Abstract
Our previous study confirmed the protective potential of Lactobacillus plantarum (L. plantarum) strains in alleviation of cadmium (Cd) toxicity in vivo and demonstrated that the observed protection largely depended on the tolerance of the strains to Cd-induced stress. It was also observed that there were significant intra-species differences in Cd tolerance of L. plantarum strains. In this study, we investigated the mechanism of Cd induced stress response of L. plantarum strains using the isobaric tags for relative and absolute quantitation (iTRAQ) based comparative proteomics. L. plantarum CCFM8610 (strongly resistant to Cd) and L. plantarum CCFM191 (sensitive to Cd) were selected as target strains, and their proteomic profiles in the presence and absence of Cd exposure were compared. We propose that the underlying mechanism of the exceptional Cd tolerance of CCFM8610 may be attributed to the following: (a) a specific energy-conservation survival mode; (b) mild induction of its cellular defense and repair system; (c) an enhanced biosynthesis of hydrophobic amino acids in response to Cd; (d) inherent superior Cd binding ability and effective cell wall biosynthesis ability; (e) a tight regulation on ion transport; (f) several key proteins, including prophage P2b protein 18, CadA, mntA and lp_3327.
Introduction
Cadmium (Cd) is a representative non-essential element and known as an environmental hazard to human health. This heavy metal can contaminate the food chain and cause cumulative toxic effects in the liver, kidney, bone and the reproductive systems of humans1–3. Cd has also been classified as a potent human carcinogen by the International Agency for Research on Cancer4. Chelation therapies, the most commonly used treatments for heavy metal toxicity, have a number of safety and efficacy concerns and as yet none of them have been approved for clinical use against Cd poisoning in humans5, 6. The development of dietary supplements against Cd toxicity represents a potential alternative strategy.
Lactic acid bacteria (LAB) are members of commensal inhabitants of the human intestinal microbiota that can confer health benefits on the host7. Among these LAB, Lactobacillus plantarum (L. plantarum) strains have been widely used in the food industry as probiotics and functional food supplements8. Our previous studies have demonstrated that L. plantarum CCFM8610 can sequester Cd in the intestines of the host, which in turn promotes fecal Cd excretion and reduction in Cd accumulation in tissues, indicating that this strain can be considered a dietary supplement for the prevention and alleviation of Cd toxicity9–11. It was further noted that CCFM8610 offers significantly better protection than some other L. plantarum strains in vivo 11, and the protective effects were dependent on bacterial cell viability9.
Cd exposure, even at low levels, can disrupt cell wall synthesis, respiratory chain function and metal ion homeostasis in microorganisms, which in turn causes oxidative stress, DNA damage and energy metabolism dysfunctions, inducing extreme cellular toxic effects12–14. As L. plantarum cells contact with Cd directly in the intestines of the host, the protective effects of the strains against Cd toxicity largely depend on their abilities to thrive after Cd exposure. Therefore, understanding the mechanism of Cd tolerance of L. plantarum strains is essential for the development of probiotic based strategy against Cd toxicity. Our previous work showed that the minimum inhibitory concentration (MIC) of L. plantarum CCFM8610 (tested on Cd-containing agar plates) is more than 1000 mg/L, while some other L. plantarum strains have MIC values 10 to 20 times lower15. A bacterium is considered tolerant to Cd if its MIC value exceeds 100 mg/L16 or 112.4 mg/L of Cd17. Based on this definition, L. plantarum CCFM8610 can be considered a highly Cd-resistant strain. This may partly explain the protection of the host against Cd toxicity in vivo offered by this strain. Therefore, it is of interest to understand the resistance mechanism of L. plantarum strains against Cd exposure, and to explore why CCFM8610 is highly tolerant to Cd.
The physiological bases of heavy metal tolerance have been well investigated in environmental and industrial microorganisms such as Pseudomonas spp., Escherichia coli, Bacillus subtilis and Saccharomyces cerevisia 12, 13, 18–20. The related metabolomic pathways and key proteins identified include sulfur assimilation/glutathione synthesis pathway, metallothionein, oxidoreductase and ion transport proteins. However, comprehensive heavy metal tolerance mechanisms have not yet been well defined in LAB strains. To our knowledge, the Cd resistance modes have only been studied in Streptococcus thermophiles and Lactococcus lactis, with a focus on two Cd resistance-associated genes, CadA and CadC21–24. Numbers of reports also indicated the presence of these genes in some strains of L. plantarum species25, 26, but the Cd stress response network in these strains is yet to be elucidated.
Proteomics has been reported to be efficient to provide global physiological profiles of bacteria in protein level27–29. The most commonly used approaches for proteomic analysis are gel-based methods, such as two-dimensional gel electrophoresis (2-DE) and two-dimensional difference gel electrophoresis (2-D DIGE). However, these methods were reported to have limitations in sensibility, reproducibility, and proteome coverage30, 31. Isobaric tags for relative and absolute quantitation (iTRAQ) is a labelling approach that allows reliable quantitative description of differentially regulated proteins in complex systems. The main advantage is to have the possibility to analyze several samples together, with biological and technical replicates (4 or 8 plex). The subsequent use of high resolution mass spectrometry analyses provides accurate relative ratio between protein concentrations, present in the different samples31. Therefore, this approach was selected in this study.
In this study, the Cd-tolerance related key proteins and pathways within the L. plantarum species were investigated by using iTRAQ based proteomic approach. L. plantarum strains CCFM8610 (strongly resistant to Cd) and CCFM191 (sensitive to Cd) were selected for comparative proteomic analysis based on their differing Cd tolerant phenotype. The comparative proteomic profiles between non-stimulating and Cd-exposed conditions were compared in the two strains. The proteomic results were further confirmed by RT-qPCR and by the measurement of several biological properties of the bacterial cells in response to Cd exposure.
Results
Cd tolerance
Twenty L. plantarum strains were cultured in MRS broth containing different concentrations of Cd, and the relative growth rates were determined (Table 1). While increasing Cd concentration caused a continuous decrease in the growth rate of all strains, significant Cd tolerance diversity could be observed, and the 20 strains could be categorized into four general groups, MIC > 50 mg/L, MIC = 50 mg/L, MIC = 20 mg/L and MIC = 10 mg/L. All tested strains showed no obvious growth since the Cd concentration reaching over 100 ppm (data not shown). L. plantarum CCFM8610 belonged to one of the eight strains with the highest MIC values, which was in agreement with our previous study on the tolerance of LAB strains tested on Cd-containing agar plates15. Based on the category of Cd tolerance of bacteria reported in a previous report32, CCFM191, one of the three strains with lowest MIC value (10 ppm), was selected as a Cd-sensitive strain for the comparative proteomic analysis with CCFM8610. As shown in Fig. 1, the dose of Cd at 5 mg/L (1/2 MIC value of the Cd-sensitive strain CCFM191) was selected for modeling a moderate and sublethal Cd exposure in the following experiments based on the previous related study33, 34.
Table 1.
