 | 20,3 | ||||||
| MSMEG_4920 | MSMEG_4920 | Acetyl-CoA acetyltransferase | 107* | B | 43,0 | 0,6 | 43,3 | |||
| 398* | B | 32,2 | 5,4 | 38,1 | ||||||
| MSMEG_5199 | MSMEG_5199 | Acetyl-CoA acetyltransferase | 55 | B | 11,8 | 4,5 | 12,2 | |||
| MSMEG_5273 | fadA3 | Acetyl-CoA acetyltransferase | 90* | B | 20,0 | 8,7 | 28,0 | |||
| 390* | B | 20,8 | 9,0 | 21,8 | ||||||
| MSMEG_5291 | MSMEG_5291 | Acyl-CoA synthase | 16 | B | 17,1 | 4,5 | 22,5 | 20,2 | 12,0 | 32,2 |
| 359 | B | 17,7 | 7,2 | 22,8 | 8,1 | 16,3 | 30,1 | |||
| Metabolism of nucleotides | ||||||||||
| MSMEG_1602 | guaB | Inosine-5′-monophosphate DH | 325* (MSH) | B | 19,3 | 5,5 | 24,1 | 22,0 | 11,1 | 30,4 |
| MSMEG_3634 | guaB2 | Inosine-5′-monophosphate DH | 302* (MSH) | B | 33,4 | 8,2 | 44,6 | 8,4 | 18,2 | 29,2 |
| 321 | B | 15,5 | 12,6 | 32,0 | 7,8 | 12,5 | 20,5 | |||
| Metabolism of cofactors | ||||||||||
| MSMEG_0789 | thiE | Thiamine-P synthase | 20 | B | 24,1 | 5,1 | 28,2 | 18,0 | 19,5 | 37,9 |
| MSMEG_0791 | thiO | Glycine oxidase | 32 | B | 44,3 | 1,2 | 42,5 | 26,1 | 9,5 | 36,3 |
| MSMEG_0793 | thiG | Thiazole synthase | 75* (MSH) | B | 10,3 | 9,1 | 17,6 | |||
| MSMEG_2671 | folA | Dihydrofolate reductase | 106 | B | 47,5 | 10,5 | 49,8 | 38,6 | 14,9 | 51,9 |
| MSMEG_3067 | ribD | Riboflavin biosynthesis protein RibD | 78* | B | 47,4 | 8,5 | 56,4 | |||
| MSMEG_3072 | ribAB | Riboflavin biosynthesis protein RibBA | 264* | B | 19,3 | 52,3 | 65,2 | 7,3 | 61,7 | 69,1 |
| MSMEG_3126 | MSMEG_3126 | Nitrogen fixation protein NifU | 38* | B | 18,1 | 22,4 | 38,9 | 7,3 | 32,8 | 41,7 |
| MSMEG_4272 | yfhF2 | HesB/YadR/YfhF family protein | 47* (MSH) | B | 11,0 | 19,5 | 28,3 | 3,8 | 29,2 | 36,9 |
| MSMEG_4827 | MSMEG_4827 | Acyl-CoA DH | 44 (MSH) | E | 32,5 | 35,4 | 66,7 | |||
| MSMEG_5698 | moaA | Cyclic pyranopterin monoP synthase | 50* | B | 24,3 | 33,2 | 54,4 | 3,7 | 42,3 | 44,0 |
| 305* | B | 19,0 | 24,0 | 44,6 | ||||||
Selected NaOCl-sensitive proteins with >10% increased thiol-oxidations in response to NaOCl stress in M. smegmatis as revealed using the OxICAT method. The M. smegmatis wild type and mshC mutant were harvested before (control) and 30 min after exposure to 1 and 0.5 mM NaOCl, respectively. Reduced and reversibly oxidized Cys residues were labelled with light and heavy ICAT, respectively, using OxICAT. Quantification of % thiol-oxidations was performed using MaxQuant software. The table includes MSMEG accessions, protein names, functions, surface accessabilities and % oxidation of Cys residues under control and NaOCl. (a) Conserved Cys are bold. (b) S-mycothiolated or S-cysteinylated Cys are marked with (+MSH) and (+Cys). (d) Relative surface accessibility (RSA) for buried (B) or exposed (E) Cys residues. (e) The % thiol-oxidation of each identified Cys peptide was calculated using MaxQuant. Based on the % thiol-oxidation of each Cys under control and NaOCl stress conditions, the % oxidation increase (% Diff NaOCl/co) was calculated under NaOCl-treatment and (f) average values are shown from at least three independent biological replicates. Selected NaOCl-sensitive peptides with >10% increased thiol-oxidation under NaOCl stress are shown here as a subset of the complete Tables S3–S4.
Figure 4.
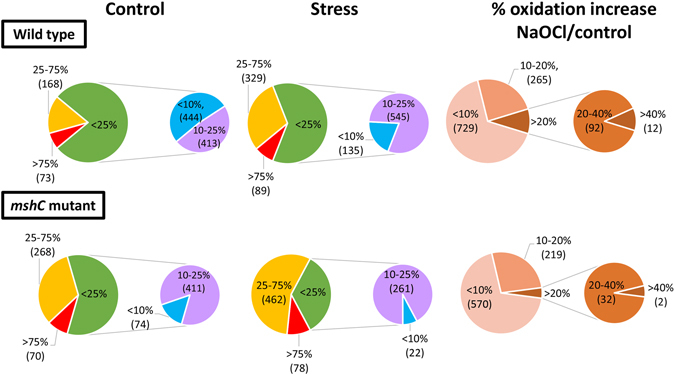
Overview of the percentages of thiol-oxidation levels of all Cys peptides identified in the redox proteome of the M. smegmatis wild type and the mshC mutant under control and NaOCl stress as revealed by OxICAT. Reduced Cys peptides with a <25% oxidation are shown in green including those <10% oxidized (blue) and 10–25% oxidized (magenta). Cys peptides with an oxidation degree of >25% and >75% are shown in yellow and red, respectively. The percentage of thiol-oxidation increase is shown with an orange-brown color gradient including Cys peptides with 10–20% and >20% increased oxidation by NaOCl stress. The mshC mutant shows a higher basal level oxidation in the control that resembles that of the wild type after NaOCl stress.
Figure 5.
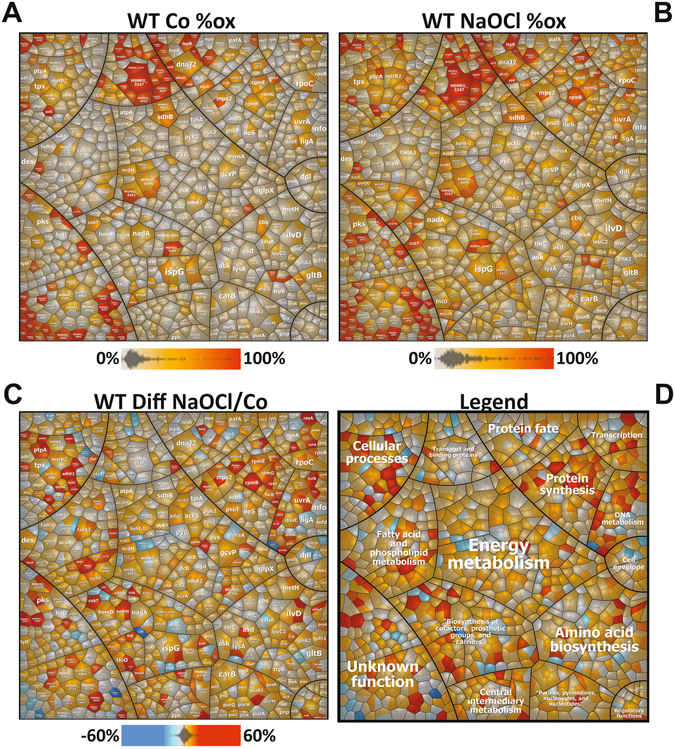
Voronoi redox treemaps show the percentages of thiol-oxidation levels of all Cys-peptides identified in the redox proteome of the M. smegmatis wild type. The “Voronoi redox treemaps” show the percentages of thiol-oxidations of 1098 Cys-residues identified using OxICAT in the wild type control (A) and 30 min after exposure to 1 mM NaOCl stress (B). The grey-yellow-red color gradient denotes 0–100% oxidation. The Voronoi redox treemap in (C) visualizes the percentages of oxidation changes under NaOCl stress using a blue-red color gradient ranging from −60 to +60% oxidation. The treemap in (D) is used as legend for the functional classification of the proteins displayed in (C). The treemaps are generated based on the OxICAT data presented in Table S4 using the Paver software (Decodon) and proteins were classified according to the M. smegmatis TIGRfam annotation.
Under NaOCl stress, 381 Cys residues (34.7%) showed a >10% increased oxidation that include 116 Cys with >20% increased oxidations (Tables 2, S3 and S4, Fig. 5B,C). Among the NaOCl-sensitive proteins identified by OxICAT are 40 of 58 S-mycothiolated proteins that were found in the shotgun approach (Tables 1 and S1A). The overlap of S-mycothiolated proteins with NaOCl-sensitive proteins is further visualized in the S-mycothiolation treemap (Fig. 2), where S-mycothiolated proteins are color-coded according to their thiol-oxidations. Of note, most S-mycothiolated proteins with >10% increased oxidations under NaOCl stress represent abundant antioxidant and metabolic enzymes in the total proteome, such as Tpx, AhpC, Adh2, GlpK3, AceA, Kgd, Ino1, UmaA, AccD5 and IlvC as well as ribosomal proteins (RpsM, RplC) and RNA polymerase subunits (RpoC) (Fig. 2, Tables S1A, S3 and S4).
NaOCl-sensitive proteins include antioxidant enzymes, Zn-containing alcohol dehydrogenases, ribosomal proteins and transcriptional regulators
Among the NaOCl-sensitive proteins are the S-mycothiolated peroxiredoxins Tpx and OsmC, the methionine-sulfoxide reductase MsrB2, the glutaredoxin NrdH and two catalases KatA2 and KatG3. The redox-sensitive low molecular weight protein-tyrosine-phosphatase PtpA showed 41.06% and 22.21% increased oxidation at Cys58 and in the Cys11-Cys15 active site motif, respectively (Tables 2, S3 and S4). PtpA was oxidized by H2O2 in its active site motif in M. tuberculosis which forms an intramolecular disulphide leading to enzyme inactivation27. In M. tuberculosis, S-nitrosylation at the non-conserved Cys53 was reported leading to a decreased enzymatic activity by interfering with protein stability and function28, 29.
Further highly oxidized Cys residues are the active site centres essential for catalysis or function in metal ion coordination (e.g. Zn, iron or FeS-clusters). These include proteins with structural or catalytic Zn-binding sites, such as Zn-finger motifs and Zn ribbons (Tables S3,S4, Fig. 5). Of note, 12 Zn-containing alcohol dehydrogenases, such as the abundant glycerol dehydrogenase MSMEG_6242 (Adh2) and 6 Adhs showed up-to 39% higher thiol-oxidation levels under NaOCl stress (AdhB1/B2/E1/E2, MSMEG_1138, MSMEG_1977 and MSMEG_4400). It is possible that these Adh enzymes participate in the glycerol oxidation pathway. These Zn-containing Adhs possess an N-terminal conserved catalytic Cys that was identified as NaOCl-sensitive in AdhE1 and MSMEG_1977. In addition, four conserved structural Cys residues are involved in Zn-binding (Figure S1D). Interestingly, AdhE1/E2/B2 and MSMEG_1138 are highly oxidized under NaOCl stress at the same Zn-binding Cys105, 106, 107 and 113, respectively. The inhibition of the yeast alcohol dehydrogenase YADH-1 due to overoxidation of its catalytic Cys has been shown previously30. Many ribosomal proteins with Zn-ribbon motifs showed >20% increased oxidation under NaOCl stress, including RpmG, RpmJ, RimL, RpmB, and RpsR2. RpsM is S-mycothiolated at the conserved Cys86 in M. smegmatis and C. glutamicum (Fig. S1E). Zn-containing ribosomal proteins are suggested to serve as reservoir for Zn-storage31.
Among the NaOCl-sensitive Zn-containing regulators are the Fur-family Zn-uptake regulator FurB (Zur), the NrdR repressor and the RshA and RseA anti-sigma factors. Zur has a CxxC Zn-redox switch motif that shows 17% increased oxidation under NaOCl stress. Zur is active as transcriptional repressor in the Zn-bound form, while Zn-deficiency leads to Zur inactivation and derepression of its regulon consisting of Zn-transporters, Zn-containing ribosomal proteins and the immunodominant ESAT-6 proteins32. The NrdR repressor is oxidized at the conserved Cys71 in its Zn ribbon motif. NrdR negatively regulates transcription of genes encoding two ribonucleotide reductases (nrdF2 and nrdF22) and NrdH-like glutaredoxins (MSMEG_1017 and MSMEG_2297) that are essential for de novo DNA synthesis33. Interestingly, the glutaredoxin NrdH is involved in reduction of NrdF and we identified both, the NrdR repressor and the glutaredoxin NrdH (MSMEG_2297) as NaOCl-sensitive proteins with >10% increased oxidations.
Two redox-regulatory anti-sigma factors RshA and RseA were identified as NaOCl-sensitive Zn-redox switches. RshA and RseA are ECF group-IV anti-sigma factors of the zinc-associated anti-sigma factor (ZAS) family (Figure S1F)34. RshA is oxidized at the conserved Cys76 with 38% increased oxidation under NaOCl stress. RseA showed 37% increased oxidation in its ZAS motif at Cys67 and Cys70 suggesting the formation of an intramolecular disulphide under NaOCl stress (Tables S3, S4, Fig. 5). The homolog of the RshA-SigH system of mycobacteria is the RsrA-SigR couple in Streptomyces coelicolor. Previous studies revealed that RsrA forms the disulphide between the N-terminal Cys11 and either Cys41 or Cys44 in the ZAS motif and the oxidized RsrA structure with the Cys11-Cys44 disulfide has been resolved recently35, 36. However, the N-terminal Cys11 of RsrA is not conserved in M. smegmatis RseA and we identified the redox switch in the ZAS motif of RseA as possible redox-signaling mechanism.
Using OxICAT, the unknown MarR family transcriptional regulator MSMEG_4471 was identified with 42% higher oxidation under NaOCl stress at Cys58 (Tables 2, S3, S4, Fig. 5). MSMEG_4471 is located adjacent to a multidrug-efflux transporter as possible target gene (MSMEG_4472) and has a homolog in M. tuberculosis (Rv2327) (Fig. S1G). MarR-family regulators often sense and respond to ROS and RES via conserved thiol-switch mechanisms (e.g. OhrR, SarZ, YodB, QsrR)37, 38. The function and redox-sensing mechanism of MSMEG_4471 and its related homolog of M. tuberculosis are subject of our current research.
We further identified many NaOCl-sensitive 4Fe4S-cluster-containing redox-switches, such as the dihydroxy-acid dehydratase IlvD, the molybdenum biosynthesis enzyme MoaA and the WhiB2 redox sensor (Fig. S1H). The FeS-cluster protein WhiB2 is essential in mycobacteria and required for septum formation and cell division39. In conclusion, Zn or FeS-cluster-binding thiol-containing proteins are often targets for oxidation under NaOCl stress and represent a large group in our list of NaOCl-sensitive proteins.
NaOCl-sensitive proteins are involved in the central carbon metabolism and in the biosynthesis of fatty acids, cofactors, nucleotides and amino acids
Our OxICAT analysis identified 23 metabolic enzymes that are involved in energy metabolism with >10% increased oxidations under NaOCl stress (Tables 2, S3,S4, Fig. 5). The glycerol kinase GlpK3 and the large subunit of the glycerol dehydratase PduC showed 10% increased thiol-oxidations under NaOCl stress. Two NaOCl-sensitive phosphoglycerate mutases GpmA and MSMEG_0970 and the fructose-1,6-bisphosphatase GlpX are involved in the gluconeogenesis. The isocitrate lyase AceA and the malate synthase GlcB function in the glyoxylate cycle and are >10% oxidized under NaOCl-stress at conserved Cys residues. AceA was also S-mycothiolated at Cys268 and oxidation of GlcB was previously reported40. Other NaOCl-sensitive TCA cycle enzymes include the citrate synthase CitA that was identified as S-mycothiolated and S-cysteinylated at the conserved Cys143.