Relative growth rate of L. plantarum strains grown in MRS broth containing different Cd concentrations.
| MIC value | Relative growth ratea of strains grown in MRS broth containing different Cd concentrations (%) | |||||
|---|---|---|---|---|---|---|
| Strains | 5 ppm | 10 ppm | 20 ppm | 50 ppm | 100 ppm | |
| MIC > 50 | CCFM11 | 96.31 ± 0.55 | 91.52 ± 1.02 | 60.47 ± 0.72 | 18.98 ± 0.18 | 13.61 ± 0.47 |
| CCFM232 | 94.92 ± 0.40 | 89.36 ± 0.35 | 63.53 ± 3.24 | 16.38 ± 0.55 | 11.13 ± 0.17 | |
| CCFM240 | 94.92 ± 0.34 | 92.49 ± 0.42 | 64.09 ± 1.31 | 17.62 ± 0.59 | 11.34 ± 0.23 | |
| CCFM8610 | 90.92 ± 0.98 | 86.75 ± 0.54 | 67.21 ± 4.00 | 20.26 ± 1.63 | 13.71 ± 0.35 | |
| CCFM405 | 101.93 ± 0.39 | 90.78 ± 2.26 | 60.30 ± 1.50 | 20.22 ± 0.90 | 11.61 ± 0.25 | |
| CCFM595 | 102.01 ± 0.07 | 101.23 ± 0.23 | 91.38 ± 1.63 | 29.37 ± 0.88 | 10.52 ± 0.45 | |
| CCFM579 | 96.81 ± 0.21 | 91.22 ± 0.17 | 64.47 ± 0.52 | 21.98 ± 0.03 | 12.88 ± 0.61 | |
| CCFM8661 | 99.01 ± 0.59 | 92.20 ± 0.88 | 60.92 ± 0.28 | 19.44 ± 0.97 | 12.69 ± 0.31 | |
| MIC = 50 | CCFM241 | 99.68 ± 0.09 | 100.04 ± 0.69 | 79.28 ± 2.49 | 8.73 ± 0.15 | 6.05 ± 0.30 |
| CCFM231 | 95.68 ± 2.11 | 92.13 ± 3.22 | 61.76 ± 2.74 | 7.61 ± 0.05 | 5.61 ± 0.09 | |
| CCFM198 | 102.46 ± 0.30 | 94.91 ± 1.06 | 25.36 ± 3.71 | 7.50 ± 0.29 | 5.79 ± 0.14 | |
| CCFM239 | 98.09 ± 0.51 | 95.67 ± 0.43 | 45.55 ± 2.47 | 7.56 ± 0.22 | 5.37 ± 0.14 | |
| MIC = 20 | CCFM308 | 79.68 ± 0.51 | 19.30 ± 0.38 | 9.42 ± 0.33 | 6.00 ± 0.36 | 3.99 ± 0.28 |
| CCFM309 | 84.95 ± 1.46 | 21.58 ± 0.86 | 10.96 ± 0.43 | 6.42 ± 0.09 | 4.62 ± 0.11 | |
| CCFM602 | 97.35 ± 0.62 | 84.03 ± 0.46 | 9.01 ± 0.56 | 4.55 ± 0.29 | 2.59 ± 0.08 | |
| CCFM605 | 96.78 ± 0.45 | 72.04 ± 1.99 | 12.11 ± 0.67 | 4.32 ± 0.20 | 3.30 ± 0.45 | |
| CCFM166 | 91.17 ± 2.12 | 37.30 ± 1.80 | 4.76 ± 0.47 | 3.20 ± 0.15 | 2.09 ± 0.03 | |
| MIC = 10 | CCFM191 | 84.63 ± 1.14 | 9.30 ± 0.06 | 6.69 ± 0.25 | 6.64 ± 0.38 | 4.76 ± 0.10 |
| CCFM436 | 95.82 ± 0.84 | 13.94 ± 0.96 | 4.81 ± 0.02 | 4.16 ± 0.13 | 3.71 ± 0.01 | |
| CCFM578 | 94.88 ± 0.48 | 9.51 ± 0.32 | 1.32 ± 0.09 | 4.56 ± 0.26 | 1.21 ± 0.25 | |
aRelative growth rate of each strain was expressed as percentage of OD600 value of control culture (without Cd exposure) which was assigned a value of 100%. Data are expressed as mean ± SEM of three independent experiments per assay.
Figure 1.
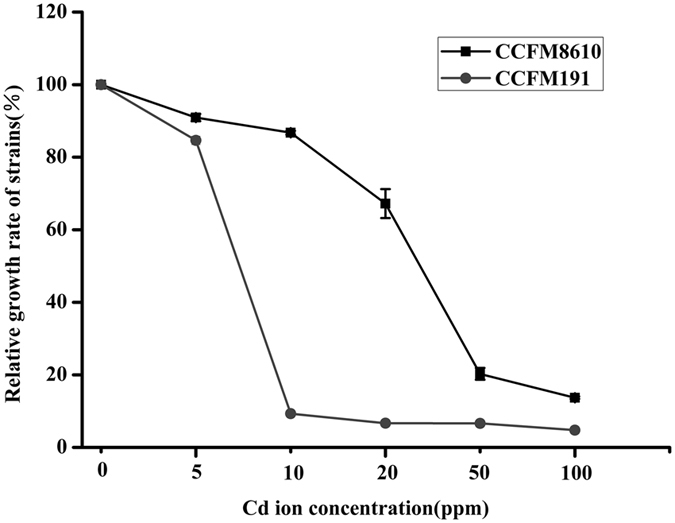
Relative growth rate of L. plantarum CCFM8610 and CCFM191 grown in MRS broth of different Cd concentrations. Relative growth rate was expressed as percentage of OD600 value of control culture (without Cd exposure) which was assigned a value of 100%. Data are expressed as mean ± SEM of three independent experiments per assay.
Proteomic characteristics of two L. plantarum strains
1592 and 1527 proteins were detected and identified in CCFM8610 (with and without Cd exposure, “A” round) and CCFM191 (with and without Cd exposure, “B” round), respectively (Fig. 2). The 1415 overlapped proteins identified in both rounds were selected for protein function analysis. The protein function annotation was conducted by Gene Ontology (GO) analysis (Fig. 3), and the related metabolomics pathways of these proteins were analyzed by KEGG classification (Fig. 4). The results indicated that the detected proteins covered a large range with functions categorizing into biological process, cellular component and molecular function. The relevant metabolic pathways were related to carbohydrate metabolism (149 proteins), amino acid metabolism (120 proteins), translation (83 proteins), membrane transport (78 proteins), lipid metabolism (49 proteins), etc.
Figure 2.
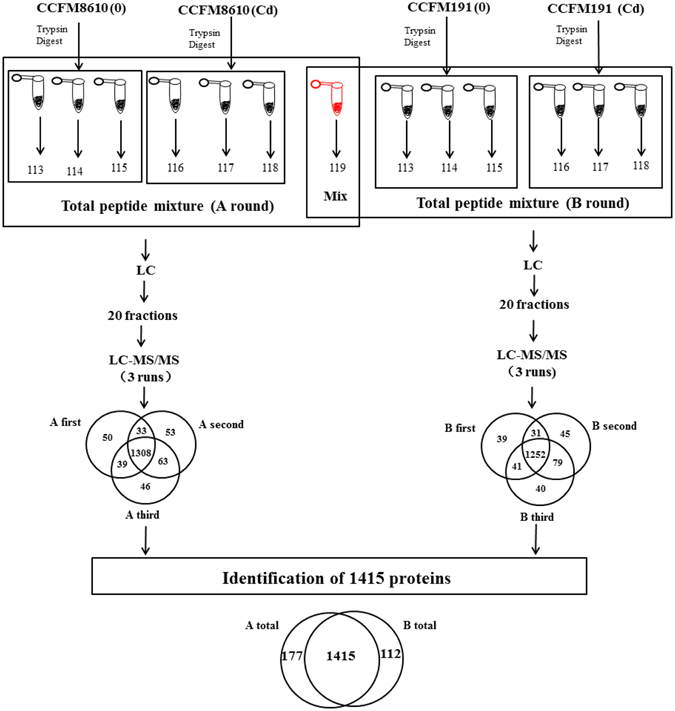
Workflow of iTRAQ experiment.