Furthermore, 27 NaOCl-sensitive enzymes function in the biosynthesis of fatty acids as precursors for mycolic acids. These include the mycothiolated acetyl-CoA carboxylases (AccD5 and AccD6) and the methoxy mycolic acid synthase (UmaA). Three acetyl-CoA acetyltransferases of the thiolase family MSMEG_4920, MSMEG_5199 and FadA3 showed >20% increased oxidations under NaOCl stress in their active sites that forms an acyl thioester intermediate during catalysis. In addition, enzymes required for the elongation of fatty acids were identified as NaOCl-sensitive, such as the 3-oxoacyl-ACP synthases KasA and KasB2 and the fatty acids synthase subunits HadA and HadC (FAS-I/II). Thus, the central carbon metabolism and the fatty acid biosynthesis pathways include many NaOCl-sensitive proteins that are important for cellular survival in mycobacteria.
We further identified 31 NaOCl-sensitive enzymes that function in cofactor biosynthesis, 11 nucleotide biosynthesis enzymes and 23 amino acid biosynthesis enzymes. Among the nucleotide biosynthesis enzymes are both S-mycothiolated IMP-dehydrogenases (GuaB and GuaB2) that showed >20% increased oxidation under NaOCl treatment at their conserved thioimidate active sites at Cys325 and Cys302, respectively.
In summary, using shotgun-LC-MS/MS and OxICAT analyses we identified 58 S-mycothiolated proteins and a >10% increased thiol-oxidation level for 33% of Cys residues under NaOCl stress that included also 40 S-mycothiolated Cys-peptides. The most interesting NaOCl-sensitive thiol-switches are the enzymes involved in energy metabolism, mycolic acid and fatty acid biosynthesis as well as the various Zn-containing alcohol dehydrogenases, ribosomal proteins and redox-sensing regulators, such as RseA, RshA, Zur and NrdR.
Loss of mycothiol leads to 2-fold increased basal oxidations for 41% of all Cys residues in the mshC mutant redox proteome
Next, we investigated whether the loss of mycothiol affects the redox state in M. smegmatis. The redox state of 823 Cys residues was quantified using OxICAT in the mshC mutant under control and NaOCl stress as shown in the Voronoi redox treemaps (Fig. 6A–D, Tables S3, S4). Under control conditions, 485 Cys residues (58.9%) showed <25% oxidation levels, while 338 Cys residues (41.1%) were significantly oxidized with >25% increased oxidations. In contrast, only 21.8% oxidized thiols with >25% increased oxidation levels were quantified in the wild type. These results indicate that the content of reduced thiols is much lower in the mshC mutant compared to the wild type. Thus, the extent of reduced and oxidized thiols in the mshC mutant control resembles that of NaOCl-exposed wild-type cells as indicated in the pie chart diagram (Fig. 4). Furthermore, NaOCl stress resulted in >10% increased oxidations of 255 Cys residues in the mshC mutant. Thus, despite the higher basal level oxidation in the mshC mutant, the thiol-oxidation increase under NaOCl stress is comparable to that of the wild type with 34.7% and 30.8% oxidation increase in the wild type and mshC mutant, respectively (Fig. 4; Tables S3, S4).
Figure 6.
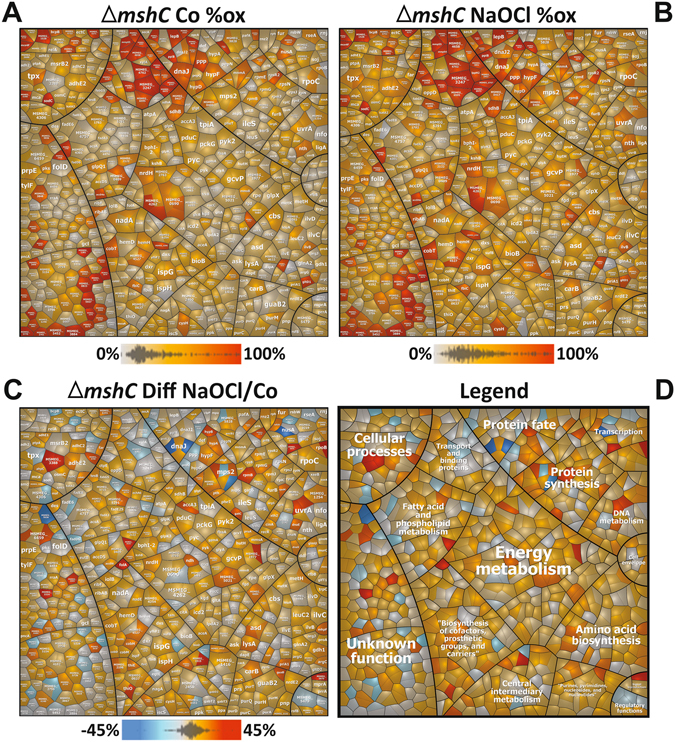
Voronoi redox treemaps show the percentages of thiol-oxidation levels of all Cys-peptides identified in the redox proteome of the M. smegmatis mshC mutant. The “Voronoi redox treemaps” show the percentages of thiol-oxidations of all 823 Cys-residues identified in the mshC mutant control (A) and 30 min after exposure to 0.5 mM NaOCl stress (B) The grey-yellow-red color gradient denotes 0–100% oxidation. The Voronoi redox treemap in (C) visualizes the percentages of oxidation changes under NaOCl stress using a blue-red color gradient ranging from −60 to +60% oxidation. The treemap in (D) is used as legend for the functional classification of the proteins displayed in (C). The treemaps are generated based on the OxICAT data presented in Table S4 using the Paver software (Decodon) and proteins were classified according to the M. smegmatis TIGRfam annotation.
To visualize the increased thiol-oxidation levels in the mshC mutant, the log2-fold-changes of the percentages in thiol-oxidations were calculated in the mshC mutant compared to the wild type (Figure S2; Tables S3, S4). It is interesting to note that especially NaOCl-sensitive and S-mycothiolated enzymes are higher oxidized in the mshC mutant and obviously vulnerable to oxidation in the absence of MSH. Proteins with 2–3 fold increased basal oxidation in the mshC mutant are involved in energy metabolism and fatty acid biosynthesis, such as TpiA, GlpK3, AceA, CitA, PckG, AccA3, AccD5, AccD6,MSMEG_0108, FadD6, FadE6, FadA3 and include many S-mycothiolated proteins. The mycothiolated Ino1 showed a 4-fold increased thiol-oxidation level at the conserved Cys18 in the NAD+ binding site in the mshC mutant. In addition, many enzymes that function in the biosyntheses pathways for cofactors and amino acids exhibit 2–3-fold increased oxidation levels in the mshC mutant.
Among the transcriptional regulators, we noticed 2–4-fold increased oxidation levels in the mshC mutant control for Cys residues of the RNA polymerase α, β and β’ subunits (RpoA/B/C), the transcription elongation and termination factors (NusA and Rho), the two-component sensor histidine kinase MSMEG_1515, the MarR-family regulator MSMEG_4471 and the FeS-cluster transcription factor WhiB1. WhiB1 is an essential DNA binding redox sensor with an NO-sensitive FeS-cluster that is nitrosylated and represses transcription of the essential chaperonin GroEL2 in M. tuberculosis under NO stress41–43. Earlier studies revealed also an interaction of WhiB1 with the alpha-1,4-glucan branching enzyme GlgB1 via the Cys residues that coordinate the FeS-cluster44. Interestingly, another FeS-cluster WhiB-family protein, WhiB3, is an important redox sensor of M. tuberculosis controlling EGT synthesis and thereby contributing to the redox and bioenergetics homeostasis8. Both, MSH and EGT are important for the redox balance, energy metabolism and virulence of M. tuberculosis 8. It is possible that WhiB-like proteins respond generally to hypochloric acid under infection conditions to module EGT biosynthesis, central carbon and fatty acid metabolism to promote intracellular survival.