Figure 3.
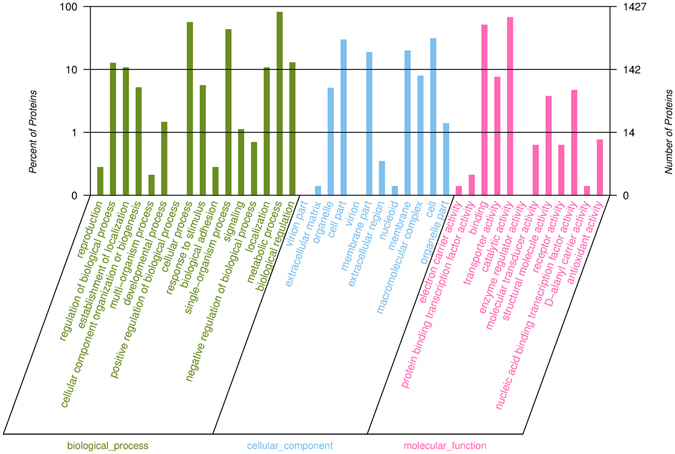
Functional categories of overlapped proteins identified in two L. plantarum strains by GO analysis. GO analysis was conducted by the software blast2go with the GO ID from the ensmbl database for each identified protein.
Figure 4.
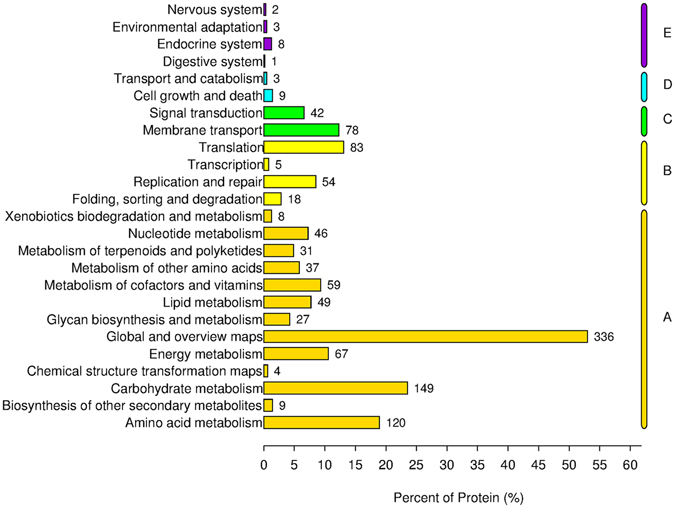
Related metabolomics pathways of overlapped proteins identified in two L. plantarum strains by KEGG classification. Each digital on the right of each bar indicates the number of proteins in each category. The letters A, B, C, D and E represent the 5 branches in KEGG pathways including Metabolism (A), Genetic Information Processing (B), Environmental Information Processing (C), Cellular Processes (D) and Organismal Systems (E).
Differentially expressed protein profiles
Intra-species difference of inherent proteomic profiles between Cd resistant and sensitive strains was evaluated, so as to pinpoint the proteins that might be implicated in the Cd stress response process. Total of 206 proteins with fold change more than 1.5 were selected as differentially expressed proteins (Table 2) for the comparison of CCFM8610 and CCFM191 in non-treated conditions (without Cd exposure). These proteins were categorized into global stress response, carbohydrate and lipid metabolism, transporters, some membrane and extracellular proteins, etc., based on the KEGG pathway analysis and their annotated functions in the Uniprot database. Compared with CCFM191, lower abundances of proteins that belong to global stress response, carbohydrate metabolism, phosphotransferase (PTS) system, two-component system, membrane protein and cell surface protein and hydrolase could be observed in CCFM8610. On the other hand, proteins involved in amino acid metabolism, nucleic acid metabolism and extracellular protein showed higher abundances in CCFM8610.
Table 2.
Differentially expressed proteins between L. plantarum CCFM8610 and CCFM191 in Cd-free conditions.
| Categorya | Accessionb | Descriptionc | FCd | |
|---|---|---|---|---|
| Global Stress Response | DNA repair, metabolism, regulation | F9UQY3 | lp_2444; Prophage P2a protein 13 | −3.30 |
| F9ULA4 | lp_0641; Prophage P1 protein 18, DNA single-strand annealing protein RecT | −1.87 | ||
| Q6LWF5 | traI; DNA topoisomerase | 6.30 | ||
| F9URZ6 | endA; DNA-entry nuclease | 1.58 | ||
| Q88V16 | mutS2; Endonuclease MutS2 | −1.70 | ||
| Q88W97 | recU; Holliday junction resolvase RecU | −1.52 | ||
| Oxidoreductase | F9UUA8 | lp_3430; Peroxidase | 5.14 | |
| F9USL4 | lp_3100; Aldo/keto reductase family protein | 3.41 | ||
| F9UPP0 | lp_1918; NAD(P)(H)-dependent oxidoreductase, quinone oxidoreductase (QOR) family | 2.10 | ||
| F9URK6 | lp_2732; NADPH-dependent FMN reductase family protein | −1.50 | ||
| F9UQD7 | lp_2212; NADH-flavin reductase | −1.51 | ||
| F9UQI9 | trxA2; Thioredoxin | −1.56 | ||
| F9UPR0 | lp_1939; Oxidoreductase, medium chain dehydrogenases/reductase (MDR)/zinc-dependent alcohol dehydrogenase-like family | −1.64 | ||
| F9USA2 | lp_2968; Nitroreductase | −1.65 | ||
| F9UTJ8 | lp_3318; Aldo/keto reductase family protein | −1.65 | ||
| F9USK5 | lp_3091; Short-chain dehydrogenase/oxidoreductase, atypical SDR family, subgroup 1 | −2.83 | ||
| F9UT37 | trxA1; Thioredoxin | 2.04 | ||
| F9UTD6 | lp_3236; Short-chain dehydrogenase/oxidoreductase, atypical SDR family, TMR-like | 1.70 | ||
| F9URB2 | lp_2604; NAD(P)-dependent oxidoreductase | 1.53 | ||
| F9US17 | nrdG; Anaerobic ribonucleoside-triphosphate reductase-activating protein | 1.55 | ||
| F9UTT1 | acdH; Acetaldehyde dehydrogenase | 1.55 | ||
| F9UNI0 | ribB; Riboflavin synthase, alpha chain | −5.89 | ||
| F9UR64 | npr2; NADH peroxidase | −4.38 | ||
| F9ULD3 | cat; Catalase | −3.76 | ||
| F9UN44 | gshR2; glutathione reductase | −4.67 | ||
| F9UUC2 | nox5; NADH oxidase | −2.30 | ||
| F9UTJ6 | pflA; Pyruvate formate-lyase-activating enzyme | −2.09 | ||
| F9UUK7 | lp_3545; D-arabitol-phosphate dehydrogenase | −3.08 | ||
| F9USK6 | gabD; succinate-semialdehyde dehydrogenase (NAD(P) + ) | −2.06 | ||
| Protein repair | F9UPH1 | msrA; Peptide methionine sulfoxide reductase MsrA | −6.02 | |
| Q88W33 | msrB; Peptide methionine sulfoxide reductase MsrB | −2.67 | ||
| Protease | F9UTF5 | lp_3259; Zinc-dependent proteinase | −1.96 | |
| F9UT31 | pepD1; Dipeptidase | −1.54 | ||
| Other | F9US30 | lp_2952; Bacteriocin immunity protein | −1.58 | |
| F9URC0 | lp_2616; Bacteriocin immunity protein | −1.96 | ||
| F9USV1 | hsp1; Small heat shock protein | −2.32 | ||
| Q88V03 | ruvB; Holliday junction ATP-dependent DNA helicase RuvB | 1.92 | ||
| Regulation network | Transcriptional regulation | F9UT59 | treR; Trehalose operon transcriptional repressor, GntR family | −1.91 |
| F9UM95 | lp_0892; Transcription regulator, MarR family | −1.70 | ||
| Q88X36 | argR1; Arginine regulator | −1.65 | ||
| F9UNC5 | lp_1360; Transcription regulator, MarR family | 2.06 | ||
| F9USP8 | lp_3138; Bifunctional protein: transcriptional antiterminator, BglG family PTS system, EIIA component | 1.87 | ||
| F9UPQ9 | lp_1938; Transcription regulator, LysR family | 1.82 | ||
| F9USH9 | lp_3060; Transcription regulator, AraC family | 1.53 | ||
| F9UMJ7 | lp_1020; Transcription regulator, TetR family | 1.57 | ||