Most oxidized Cys residues in the OxICAT dataset are not surface-exposed
Next, we were interested if the Cys residues are oxidized since they are accessible and surface-exposed or if they are buried in the predicted secondary protein structure. We used the program NetSurfP45 (http://www.cbs.dtu.dk/services/NetSurfP/) to calculate the relative surface accessibilities (RSA) of all 1332 Cys residues identified in the M. smegmatis wild type and in the mshC mutant (Tables S3, S4). However, only 180 Cys residues (13.5%) have RSA values of >20% and are predicted as surface-exposed while most Cys residues are not predicted as surface-accessible. This is visualized in a Surface accessibility treemap where the RSA values of all Cys peptides are presented as white-red color gradient (Fig. 7). Moreover, among the 370 NaOCl-sensitive thiols, only 29 exposed Cys are predicted that include the S-mycothiolated proteins Rnz2, RpsR2, CitA, MSMEG_2799, LysA, AccD6 and MSMEG_4827. In contrast, highly redox-sensitive and nucleophilic active site Cys residues, such as Cys 302 and Cys325 of GuaB and GuaB2 are buried in the protein structure. Similarly, the S-mycothiolated active site Cys of the peroxiredoxins Tpx, AhpC and OsmC or the catalytic and Zn-binding sites of the Adhs are not surface-exposed, although these are major targets for NaOCl-induced oxidation. This indicates that the majority (86.5%) of the Cys residues in the M. smegmatis redox proteome are buried in the predicted secondary protein structure. These results are in agreement with previous predictions about the accessible surface area of Cys residues in the human, yeast and E. coli proteomes46.
Figure 7.
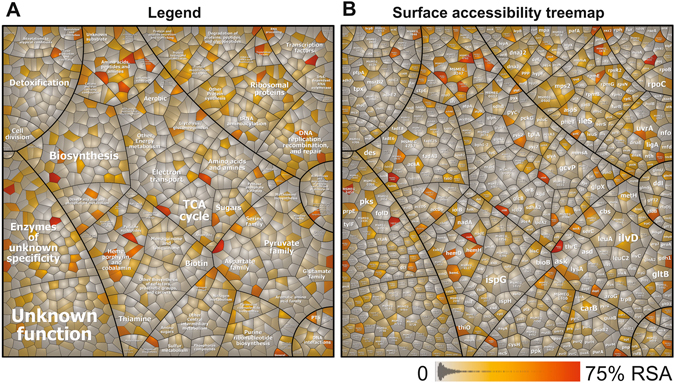
Relative surface accessibility (RSA) treemap of Cys residues identified in the redox proteome of M. smegmatis using OxICAT. The surface accessibilities of Cys residues were predicted using the NetSurfP server ver. 1.1 http://www.cbs.dtu.dk/services/NetSurfP/. The RSA treemap is composed of 1332 Cys residues identified in the redox proteome of the M. smegmatis wild type and the mshC mutant. Proteins are classified according to the M. smegmatis TIGRfam annotation. The treemap in (A) serves as legend for the functional classification of the proteins shown in the treemap in (B). The cells in the treemap represent Cys residues that are color-coded according to their relative surface accessibilities (RSA) with a white-orange-red color gradient ranging from 0–75% RSA. Exposed Cys are orange and buried Cys are shown in white-grey. In total, about 180 Cys residues (13.5%) in 163 proteins have a RSA value of >20% indicating that a majority of Cys residues are not surface-exposed in the predicted secondary structure of the proteins.
Thiol-oxidation of redox-sensitive regulators leads to increased transcription of the SigH, SigE, Zur and NrdR regulons in the transcriptome
The OxICAT results revealed an increased oxidation of many redox-sensitive transcriptional regulators under NaOCl stress, such as RseA, RshA, Zur and NrdR. Thiol-oxidation should lead to inactivation of these transcription factors resulting in transcriptional induction of the corresponding regulon members. To prove this hypothesis, we performed a RNA-seq transcriptome analysis of M. smegmatis after exposure to NaOCl stress. The m-value cut-off (log2-fold-change) for significant expression changes was defined as +/−4.47 (99% confidence, 3 bioreplicates, P < 0.001). In total, about 251 transcripts were significantly up-regulated under NaOCl stress in the transcriptome dataset including also the sigH-rshA operon (Table S5). This confirms previous transcriptome results in C. glutamicum where the SigH regulon was strongly induced under NaOCl stress9. The comprehensive SigH regulon has been recently defined in M. tuberculosis using ChIP-Seq analysis and 25 SigH-dependent promoter sequences were identified with the GGAAY-(N18/19)-GTT consensus47. Thus, we used this consensus to define the SigH regulon in M. smegmatis based on the RNA-seq data. First, we analyzed the upstream promoter sequences of the identified transcription start sites (TSS) among the NaOCl-induced transcriptional units for the SigH consensus using the MEME suite software. In total, 124 SigH-dependent promoter sequences were identified with the consensus sequence GGAAY-N18/19-GTT (p < 0.0001) and at least two-fold induction under NaOCl stress. SigH-dependent promoters were identified upstream upstream of 84 genes with m-values of ≥4.47 under NaOCl stress (Tables S6, S7). In total, 203 genes were identified that are transcribed from 124 SigH-dependent promoters either directly or as part of an operon.
Among these are 36 promoters that match the SigE promoter consensus GGAACY-N16/17-CGTT which could be recognized by both SigH and SigE-containing RNA polymerases48. The identified SigH/SigE-regulon members encode thioredoxins and thioredoxin reductases (Trx, Trx2, TrxB), chaperones and proteases (DnaK, DnaJ, GrpE, ClpB, Lon2, MSMEG_0424) and peroxiredoxins (AhpD). The comparison of the SigH-promoters identified in M. tuberculosis 47 with our list revealed only an overlap of 15 conserved SigH-promoter sequences. Thus, our in silico promoter analysis combined with NaOCl stress as inducing condition identified 109 new SigH-dependent promoters unique to M. smegmatis.
The gene expression data of the transcription factor regulons of M. smegmatis are visualized in a Voronoi transcriptome treemap using a blue-orange color gradient (Fig. 8). The SigH and SigE regulons represent the most strongly induced regulons under NaOCl treatment in this transcriptome treemap. In addition, expression of the NrdR regulon was increased under NaOCl stress, including the genes for the ribonucleotide reductases (nrdF2 and nrdF22) and nrdH glutaredoxins (MSMEG_1017 and MSMEG_2297). Thus, oxidation of NrdR leads to its inactivation and derepression of the NrdR regulon. In addition, the Zur repressor was identified as another NaOCl-sensitive Zn-redox-switch. Zur oxidation leads to derepression of the Zur regulons under NaOCl stress, including the MSMEG_4486-zur operon, genes for ribosomal proteins (rpsR, rpsN2, rpmG2/B2/E2) and zinc uptake transporters.
Figure 8.
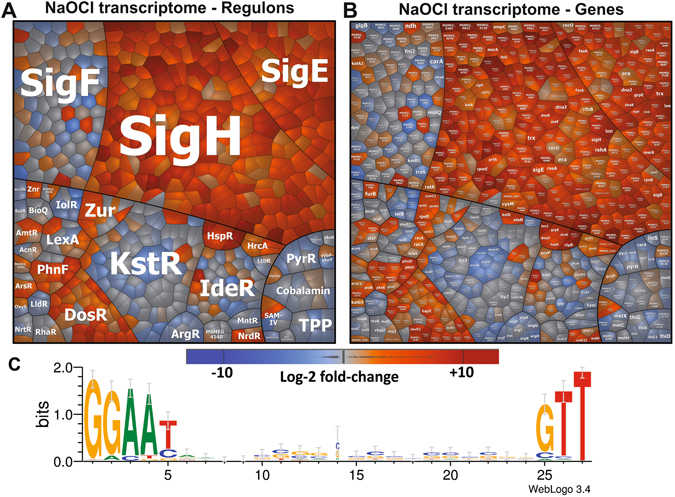
The transcriptome treemap indicates the strong induction of the SigH and SigE oxidative stress regulons under NaOCl stress in M. smegmatis. (A,B) The transcriptome treemap shows changes in gene expression of the M. smegmatis wild type under 1 mM NaOCl stress as log2-fold changes (m-values). The genes are classified into operons and regulons based on the RegPrecise database (http://regprecise.lbl.gov/RegPrecise/index.jsp) and the newly defined SigH and SigE regulons of this work. The regulon classification is used as legend (A) for the gene expression treemap in (B). Differential gene expression is visualized using an orange-blue color code where orange indicates log2-fold induction and blue repression of transcription under NaOCl stress. The oxidative stress-specific SigH and SigE-regulons are most strongly up-regulated under NaOCl stress. The Web-logo of the SigH-promoter consensus sequence (C) was created using WebLogo 3.078 based on the alignment of 124 SigH-promoters identified in this work.