| F9UL40 | tex; Transcription accessory protein, contains S1 RNA binding domain | −1.82 | ||
| Other regulation proteins | Q88XG5 | recX; Regulatory protein RecX | 2.02 | |
| F9UM52 | spx1; RNA polymerase (RNAP)-binding regulatory protein, arsenate reductase (ArsC) family, Spx subfamily | −1.59 | ||
| F9ULW6 | lp_0737; Sigma 54 modulation protein/ribosomal protein S30EA | −1.53 | ||
| Carbohydrate metabolism | TCA cycle | F9UMS1 | fum; fumarate hydratase | −3.81 |
| F9UQ90 | pdhD; pyruvate dehydrogenase complex, E3 component; dihydrolipoamide dehydrogenase | −1.90 | ||
| F9UQ91 | pdhC; pyruvate dehydrogenase complex, E2 component; dihydrolipoamide S-acetyltransferase | −2.30 | ||
| F9UQ92 | pdhB; pyruvate dehydrogenase complex, E1 component, beta subunit | −2.21 | ||
| F9UQ93 | pdhA; pyruvate dehydrogenase complex, E1 component, alpha subunit | −2.12 | ||
| Pyruvate metabolism | F9UTR4 | ack2; acetate kinase | −1.59 | |
| F9UM63 | pox1; pyruvate oxidase | −2.79 | ||
| P59390 | ldhL2; L-lactate dehydrogenase | −3.36 | ||
| Q88VJ2 | ldhD; D-lactate dehydrogenase | 1.69 | ||
| F9URC8 | pox3; pyruvate oxidase | −8.87 | ||
| F9UTJ5 | pflB; formate C-acetyltransferase | −1.85 | ||
| P37063 | pox5; pyruvate oxidase | −4.19 | ||
| Pentose phosphate pathway | F9UN42 | gntK; gluconokinase | −1.99 | |
| F9URA8 | tal1; Transaldolase | −5.21 | ||
| F9UN43 | lp_1251;6-phosphogluconate dehydrogenase | −7.13 | ||
| Q88S87 | xfp; xylulose-5-phosphate phosphoketolase | −1.64 | ||
| F9ULK7 | rbsK1; ribokinase | −1.72 | ||
| Glycolysis | Q88YY8 | pgm2; phosphoglycerate mutase family protein | 2.56 | |
| F9URP6 | pbg4; 6-phospho-beta-glucosidase | −1.94 | ||
| F9URP7 | pbg5; 6-phospho-beta-glucosidase | −2.03 | ||
| Other | F9US84 | pgmB2; Beta-phosphoglucomutase | −2.71 | |
| F9USZ1 | malS; Alpha-amylase, maltodextrins and cyclomaltodextrins | −1.61 | ||
| F9USY2 | dak3; Dihydroxyacetone phosphotransferase, phosphoryl donor protein | −1.66 | ||
| F9UP85 | mapA; Maltose phosphorylase | −2.73 | ||
| Q88RZ2 | rbsD; D-ribose mutarotase | −1.78 | ||
| Q88S51 | rhaA; L-rhamnose isomerase | −1.56 | ||
| PTS system | PTS system | F9UT61 | pts4ABC; PTS system trehalose-specific transporter subunit IIBC | −1.86 |
| F9UL45 | pts9AB; PTS system, mannose-specific EIIAB component | 1.73 | ||
| F9UL47 | pts9D; PTS system, mannose-specific EIID component | 1.82 | ||
| F9UL56 | pts10A; PTS system, mannose-specific EIIA component | −1.52 | ||
| F9UL57 | pts10B; PTS system, mannose-specific EIIB component | −3.91 | ||
| F9URE1 | pts19D; PTS system, N-acetylglucosamine–specific EIID component | −1.79 | ||
| F9URE3 | pts19B; PTS system, N-acetylglucosamine–specific EIIB component | −1.70 | ||
| F9URP8 | pts20A; PTS system, cellobiose-specific EIIA component | −2.47 | ||
| F9URP9 | pts20B; PTS system, cellobiose-specific EIIB component | −2.99 | ||
| F9UUH8 | pts30BCA; PTS system, beta-glucoside–specific EIIBCA component | −1.71 | ||
| F9UUK9 | pts35B; PTS system, galactitol-specific EIIB component | −2.50 | ||
| F9ULA5 | pts35A; PTS system, galactitol-specific EIIA component | −1.54 | ||
| F9UUH6 | lp_3510; PTS-associated protein | −1.80 | ||
| Amino acid metabolism | Lysine biosynthesis | F9UPP5 | dapE1; succinyl-diaminopimelate desuccinylase | 2.86 |
| F9URV7 | dapE2; succinyl-diaminopimelate desuccinylase | 1.95 | ||
| F9UT53 | cblB; cystathionine beta-lyase/cystathionine gamma-lyase | −3.54 | ||
| Sulfur amino acid metabolism | F9UQB3 | iscS; Cysteine desulfurase | 1.64 | |
| F9UTH2 | lp_3283; Methonine synthase (Cobalamine-independent), C-terminal domain | −2.40 | ||
| Q88UW5 | gshAB; glutathione biosynthesis bifunctional protein: glutamate-cysteine ligase; glutathione synthetase | 1.50 | ||
| F9UR58 | oahS; O-acetylhomoserine sulfhydrylase | −2.44 | ||
| Other | Q88UT5 | glyA; glycine hydroxymethyltransferase | 1.53 | |
| Q88WI3 | trpD; anthranilate phosphoribosyltransferase | 4.58 | ||
| Nucleic acid metabolism | F9UT41 | ndk; nucleoside-diphosphate kinase | −1.72 | |
| F9UM77 | gmk2; guanylate kinase | 1.64 | ||
| F9URN3 | lp_2762; phosphohydrolase | 1.87 | ||
| F9US18 | nrdD; anaerobic ribonucleoside-triphosphate reductase | 2.16 | ||
| P71479 | pyrR1; pyrimidine operon regulator | 1.62 | ||
| F9UNA0 | dgk2; Deoxynucleoside kinase | 2.04 | ||
| Lipid metabolism | Fatty acid biosynthesis | F9UP42 | accC2; acetyl-CoA carboxylase, biotin carboxylase subunit | 1.90 |
| Q88WG0 | accD2; acetyl-CoA carboxylase, carboxyl transferase subunit beta | 1.84 | ||
| F9UP44 | accA2; acetyl-CoA carboxylase, carboxyl transferase subunit alpha | 1.98 | ||
| Glycerol lipid metabolism | F9USY0 | dak1B; dihydroxyacetone phosphotransferase, dihydroxyacetone binding subunit | −1.70 | |
| F9USY1 | dak2; dihydroxyacetone phosphotransferase, ADP-binding subunit | −1.70 | ||
| Q88ZF1 | glpK; glycerol kinase | −5.69 | ||
| F9UTW9 | glpF3; Glycerol uptake facilitator protein | −3.30 | ||
| Q88YD9 | glpK2; Glycerol kinase 2 | −3.04 | ||
| F9UT65 | tagF1; CDP-glycerol glycerophosphotransferase | −2.15 | ||
| F9UT64 | tagD1; glycerol-3-phosphate cytidylyltransferase | −5.85 | ||
| F9UPG2 | tarL; Ribitolphosphotransferase | 4.22 | ||
| F9UTW8 | glpD; glycerol-3-phosphate dehydrogenase, FAD-dependent | −6.09 | ||
| Terpenoid backbone biosynthesis | Q88W46 | tarI; D-ribitol-5-phosphate cytidylyltransferase | 6.81 | |