Transcription of the DosR dormancy regulon was elevated under NaOCl stress. The DosR regulon of M. smegmatis consists of dosR and a dosR paralog, universal stress proteins (MSMEG_5245, MSMEG_3945 and MSMEG_3950), a ribosome stabilizing-factor, diacylglycerol acyltransferases and nitroreductases and the [Ni-Fe]-hydrogenase Hyd3 (MSMEG_3931-3928). Hyd3 plays a role in hydrogen production under fermentation conditions to allow survival of M. smegmatis in oxygen-deprived environments49. The DosR regulon is induced in M. tuberculosis under dormany conditions by the gases NO, CO and hypoxia, which are sensed by the DosS and DosT heme sensor kinases50.
Furthermore, NaOCl stress leads to induction of two high-affinity phosphate uptake systems, the PstSCAB and the PhnDCE systems in the transcriptome. It has been shown in E. coli that NaOCl-treated cells encounter phosphate-starvation that is linked to the accumulation of polyphosphate as primordial chaperone in protection of proteins against NaOCl-induced unfolding51, 52. Thus, polyphosphate could also play an important role in the defense of mycobacteria against hypochloric acid under infection conditions which remains to be elucidated.
Discussion
Protein S-mycothiolation is an emerging and widespread redox modification in Actinomycetes that particularly occurs under hypochlorite stress and functions in redox regulation and thiol-protection against overoxidation to sulphonic acids. Here, we aimed (i) to identify S-mycothiolated proteins using shotgun proteomics in M. smegmatis, (ii) to analyze the global thiol-oxidation state using OxICAT and (iii) to demonstrate the changes in gene expression due to thiol-oxidation in the RNA-seq transcriptome. We have selected NaOCl as infection-relevant condition for S-mycothiolation since our previous results revealed strongly increased S-thiolation in several bacteria under NaOCl1. Hypochloric acid (HOCl) is a strong thiol-oxidant with a high redox potential that targets Cys with a second-order rate constant of k = 3 × 107 M−1 s−1 53. The thiol group is first chlorinated to the unstable sulfenylchloride (-SCl) intermediate that reacts further with LMW thiols leading to S-thiolations, such as S-mycothiolations (-SSM) (Fig. 2)53. Protein S-mycothiolation was previously shown to function in thiol-protection and redox regulation in C. glutamicum under NaOCl stress.
Using the shotgun proteomics approach, we identified 58 S-mycothiolated proteins under NaOCl stress in M. smegmatis that participate in many essential cellular pathways, including energy metabolism, fatty acid and mycolic acid biosynthesis, nucleotide, cofactor and amino acid biosynthesis, redox regulation, transcription and translation to ensure survival and redox homeostasis under oxidative stress. Many S-mycothiolated proteins are conserved and essential targets for S-thiolation across Gram-positive bacteria, including the thiol-peroxidase Tpx, ribosomal proteins (RpsM), two IMP dehydrogenases (GuaB and GuaB2) and the myo-inositol-1-phosphate synthase (Ino1). Among the 58 targets for S-mycothiolation, 39 have Cys residues that are conserved also in the pathogen M. tuberculosis (Table S1A). Since these S-mycothiolated proteins were observed under infection-related conditions upon hypochlorite stress, they could be important to provide protection against the host immune defense in M. tuberculosis.
The quantitative redox proteomics approach OxICAT revealed an >10% increased thiol-oxidation level for 381 Cys residues (33.6%) under NaOCl stress. The 381 NaOCl-sensitive Cys-peptides overlap with 40 of 58 S-mycothiolated proteins which are also present at significant amounts in the proteome (Fig. 2). This indicates that protein S-mycothiolation is probably more abundant in M. smegmatis and that many more of the 381 NaOCl-sensitive proteins could represent S-mycothiolated proteins. However, due to the unstable nature of the MSH modification, the shotgun approach has limitations to detect only the tip-of-the-iceberg. Of note, 227 identified NaOCl-sensitive Cys residues are conserved in M. tuberculosis (Table S3) and could represent possible future drug-targets. This would be particularly attractive in combination with inhibitors of MSH biosynthesis to combat tuberculosis (TB) disease since MshB inhibitors are successfully applied in the clinical practice54, 55.
The conserved NaOCl-sensitive thiol-switches include Zn-containing proteins, such as alcohol dehydrogenases, ribosomal proteins, the ZAS anti sigma factors RseA and RshA and the transcriptional repressors Zur and NrdR. Using transcriptome analysis, we were able to demonstrate that thiol-oxidation leads to inactivation of NaOCl-sensitive transcriptional regulators resuling in changes of gene expression. Oxidation of both ZAS anti sigma factors (RshA and RseA) and the Zur and NrdR repressors was detected by OxICAT which is accompanied by the up-regulation of the corresponding regulons in the RNA-seq transcriptome. The oxidative stress responsive SigH and SigE regulons are major defense mechanisms and mediate redox homeostasis and protein quality control in mycobacteria and other actinomycetes47, 48, 56, 57. Moreover, based on the strong up-regulation of the SigH and SigE regulons in the transcriptome, we could identify 124 new SigH-dependent promoters that are transcribed under NaOCl stress and share the SigH promoter consensus sequence. Thus, our combined redox proteome and transcriptome results provide novel insights into the redox-signaling mechanisms of the RshA and RseA ZAS anti sigma factors and identified Zur and NrdR as new Zn-redox-sensors in mycobacteria. In E. coli, the chaperone holdase Hsp33 represents a NaOCl-sensitive Zn-redox switch that protects oxidatively damaged proteins against aggregation under oxidative stress25, 58–61. It will be interesting to elucidate if the newly identified conserved Zn-containing NaOCl-sensitive proteins function in protection against oxidative stress in M. tuberculosis during the infection cycle.
Our list of NaOCl-sensitive proteins includes many enzymes that have antioxidant functions or are involved in energy metabolism, such as glycerol catabolism and gluconeogenesis as well as in the biosynthesis pathways for fatty acids, mycolic acids, nucleotides and cofactors in M. smegmatis (Tables 2, S3 and S4). Some proteins that are susceptible to reversible thiol-oxidation by NaOCl stress in M. smegmatis were previously found as targets for S-nitrosylation in the S-nitrosoproteome of M. tuberculosis using the biotin-switch method62, 63. Of note, among the 29 S-nitrosylated proteins are also many antioxidant proteins and enzymes essential for the intermediary and fatty acid metabolism in M. tuberculosis 63. Specifically, 14 S-nitroslyated proteins overlap with the targets for protein thiol-oxidation in M. smegmatis, including the PEP carboxykinase PckG, the malate synthase GlcB and the citrate synthase AcnA and LpdA2, important for anaplerosis, gluconeogenesis and the TCA cycle. Common targets for thiol-oxidation and nitrosylation are further AtpA, SerA, PepN, GlnA2, FadD32, RpoB/C, KatG3 and Mpa. Using a transposon mutant screen, the proteasome was identified as major component in the defense against nitrosative stress including the ATPase Mpa that is required for NO-resistance and virulence in mice64. Another interesting target for S-nitrosylation and NaOCl-induced thiol-oxidation is the protein-tyrosine-phosphatase PtpA that is required for virulence in M. tuberculosis and was strongly oxidized in its active site motif under NaOCl stress65. This indicates that NO and NaOCl may target similar specific thiols in mycobacteria that functions in the protection against ROS and RNS to ensure cellular survival and virulence under infection conditions. In support of this notion, the transcriptome analysis revealed a strongly up-regulated DosR regulon under both, NaOCl and NO stress conditions suggesting that the DosS sensor kinase may function as redox sensor of NaOCl and NO50, 66. Thus, future studies should be directed to apply the NOxICAT and OxICAT approach25, 26 under NO and NaOCl stress in M. tuberculosis for more detailed comparison of the targets for nitrosylation and thiol-oxidation using similar methods.
The OxICAT data further revealed that the majority of NaOCl-sensitive thiols (24%) display only 10–20% increased thiol-oxidation levels under NaOCl stress. This is in agreement with the OxICAT data obtained in E. coli where only a subset of thiols was strongly oxidized under NaOCl treatment25. Using NetSurfP, we confirmed that only a minor part of 13.5% of all identified Cys residues are surface-exposed. The majority of NaOCl-sensitive thiols and S-mycothiolated proteins represent nucleophilic, catalytic or structural Cys residues that are buried and not solvent accessible in M. smegmatis further supporting that NaOCl targets specific redox-sensitive Cys residues with regulatory consequences resulting in changes of gene expression in the transcriptome.
To analyse the role of MSH for the thiol-oxidation state in M. smegmatis, we further analysed the changes in the thiol-redox proteome in the mshC mutant under NaOCl stress. Our results revealed that the absence of MSH leads to an increased basal level oxidation of 41.1% Cys residues that are >25% oxidized in the mshC mutant control compared to only 21.9% Cys residues that are >25% oxidized in the wild type control. NaOCl stress resulted in a further oxidation increase in the mshC mutant for 30.9% Cys residues that showed >10% oxidation increase. Thus, the level of 59% reduced and 41% oxidized thiols under control conditions in the mshC mutant resembled that of the wild type under NaOCl stress (Fig. 4). This difference in the thiol-redox state was even enhanced under NaOCl stress in the mshC mutant, with 34.4% reduced and 65.6% oxidized thiols. These results demonstrate in a quantitative manner the importance of MSH to maintain the reduced state of protein thiols and that protein thiols are more sensitive to NaOCl-induced oxidation in the absence of MSH.
The final question remains about the nature of the reversible thiol-modifications in the absence of MSH in M. smegmatis. Increased levels of EGT were previously reported in the M. smegmatis mshA mutant67. Both, MSH and EGT have been shown to be critical for redox homeostasis, energy metabolism and virulence in M. tuberculosis and mutants disrupted in MSH and EGT biosynthesis showed overlapping responses in the transcriptome8, 15. Thus, EGT might compensate for the absence of MSH and it will be subject of future studies to investigate whether the increased reversible thiol-oxidations in the mshC mutant represent S-ergothioneinylations.
Methods
Bacterial strains and growth conditions
Mycobacterium smegmatis mc2155 wild type and its isogenic ∆mshC mutant68 were cultivated in Hartmans-de Bont minimal medium (HdB) at 37 °C under vigorous agitation as described16. Cells were exposed to 0.5–1 mM NaOCl during the exponential growth phase at an optical density at 500 nm (OD500) of 0.4. Sodium hypochlorite (NaOCl) (15%) and N-ethylmaleimide (NEM) were purchased from Sigma Aldrich.
Non-reducing MSH specific immunoblotting
About 25 µg of M. smegmatis protein extract was separated by non-reducing 12% SDS-PAGE and subjected to MSH specific immunoblot analysis using a polyclonal rabbit MSH antibody at dilution 1:1000 as described9, 69.
Monobromobimane-labelling and HPLC-thiol metabolomics analysis
Thiol-labelling using monobromobimane (mBBr) was performed as described10. The mBBr-labelled thiols were separated by reverse phase chromatography and quantified by fluorescence detection using the same HPLC system as described70. The following gradient method was applied: 10 min 92% buffer A (10% methanol, 0.25% acetic acid, pH 3,9) supplemented with 8% buffer B (90% methanol, 0.25% acetic acid, pH 3,9), linear increase to 40% buffer B in 10 min, constant flow of 40% buffer B for 5 min, linear increase to 90% buffer B in 5 min, washing with 100% buffer B for 2 min followed by re-equilibration with 8% buffer B for 8 min. The flow rate was constantly set to 1.5 ml min−1.
Identification of S-mycothiolated peptides using LTQ-Orbitrap Velos mass spectrometry
NEM-alkylated protein extracts from cells exposed to 1 mM NaOCl for 30 min were separated by 15% non-reducing SDS-PAGE followed by tryptic in-gel digestion and LTQ-Orbitrap-Velos mass spectrometry as described9. Post-translational thiol-modifications of proteins were identified by searching all MS/MS spectra in “dta” format against the M. smegmatis mc2155 target-decoy protein sequence database extracted from UniprotKB release 12.7 (UniProt Consortium, Nucleic acids research 2007, 35, D193-197) using Sorcerer™-SEQUEST® (Sequest v. 2.7 rev. 11, Thermo Electron including Scaffold 4.0, Proteome Software Inc., Portland, OR). The SEQUEST search parameters and thiol-modifications were used as described9. The MS proteomics data (raw files and Scaffold files) are deposited into the ProteomeXchange database via the PRIDE partner repository with the dataset identifier PXD003303.
Mass spectrometry (MS)-based thiol-redox proteomics using the OxICAT approach
To obtain 100 µg protein extract, 7–9 ml of the M. smegmatis wild type and mshC mutant cultures were harvested by centrifugation before and 30 min after treatment with 1 mM and 0.5 mM NaOCl, respectively. The OxICAT method was performed according to the protocol of Lindemann and Leichert25, 26 with the modification that cells were disrupted using a ribolyzer. The ICAT-labelled peptides were dissolved in 0.1% (v/v) acetic acid and loaded onto self-packed LC columns with 10 μl of buffer A (0.1% (v/v) acetic acid) at a constant pressure of 220 bar without trapping. Peptides were eluted using a non-linear 85 min gradient from 1 to 99% buffer B (0.1% (v/v) acetic acid in acetonitrile) with a constant flow rate of 300 nl/min and measured using Orbitrap mass spectrometry as described71.
Quantification of thiol-oxidation using the MaxQuant software
The M. smegmatis mc2155 sequence database (accession CP000480 http://www.ncbi.nlm.nih.gov/nuccore/118168627) was used by the search engine Andromeda associated with the MaxQuant software (version 1.5.1.2) to quantify the ICAT-labelled Cys peptides. Two miscleavages were allowed, the parent ion mass tolerance was 10 ppm and the fragment ion mass tolerance was 1.00 Da. The average percentage of oxidation of each Cys peptide and the percentage change under NaOCl stress were calculated from three independent biological replicates using the intensity values provided by MaxQuant. Voronoi treemaps were generated using the Paver software to visualize the percentage oxidation of all identified ICAT-labelled peptide pairs. The MS raw files and MaxQuant search files are deposited into the ProteomeXchange database via the PRIDE partner repository with the dataset identifier PXD003303.
RNA isolation, library preparation and next generation cDNA sequencing
M. smegmatis mc2155 wild-type cells were grown in 3 biological replicates, harvested before and 30 min after exposure to 1 mM NaOCl stress and disrupted in RNA lysis buffer containing 3 mM EDTA and 200 mM NaCl with a Precellys24 Ribolyzer. RNA isolation was performed using the acid phenol extraction protocol as described9. The RNA quality was checked by Trinean Xpose (Gentbrugge,Belgium) and the Agilent RNA Nano 6000 kit using an Agilent 2100 Bioanalyzer (Agilent Technologies, Böblingen, Germany). Ribo-Zero rRNA Removal Kit (Bacteria) from Illumina (San Diego, CA, USA) was used to remove the rRNA. TruSeq Stranded mRNA Library Prep Kit from Illumina (San Diego, CA, USA) was used to prepare the cDNA libraries. The resulting cDNAs were sequenced paired end on an Illumina HiSeq 1500 and MiSeq system (San Diego, CA, USA) using 75 bp read length. The transcriptome sequencing raw datafiles are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number: E-MTAB-4522.
Bioinformatics data analysis, read mapping, data visualization and analysis of differential gene expression
Trimmed reads (26 nt) were mapped to the M. smegmatis mc2155 genome sequence (accession number NC_008596) using SARUMAN72, allowing one error per read. The forward and reverse reads, if both present and with a maximum distance of 1 kb, were combined to one read containing the reference sequence as insert. Paired mappings with a distance >1 kb were discarded, and paired reads with either only the forward or the reverse read mapping were retained as single mapping reads. For the visualization and counting of short read alignments, ReadXplorer v2.273 was used.