| F9URB5 | dxs; 1-deoxy-D-xylulose-5-phosphate synthase | 2.46 | ||
| Other | F9UT87 | cfa2; Cyclopropane-fatty-acyl-phospholipid synthase | 1.67 | |
| F9UMC4 | lp_0925; Acyltransferase | 2.30 | ||
| Two-component system | F9UMR7 | citC; [citrate (pro-3S)-lyase] ligase | −1.52 | |
| Q88XS8 | citD; citrate lyase, gamma chain, acyl carrier protein | −2.43 | ||
| F9UMR9 | citE; citrate lyase, beta chain | −1.82 | ||
| F9UMS0 | citF; citrate lyase, alpha chain | −1.95 | ||
| Q88VM8 | dltC1; D-alanine–poly(phosphoribitol) ligase subunit 2-1 | −1.65 | ||
| Transporter | F9USY7 | mdxE; maltodextrin ABC transporter, substrate binding protein | −1.91 | |
| F9USZ2 | msmX; maltodextrin ABC transporter, ATP-binding protein | −2.23 | ||
| F9USG5 | lp_3042; Multidrug ABC transporter, ATP-binding and permease protein | −1.97 | ||
| F9UM05 | lp_0783; Oligopeptide ABC transporter, substrate binding protein | 2.77 | ||
| F9UR50 | lp_2525; ABC transporter, ATP-binding protein | 1.75 | ||
| F9UPR5 | lp_1945; Multidrug ABC transporter, ATP-binding protein | 1.65 | ||
| F9USL7 | fhuD; iron chelatin ABC transporter, substrate binding protein | 1.74 | ||
| F9UMR4 | citP; Citrate transport protein | −2.16 | ||
| F9UP84 | malT; Carbohydrate (Maltose)/proton symporttransporter, GPH family | −2.12 | ||
| Membrane protein and cell surface protein | F9URF3 | lp_2663; Hypothetical membrane protein | −1.53 | |
| F9UU92 | lp_3413; Cell surface protein, CscA/DUF916 family | −1.69 | ||
| F9UTN0 | lp_3359; Hypothetical membrane protein, DUF125 family | −2.24 | ||
| F9ULS6 | lp_0689; Cell surface protein, lipoprotein | 2.05 | ||
| F9URZ3 | lp_2901; Hypothetical membrane protein | 1.73 | ||
| F9UU47 | lp_3360; Hypothetical membrane protein, DUF125 family | −3.35 | ||
| Extracellular protein | F9USP4 | lp_3134; Extracellular protein, DUF 1093 family, membrane-bound | −2.04 | |
| F9UR45 | lp_2520; Extracellular protein, NlpC/P60 family, gamma-D-glutamate-meso-diaminopimelate muropeptidase | −1.79 | ||
| F9UQA0 | lp_2162; Extracellular protein, NlpC/P60 family, gamma-D-glutamate-meso-diaminopimelate muropeptidase | 1.55 | ||
| F9URD4 | lp_2636; Extracellular protein | 1.55 | ||
| F9UUA0 | lp_3421; Extracellular protein, gamma-D-glutamate-meso-diaminopimelate muropeptidase | 2.39 | ||
| F9USJ3 | lp_3077; Extracellular protein | 2.37 | ||
| F9UQ85 | lp_2145; Extracellular protein, cell wall–anchored | 1.90 | ||
| F9UNC2 | lp_1357; Extracellular protein, membrane-anchored | 1.88 | ||
| F9UL67 | zmp2; Extracellular zinc metalloproteinase, M10 family | 1.96 | ||
| F9USE1 | lp_3014; Extracellular transglycosylase, with LysM peptidoglycan–binding domain | 1.62 | ||
| F9USH2 | lp_3050; Extracellular transglycosylase, membrane-bound | 3.29 | ||
| Hydrolase | F9UTL5 | lp_3341; Cell surface hydrolase, DUF915 family, membrane-bound | 2.01 | |
| F9UM81 | gph1; Phosphohydrolase | 1.58 | ||
| F9UM42 | lp_0824; Hydrolase, HAD superfamily, Cof family | −3.78 | ||
| F9URL1 | lp_2737; Cell surface hydrolase, DUF915 family, membrane-bound | −2.25 | ||
| F9URQ4 | lp_2787; Hydrolase, HAD superfamily, Cof family | −1.96 | ||
| F9URF0 | xynC; Acetyl xylosidase (Promiscuous) | 1.51 | ||
| F9USG7 | amd; Aminohydrolase/peptidase, M20D family | −2.33 | ||
| Q06115 | cbh; Choloylglycine hydrolase | −2.05 | ||
| F9UTI3 | folQ; Dihydroneopterin triphosphate pyrophosphohydrolase | −1.98 | ||
| F9UPB8 | lp_1767; Glycosyl hydrolase, family 25 | 1.84 | ||
| F9UQI5 | lp_2266; Phosphoesterase | 1.75 | ||
| F9UL25 | lp_0552; Phosphoesterase | −1.62 | ||
| Other | Galactose metabolism and cell wall synthesis | F9UMX4 | glf1; UDP-galactopyranose mutase | 1.72 |
| F9URD9 | acm2; Cell wall hydrolase/muramidase | 1.51 | ||
| Inositol phosphate metabolism | Q88S38 | iolG; myo-inositol 2-dehydrogenase (promiscuous) | −6.86 | |
| Q88S37 | iolE; 2-keto-myo-inositol dehydratase (promiscuous) | −2.18 | ||
| F9ULG2 | lp_3608; myo-inositol 2-dehydrogenase-like (promiscuous) | −3.25 | ||
| F9ULG4 | lp_3612; myo-inositol 2-dehydrogenase-like (promiscuous) | −2.95 | ||
| Amino sugar and nucleotide sugar metabolism | F9USZ6 | sacK1; fructokinase | 1.61 | |
| Q88SC3 | murQ1; N-acetylmuramic acid 6-phosphate etherase | −1.80 | ||
| Oxidative phosphorylation | Q88UT8 | atpE; H(+)-transporting two-sector ATPase, C subunit | 1.62 | |
| F9UQR9 | atpB; H(+)-transporting two-sector ATPase, A subunit | 1.73 | ||
| Riboflavin metabolism | F9UNI1 | ribA; 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II | −4.71 | |
| Q88X16 | ribH; riboflavin synthase, beta chain | −4.63 | ||
| Other | F9UQZ9 | lp_2463; Prophage P2b protein 18, major capsid protein | 6.88 | |
| F9UQX7 | lp_2437; Prophage P2a protein 20, replication protein DnaD domain | 1.95 | ||
| F9US31 | lp_2953; Esterase | 1.68 | ||
| F9UNU9 | fthC; 5-formyltetrahydrofolate cyclo-ligase | 1.98 | ||
| Q890D7 | lp_0089; UPF0246 protein | 1.71 | ||
| Q88V85 | sepF; Cell division protein SepF | 2.24 | ||
| F9UPG0 | tarJ; ribitol-5-phosphate 2-dehydrogenase | 10.23 | ||
| F9URL0 | cah; Carbonate dehydratase | 1.51 | ||
| F9URA1 | rnh; Ribonuclease H | 1.62 | ||
| F9UPE3 | lp_1796; DegV family protein | 1.50 | ||
| Q6LWH3 | repB; Copy number control protein | −4.51 | ||
| Q88VB0 | lp_2157; UPF0356 protein | −1.52 | ||