Differential gene expression analysis was performed using the software DEseq274 included in the ReadXplorer v2.2 software73. The signal intensity value (a-value) was calculated by log2 mean of normalized read counts and the signal intensity ratio (m-value) by log2 fold change. The evaluation of the differential RNAseq data was performed using an adjusted p-value cut-off of P ≤ 0.01 and a signal intensity ratio (m-value) cut-off of ≥4.47 or ≤−4.47. The latter was determined by applying a significance level of 1% to the experiment with the assumption that the majority of genes are not differentially transcribed. Thus, 99% of all m-values should fall within this range. Therefore, the standard deviation (STDEV) for all m-values was calculated and the cut-off was set to m = 2.58 * STDEV. Genes with an m-value outside this interval and P ≤ 0.01 were considered as differentially transcribed. For the identification of operons, ReadXplorer v2.2 was used73. Therefore, all mapped reads were combined and if two neighbouring genes were connected by at least 20 spanning reads, the genes were considered as an operon.
Identification of SigH- and SigE-regulated promoters with increased transcription under NaOCl stress
The upstream regions of 25 genes with the highest induction under NaOCl stress were selected for manual prediction of promoters. In case of accumulation of mapped reads as visualized by ReadXplorer v2.2, the region −5 nt to −55 nt was defined as promoter region73. For 20 genes, a stack of mapped reads and the corresponding promoter could be identified. These identified promoter regions were analyzed using the MEME algorithm75 and the MEME Suite76 to identify conserved promoter sequences. The identified conserved motif showed high similarity to SigH/SigE-dependent −35 (GGAAY) and −10 (GTT) promoter motifs separated by 17 or 18 bases48. The original MEME motif and an artificial MEME motif with a spacer length of 18 bases were used in the search for additional SigH-dependent promoter sequences. The 5′-regions (−300 nt–+100 nt) of all genes with 2-fold higher expressions after NaOCl treatment (m-value ≥ 1) (P ≤ 0.01) were scanned for both MEME motifs using FIMO77 with a p-value < 0.0001 of the MEME Suite software76. The identified promoter motifs were manually checked for the presence of the highly conserved SigH/SigE-dependent −35 (GGAAY) and −10 (GTT) promoter motifs48. One mismatch was allowed at position 1 or 2 of the −35 motif and another one within the −10 motif. Finally, the clear accumulation of mapped reads downstream of the identified promoters was validated using ReadXplorer v2.273.
Electronic supplementary material
Acknowledgements
We would like to thank the Metabolomics Core Technology Platform of the Excellence cluster “CellNetworks” (University of Heidelberg) for support with LC-based metabolite quantification and Anika Winkler (CeBiTec, Bielefeld University) for excellent technical assistance in transcriptome sequencing. This work was supported by an ERC Consolidator grant (GA 615585) MYCOTHIOLOME and the DFG Research Training Group GRK1947, project [C1] to H.A. We further acknowledge support from the DFG priority program SPP1710 on “Thiol-based redox switches”, projects AN746/4-1 to H.A. and project WI 3560/2-1 and HE 1848/16-1 to M.W. and R.H.
Author Contributions
M.H. and H.A. designed the experiments of this study. M.H. and M.R. performed the redox proteomics experiments, processed and analyzed the redox proteomics data. M.H. and J.B. performed the bioinformatics analysis and treemap constructions. T.B., C.R. and J.K. performed transcriptome sequencing, data processing, gene expression analysis and promoter identification. S.M. and D.B. measured the peptides using mass spectrometry. M.W. and R.H. performed the thiol-metabolomics analysis. M.R. contributed the mshC mutant. M.H. and H.A. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01179-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Loi VV, Rossius M, Antelmann H. Redox regulation by reversible protein S-thiolation in bacteria. Front Microbiol. 2015;6:187. doi: 10.3389/fmicb.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–94. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jothivasan VK, Hamilton CJ. Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat Prod Rep. 2008;25:1091–117. doi: 10.1039/b616489g. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier NA, Newton GL, Fahey RC. A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. J Bacteriol. 2006;188:6245–52. doi: 10.1128/JB.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier NA, Newton GL, Koledin T, Fahey RC. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol Microbiol. 2003;47:1723–32. doi: 10.1046/j.1365-2958.2003.03416.x. [DOI] [PubMed] [Google Scholar]
- 6.Rawat M, Johnson C, Cadiz V, Av-Gay Y. Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem Biophys Res Commun. 2007;363:71–6. doi: 10.1016/j.bbrc.2007.08.142. [DOI] [PubMed] [Google Scholar]
- 7.Liu, Y. B. et al. Physiological roles of mycothiol in detoxification and tolerance to multiple poisonous chemicals in Corynebacterium glutamicum. Arch Microbiol (2013). [DOI] [PubMed]
- 8.Saini V, et al. Ergothioneine Maintains Redox and Bioenergetic Homeostasis Essential for Drug Susceptibility and Virulence of Mycobacterium tuberculosis. Cell Rep. 2016;14:572–85. doi: 10.1016/j.celrep.2015.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi BK, et al. Protein S-mycothiolation functions as redox-switch and thiol protection mechanism in Corynebacterium glutamicum under hypochlorite stress. Antioxid Redox Signal. 2014;20:589–605. doi: 10.1089/ars.2013.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi BK, et al. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid Redox Signal. 2013;18:1273–95. doi: 10.1089/ars.2012.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Laer K, et al. Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol Microbiol. 2012;86:787–804. doi: 10.1111/mmi.12030. [DOI] [PubMed] [Google Scholar]
- 12.Si MR, et al. Corynebacterium glutamicum Methionine Sulfoxide Reductase A Uses both Mycoredoxin and Thioredoxin for Regeneration and Oxidative Stress Resistance. Applied and Environmental Microbiology. 2015;81:2781–2796. doi: 10.1128/AEM.04221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tossounian MA, et al. Corynebacterium diphtheriae Methionine Sulfoxide Reductase A Exploits a Unique Mycothiol Redox Relay Mechanism. Journal of Biological Chemistry. 2015;290:11365–11375. doi: 10.1074/jbc.M114.632596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugo M, et al. Mycothiol/mycoredoxin 1-dependent reduction of the peroxiredoxin AhpE from Mycobacterium tuberculosis. J Biol Chem. 2014;289:5228–39. doi: 10.1074/jbc.M113.510248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smeulders MJ, Keer J, Speight RA, Williams HD. Adaptation of Mycobacterium smegmatis to stationary phase. Journal of Bacteriology. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton GL, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–5. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–7. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 19.D’Aquino JA, Tetenbaum-Novatt J, White A, Berkovitch F, Ringe D. Mechanism of metal ion activation of the diphtheria toxin repressor DtxR. Proc Natl Acad Sci USA. 2005;102:18408–13. doi: 10.1073/pnas.0500908102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenzie JL, et al. A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux in mycobacteria. J Bacteriol. 2012;194:2189–204. doi: 10.1128/JB.06790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–44. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bazet Lyonnet B, et al. Pleiotropic effect of AccD5 and AccE5 depletion in acyl-coenzyme A carboxylase activity and in lipid biosynthesis in mycobacteria. PLoS One. 2014;9:e99853. doi: 10.1371/journal.pone.0099853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurth DG, et al. ACCase 6 is the essential acetyl-CoA carboxylase involved in fatty acid and mycolic acid biosynthesis in mycobacteria. Microbiology. 2009;155:2664–75. doi: 10.1099/mic.0.027714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ta P, Buchmeier N, Newton GL, Rawat M, Fahey RC. Organic Hydroperoxide Resistance Protein and Ergothioneine Compensate for Loss of Mycothiol in Mycobacterium smegmatis Mutants. Journal of Bacteriology. 2011;193:1981–1990. doi: 10.1128/JB.01402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leichert LI, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindemann C, Leichert LI. Quantitative redox proteomics: the NOxICAT method. Methods Mol Biol. 2012;893:387–403. doi: 10.1007/978-1-61779-885-6_24. [DOI] [PubMed] [Google Scholar]