| F9UNZ6 | tpk; Thiamin pyrophosphokinase | 2.12 | ||
| F9ULU2 | rsmI; Ribosomal RNA small subunit methyltransferase I | 1.74 | ||
| Uncharacterized protein | F9UQ40 | lp_2093; Uncharacterized protein | −1.51 | |
| F9UNU5 | lp_1566; Uncharacterized protein | −1.61 | ||
| Q6LWD6 | orf41; Uncharacterized protein | −1.79 | ||
| F9UPK2 | lp_1872; Uncharacterized protein | −1.56 | ||
| F9UU56 | lp_3372; Uncharacterized protein | −2.05 | ||
| F9UM53 | lp_0837; Uncharacterized protein | −1.63 | ||
| F9UTZ1 | lp_0402; Uncharacterized protein | −1.69 | ||
| F9UKY5 | lp_0507; Uncharacterized protein | −1.88 | ||
| Q6LWD8 | orf39; Uncharacterized protein | 2.86 | ||
| Q6LWD7 | orf40; Uncharacterized protein | 2.66 | ||
| F9UT92 | lp_3179; Uncharacterized protein | 1.78 | ||
| F9URF7 | lp_2667; Uncharacterized protein | 1.67 | ||
| F9UQN8 | lp_2333; Uncharacterized protein | 1.54 | ||
| F9UN47 | lp_1257; Uncharacterized protein | 1.54 | ||
| F9USX1 | lp_0158; Uncharacterized protein | 1.51 | ||
| F9UQ59 | lp_2112; Uncharacterized protein | 1.66 | ||
| F9UQ60 | lp_2113; Uncharacterized protein | −4.47 | ||
| F9UTE8 | lp_3250; Uncharacterized protein | −3.03 |
aCategory of differently expressed proteins was based on their functions annotated in the database of Uniprot and KEGG. bAccession number of each protein in Uniprot database. cDescription of each differently expressed protein, including corresponding gene name of each protein and full protein name. dFC indicates fold change of each differently expressed protein in the comparison of CCFM8610/CCFM191. Negative values indicate down-regulation of proteins, and positive values indicate up-regulation.
The proteomic dynamic changes of CCFM8610 and CCFM191 after Cd exposure were also evaluated to analyze the possible Cd tolerance mechanisms of L. plantarum strains (Tables S2 and S3). For CCFM8610, twenty-seven proteins that changed significantly (i.e., a fold change >1.5 or <−1.5, and P value < 0.05) by Cd stress were categorized into biological processes including global stress response, transportation, lipid, amino acid and pyrimidine metabolism and cell wall biosynthesis. For CCFM191, the abundances of 111 proteins were markedly changed after Cd exposure (i.e., a fold change >1.5 or <−1.5, and P value < 0.05) (Table S3). These proteins were associated with global stress response, cell wall biosynthesis and adhesion, transporters, amino acid, lipid, pyrimidine and energy metabolism, membrane proteins and extracellular proteins. It is noted that one protein with the greatest abundance change (4.45 fold up-regulation) in CCFM8610 after Cd exposure was prophage P2b protein 18, major capsid protein (lp_2463). This protein was also in higher abundance (6.88 fold) in CCFM8610 than that in CCFM191 in the untreated condition, while the abundance of this protein was not changed in CCFM191 after Cd exposure. In addition, the abundances of prophage proteins lp_0641 and lp_2444 were significantly up-regulated in CCFM191 after Cd exposure, but remained unaffected in CCFM8610. It was also observed that these two proteins were in lower abundances in CCFM8610 than CCFM191 in non-treated conditions (with fold changes of −1.87 and −3.30, respectively). This may indicate that the up-regulation of lp_0641 and lp_2444 should be considered a specific Cd response mechanism of CCFM191 itself. Such mechanism seems insufficient to protect the bacterial cell against Cd stress, as CCFM191 showed poorer Cd tolerant ability than CCFM8610.
The protein-protein interaction networks were constructed for further comparative proteomic analysis (Figs S1–S3). For the comparison between CCFM8610 and CCFM191 in natural conditions (Fig. S1), the differentially expressed proteins were categorized into 3 main clusters, including PTS system, carbohydrate metabolism and glycerol lipid metabolism. It was observed that some global stress-related proteins (including trxA1, trxA2, gshR2, msrB and msrA2) and some nucleic acid metabolism-related proteins (including pyrR1, gmk2 and ndk) were implicated in the tricarboxylic acid (TCA) sub-cluster (including fum, pdhA, pdhB, pdhC and pdhD) of carbohydrate metabolism. Since only 27 proteins changed after Cd exposure in CCFM8610, the available protein-protein interaction network is transparent (Fig. S2). The possible interaction between lp_2993 (a global stress response protein) and pdc (PadA) was observed, and the two proteins (dnaE and lp_0811) involved in pyrimidine metabolism interacted with each other. For altered protein profiles of CCFM191 after Cd exposure (Fig. S3), proteins related to glycerol lipid metabolism, pyrimidine metabolism and global stress response clustered respectively. Some differentially expressed protein profiles between CCFM8610 and CCFM191 in non-treated conditions (CCFM8610(0)/CCFM191(0); Fig. S1 and Table 2) were also altered in CCFM191 after Cd exposure (CCFM191(Cd)/CCFM191(0); Fig. S3 and Table S3), such as carbohydrate metabolism and concomitant transportation (PTS system), and a cluster of enzymes involved in glycerolipid metabolism. This might indicate that CCFM8610 exhibits an inherent resistant status to Cd even in the absence of Cd exposure, while CCFM191 displays a similar response only after Cd exposure.
Transcription confirmation and biological phenomena
Based on the genome sequence data of L. plantarum strains (https://www.ncbi.nlm.nih.gov/genome/?term=lactobacillus%20plantarum) and some well-studied Cd-tolerant microorganisms19, 35, 36, Cd tolerance related proteins Cd-/zinc-/cobalt-transporting ATPase (lp_3327), Cd-transporting P-type ATPase (CadA) and Cd-/manganese-transporting P-type ATPase (mntA) were further analyzed by RT-qPCR assay, as the information of these low-abundance membrane proteins is easily to be lost during proteomic analysis. Carbamoyl-phosphate synthase, pyrimidine-specific, large chain (pyrAB), Carbamoyl-phosphate synthase, pyrimidine-specific, small chain (pyrAA), D-alanine-poly (phosphoribitol) ligase subunit 2-1 (dltC1), D-alanine-poly(phosphoribitol) ligase subunit 2-2 (dltC2), Transcription regulator of CopAB ATPases (copR), Prophage P2a protein 13 (lp_2444), Nucleotide-binding protein, universal stress protein UspA family (lp_2993), DNA-directed DNA polymerase III subunit epsilon (lp_0811) and Cold shock protein 1 (CspP), were randomly selected for RT-qPCR assay to confirm the reliability of proteomic results. For this reason, the proteins in the same operon are preferred, such as pyrAA and pyrAB, and dltC1 and dltC2. The mRNA expressions of Cd tolerance-related proteins, CadA, mntA and lp_3327, showed clear up-regulation in CCFM8610 after Cd exposure, while these genes (lp_3327 and mntA) were down-regulated or remained unchanged (CadA) in CCFM191 after Cd exposure (Fig. 5). The alterations in the mRNA expressions of Csp P and lp_0811 in CCFM8610 after Cd exposure and those of PyrAA, PyrAB, dltC1, dltC2, copR, lp_2444 and lp_2993 in CCFM191 were in accordance with the corresponding changes in protein level, which verified the results of the proteomic study.
Figure 5.
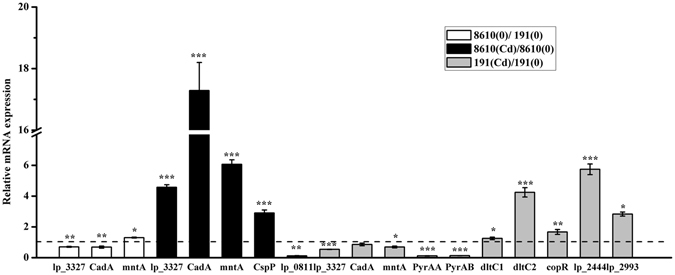
Relative mRNA expressions of genes encoding proteins altered in three comparisons, including CCFM8610(0)/CCFM191(0), CCFM8610(Cd)/CCFM8610(0) and CCFM191(Cd)/CCFM191(0). Values are expressed as mean ± SEM of 4 independent replicates. mRNA expression of denominator in each comparison is set as reference value of 1. *P < 0.05; **P < 0.01; ***P < 0.001.
Compared with that of CCFM191, CCFM8610 showed more obvious enhancement of surface hydrophobicity after Cd exposure (Fig. S4). The autoaggregation ability of non-Cd-treated CCFM8610 was significantly greater than that of CCFM191 (P < 0.05, Fig. S5). Cd stress increased the autoaggregation ability of the former strain, but exhibited no marked effects on the latter. The scanning electron microscope (SEM) micrographs further confirmed these results (Fig. S6). The Cd binding ability of CCFM8610 was more than 2-fold higher than that of CCFM191 (Fig. S7). After Cd stress, the greatest amount of Cd accumulated on the external surface of the cell wall (59.22% ± 7.94% for CCFM8610 and 45.90% ± 2.03% for CCFM191, respectively) and in the space between the cell wall and the plasma membrane (12.01% ± 2.23% for CCFM8610 and 4.41% ± 0.64% for CCFM191, respectively, Fig. S8). Moreover, there is a significantly higher amount of Cd accumulated in CCFM8610 than that in CCFM191 after Cd exposure (14 times higher in the relative amount; Table S4). The contents of the other metal ions including manganese (Mn), zinc (Zn), potassium (K), sodium (Na) and magnesium (Mg) were either unchanged or showed insignificant perturbations after Cd exposure in both strains.
We also determined the intracellular reactive oxygen species (ROS) levels of two strains before and after Cd exposure (Fig. S9). The results showed that in the non-treated conditions (without Cd exposure), the intracellular ROS level of CCFM8610 was significantly lower than that of CCFM191. The former strain could also survive Cd stress with less drastic cellular response than the latter, as CCFM8610 showed a less significant increase in ROS level after Cd exposure. Seven detected hydrophobic amino acids were significantly up-regulated in CCFM8610 and CCFM191 after Cd exposure, with an exception of proline in CCFM8610 (Fig. S10). Compared with CCFM191, CCFM8610 consumed significantly less glucose in natural conditions (Fig. S11). Meanwhile, this strain showed less significant fluctuation in glucose consumption than CCFM191 after Cd exposure.
Discussion
In order to explore the underlying mechanism of the intra-species differences in Cd tolerance of L. plantarum strains, we examined the proteomic profiles of CCFM8610 (strongly resistant to Cd) and CCFM191 (sensitive to Cd) in non-stimulating conditions as well as after Cd exposure using the iTRAQ approach. The results revealed that L. plantarum CCFM8610 displayed a complex biological network to tackle Cd stress, which may be related to carbohydrate, purine and pyrimidine metabolism, global stress responses, lipid and amino acid metabolism, metal binding properties, cell wall biosynthesis and transporters of the bacterial cell (Fig. 6). The Cd resistant mechanism of this strain involves a specific energy conservation survival mode, a mild induction of cellular defense and repair systems, an enhanced biosynthesis of hydrophobic amino acids, a promoted tolerance against osmotic stress and an inherent superior Cd-binding ability and effective cell wall biosynthesis ability in response to Cd stress. Potential key proteins that are important in protection of the bacterial cells against Cd toxicity were also identified.
Figure 6.