- 27.Caselli A, et al. The inactivation mechanism of low molecular weight phosphotyrosine-protein phosphatase by H2O2. Journal of Biological Chemistry. 1998;273:32554–32560. doi: 10.1074/jbc.273.49.32554. [DOI] [PubMed] [Google Scholar]
- 28.Ecco G, et al. Mycobacterium tuberculosis tyrosine phosphatase A (PtpA) activity is modulated by S-nitrosylation. Chem Commun (Camb) 2010;46:7501–3. doi: 10.1039/c0cc01704c. [DOI] [PubMed] [Google Scholar]
- 29.Matiollo C, et al. S-nitrosylation of Mycobacterium tuberculosis tyrosine phosphatase A (PtpA) induces its structural instability. Biochim Biophys Acta. 2013;1834:191–6. doi: 10.1016/j.bbapap.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Men L, Wang YS. The oxidation of yeast alcohol dehydrogenase-1 by hydrogen peroxide in vitro. Journal of Proteome Research. 2007;6:216–225. doi: 10.1021/pr0603809. [DOI] [PubMed] [Google Scholar]
- 31.Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biology. 2001;2:14. doi: 10.1186/gb-2001-2-9-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maciag A, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. Journal of Bacteriology. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mowa MB, Warner DF, Kaplan G, Kana BD, Mizrahi V. Function and Regulation of Class I Ribonucleotide Reductase-Encoding Genes in Mycobacteria. Journal of Bacteriology. 2009;191:985–995. doi: 10.1128/JB.01409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paget MS, Kang JG, Roe JH, Buttner MJ. sigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2) EMBO J. 1998;17:5776–82. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang JG, et al. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999;18:4292–8. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajasekar KV, et al. The anti-sigma factor RsrA responds to oxidative stress by reburying its hydrophobic core. Nat Commun. 2016;7:12194. doi: 10.1038/ncomms12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14:1049–63. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hillion, M. & Antelmann, H. Thiol-based redox switches in prokaryotes. Biol Chem (2015). [DOI] [PMC free article] [PubMed]
- 39.Raghunand TR, Bishai WR. Mycobacterium smegmatis whmD and its homologue Mycobacterium tuberculosis whiB2 are functionally equivalent. Microbiology-Sgm. 2006;152:2735–2747. doi: 10.1099/mic.0.28911-0. [DOI] [PubMed] [Google Scholar]
- 40.Anstrom DM, Kallio K, Remington SJ. Structure of the Escherichia coli malate synthase G: pyruvate: acetyl-coenzyme A abortive ternary complex at 1.95 angstrom resolution. Protein Science. 2003;12:1822–1832. doi: 10.1110/ps.03174303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stapleton MR, Smith LJ, Hunt DM, Buxton RS, Green J. Mycobacterium tuberculosis WhiB1 represses transcription of the essential chaperonin GroEL2. Tuberculosis (Edinb) 2012;92:328–32. doi: 10.1016/j.tube.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith LJ, Stapleton MR, Buxton RS, Green J. Structure-function relationships of the Mycobacterium tuberculosis transcription factor WhiB1. PLoS One. 2012;7:e40407. doi: 10.1371/journal.pone.0040407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith LJ, et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem J. 2010;432:417–27. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garg S, Alam MS, Bajpai R, Kishan KR, Agrawal P. Redox biology of Mycobacterium tuberculosis H37Rv: protein-protein interaction between GlgB and WhiB1 involves exchange of thiol-disulfide. BMC Biochem. 2009;10:1. doi: 10.1186/1471-2091-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. Bmc Structural Biology. 2009;9:10. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leichert LI, Dick TP. Incidence and physiological relevance of protein thiol switches. Biol Chem. 2015;396:389–99. doi: 10.1515/hsz-2014-0314. [DOI] [PubMed] [Google Scholar]
- 47.Sharp JD, et al. Comprehensive Definition of the SigH Regulon of Mycobacterium tuberculosis Reveals Transcriptional Control of Diverse Stress Responses. PLoS One. 2016;11:e0152145. doi: 10.1371/journal.pone.0152145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song T, Song SE, Raman S, Anaya M, Husson RN. Critical role of a single position in the −35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J Bacteriol. 2008;190:2227–30. doi: 10.1128/JB.01642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berney M, Greening C, Conrad R, Jacobs WR, Jr., Cook GM. An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci USA. 2014;111:11479–84. doi: 10.1073/pnas.1407034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chinta KC, et al. The emerging role of gasotransmitters in the pathogenesis of tuberculosis. Nitric Oxide. 2016;59:28–41. doi: 10.1016/j.niox.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gray MJ, Jakob U. Oxidative stress protection by polyphosphate–new roles for an old player. Curr Opin Microbiol. 2015;24:1–6. doi: 10.1016/j.mib.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray MJ, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53:689–99. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hawkins CL, Pattison DI, Davies MJ. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids. 2003;25:259–74. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 54.Hernick M. Mycothiol: a target for potentiation of rifampin and other antibiotics against Mycobacterium tuberculosis. Expert Rev Anti Infect Ther. 2013;11:49–67. doi: 10.1586/eri.12.152. [DOI] [PubMed] [Google Scholar]
- 55.Nilewar SS, Kathiravan MK. Mycothiol: a promising antitubercular target. Bioorg Chem. 2014;52:62–8. doi: 10.1016/j.bioorg.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Song TS, Dove SL, Lee KH, Husson RN. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Molecular Microbiology. 2003;50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 57.Barik S, Sureka K, Mukherjee P, Basu J, Kundu M. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Molecular Microbiology. 2010;75:592–606. doi: 10.1111/j.1365-2958.2009.07008.x. [DOI] [PubMed] [Google Scholar]
- 58.Graumann J, et al. Activation of the redox-regulated molecular chaperone Hsp33–a two-step mechanism. Structure. 2001;9:377–87. doi: 10.1016/S0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 59.Ilbert M, et al. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–63. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakob U, Eser M, Bardwell JC. Redox switch of hsp33 has a novel zinc-binding motif. J Biol Chem. 2000;275:38302–10. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- 61.Wholey WY, Jakob U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol Microbiol. 2012;83:981–91. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 63.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci USA. 2005;102:467–72. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–6. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 65.Ecco G, et al. Mycobacterium tuberculosis tyrosine phosphatase A (PtpA) activity is modulated by S-nitrosylation. Chemical Communications. 2010;46:7501–7503. doi: 10.1039/c0cc01704c. [DOI] [PubMed] [Google Scholar]
- 66.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr., Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104:11568–73. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ta P, Buchmeier N, Newton GL, Rawat M, Fahey RC. Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J Bacteriol. 2011;193:1981–90. doi: 10.1128/JB.01402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vargas, D., Hageman, S., Gulati, M., Nobile, C. J. & Rawat, M. S-nitrosomycothiol reductase and mycothiol are required for survival under aldehyde stress and biofilm formation in Mycobacterium smegmatis. IUBMB Life (2016). [DOI] [PMC free article] [PubMed]
- 69.Pedre B, et al. The Corynebacterium glutamicum mycothiol peroxidase is a reactive oxygen species-scavenging enzyme that shows promiscuity in thiol redox control. Molecular Microbiology. 2015;96:1176–1191. doi: 10.1111/mmi.12998. [DOI] [PubMed] [Google Scholar]
- 70.Wirtz M, Droux M, Hell R. O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot. 2004;55:1785–98. doi: 10.1093/jxb/erh201. [DOI] [PubMed] [Google Scholar]
- 71.Bonn F, et al. Picking Vanished Proteins from the Void: How to Collect and Ship/Share Extremely Dilute Proteins in a Reproducible and Highly Efficient Manner. Analytical Chemistry. 2014;86:7421–7427. doi: 10.1021/ac501189j. [DOI] [PubMed] [Google Scholar]
- 72.Blom J, et al. Exact and complete short-read alignment to microbial genomes using Graphics Processing Unit programming. Bioinformatics. 2011;27:1351–8. doi: 10.1093/bioinformatics/btr151. [DOI] [PubMed] [Google Scholar]
- 73.Hilker R, et al. ReadXplorer–visualization and analysis of mapped sequences. Bioinformatics. 2014;30:2247–54. doi: 10.1093/bioinformatics/btu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 76.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