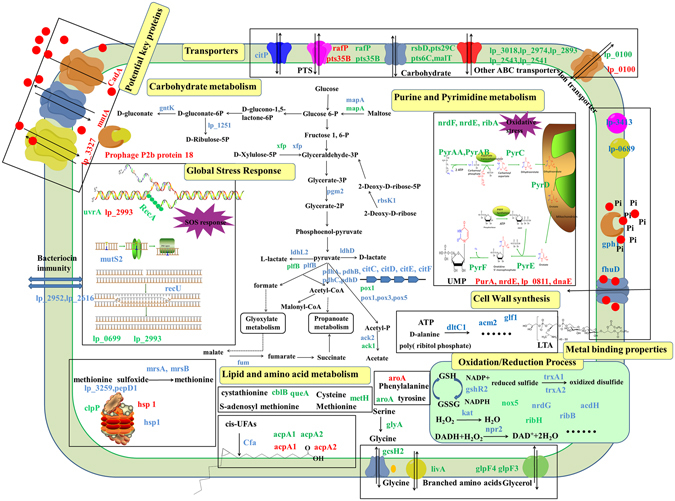
Proposed model for response mechanisms of CCFM8610 to Cd stress. Altered proteins (fold change >1.5 or <−1.5 and P < 0.05) in three comparisons of CCFM8610(0)/CCFM191(0), CCFM8610(Cd)/CCFM8610(0) and CCFM191(Cd)/CCFM191(0) are presented in the color of blue, red and green, respectively. Red dots represent Cd ions.
Carbohydrate metabolism
The comparative proteomic profiles between the two strains in the absence of Cd treatment showed that CCFM8610 has lower abundance of 24 proteins and higher abundance of only 2 proteins associated with carbohydrate metabolism (Table 2). Five enzymes (fum, pdhA, pdhB, pdhC, and pdhD) involved in TCA cycle were in lower abundance in CCFM8610. The genes that encode Pdh proteins are known to share the same operon. The repression of the pdh operon has been reported to switch TCA cycle to a branched or noncyclic anaerobic form, which is regarded as an energy conservation strategy in E. coli 12 and P. brassicacearum 19 during Cd stress. This reduces intracellular free radicals and thus protects the bacteria against Cd-induced cytotoxicity. Citrate transport protein (citP) and the four citrate lyases (citC, citD, citE and citF) were in lower abundance in CCFM8610. This corresponded to the lower citrate concentration in this strain (unpublished data). We also observed reduced abundance of 11 proteins involved in the PTS system in CCFM8610, further indicating repression of carbohydrate metabolism even in the absence of Cd stress.
The differentially expressed proteomic profiles in the two strains after Cd exposure also confirmed this self-protection mechanism of CCFM8610. No significant changes in the proteins involved in carbohydrate metabolism were found in CCFM8610 after Cd stress (Table S2), indicating that the energy conservation survival mode of this strain is beneficial for tackling Cd toxicity. In contrast, carbohydrate metabolism was markedly down-regulated in CCFM191 after Cd exposure (Table S3). This is further validated by a relative “poised” status of glucose consumption of CCFM8610 both in the presence and absence of Cd stress (Fig. S11). Such energy conservation and survival can be regarded as an inherent “poised” physiological status of the strain against environmental stresses, including Cd exposure. A previous study on the resistant mechanism of P. pseudoalcaligenes KF707 and its more resistant mutant (T5) to tellurite exposure indicated that even in the absence of tellurite, T5 cells displayed a “poised” status with altered intracellular levels of glutathione and branched-chain amino acids, along with increased resistance to other toxic metals and metabolic inhibitors37. Such a mechanism was believed as an inherent tolerance of the mutant strain primed for tellurite exposure, which further supports our hypothesis.
Purine and pyrimidine metabolism
Very limited effect on pyrimidine metabolism-related proteins was observed after Cd exposure in CCFM8610. PurA, an important enzyme in the de novo pathway of purine nucleotide biosynthesis, was down-regulated in CCFM8610 after Cd exposure. NrdE, lp_0811 and dnaE, three catalytic proteins involved in DNA replication, were also changed by Cd stress. In contrast, nine purine and pyrimidine metabolism-related proteins were altered during the Cd stress responses of CCFM191 (Table S3), most of which (pyrE, pyrF, pyrD, pyrAB, pyrAA, and pyrC) showed down-regulation after Cd exposure. A similar alteration was also observed in other LAB strains in response to acid and bile stress38, 39. The superior Cd tolerance of CCFM8610 may be in part due to its ability to maintain a steady physiological status, including purine and pyrimidine metabolism, after Cd exposure, which can be further validated in the following analysis of its global stress responses.
Global stress responses
Induction of global stress responses is an important strategy for bacteria to endure harsh environmental conditions. The comparative proteomic profiles between native and Cd-treated CCFM8610 showed that Cd stress caused fluctuations of only two proteins that are involved in the universal stress response, lp_2993 (up-regulated) and hsp1 (down-regulated) (Table S2). The former has also been reported to play a putative role against Cd stress in P. brassicacearum 19.
In contrast, CCFM191 showed more marked changes with 16 significantly altered proteins related to stress response after Cd shock (Table S3). These proteins include a key protease (Clp) functioning in maintaining cytoplasmic protein quality, four universal stress related proteins (uvrA, recA, lp_0699 and lp_2993) involving in DNA repair, 10 proteins belonging to oxidoreductases. An increase in the level of Clp, a key protease that functions in maintaining quality of cytoplasmic proteins40, was also found after environmental stress in other microorganisms, such as L. reuteri 41, L. acidophilus 42, and Oenococcus oeni 43.
In non-stimulating conditions, CCFM191 showed higher abundance (compared to CCFM8610) of two proteins (mutS2 and recU) involved in DNA repair, four proteins (msrA, msrB, lp_3259 and pepD1) related to protein repair, two proteins (lp_2952 and lp_2616) related to bacteriocin immunity, and one protein (hsp1) involved in stress response. In addition, a marked distinction was observed between CCFM8610 and CCFM191 in 23 oxidoreductases, indicating that these two strains display different oxidative stress status even in the absence of Cd exposure.
However, not all repair or defense proteins are stress-inducible44. Looking at the different nucleic acid metabolism–related and global stress response-related proteomic profiles between these two strains in non-stimulating conditions, as well as in response to Cd exposure, we conclude that compared with CCFM191, CCFM8610 is able to respond to environmental stress with weaker induction of the cellular defense and repair system. The intracellular ROS level of CCFM8610 was significantly lower than that of CCFM191 both in the presence and absence of Cd stress, which further supported our conclusion (Fig. S9).
Lipid and amino acid metabolism
In non-stimulating conditions, cyclopropane-fatty-acyl-phospholipid synthase (cfa2) was in a higher abundance in CCFM8610 than that in CCFM191. This protein is responsible for the methylation reaction that translates preexisting cis-UFAs to cyclopropane fatty acids45. As cyclopropane fatty acids have been reported to increase acid resistance in E. coli 46, the relatively high expression of Cfa in CCFM8610 may be a determinant for its stress tolerance.
A chorismate mutase (aroA) and two acyl carrier proteins (acpA1 and acpA2) were up-regulated in both CCFM8610 and CCFM191 after Cd exposure (Tables S2 and S3). AroA can trigger the biosynthesis of phenylalanine and tyrosine, which in turn increases the hydrophobicity of the bacterial cell surface to prevent Cd-induced protein damage47. Our in vitro assays demonstrated a significant up-regulation of hydrophobic amino acids and surface hydrophobicity in both strains (Figures S4 and S10), which further supported this hypothesis. AcpA1 and acpA2 are well-known proteins involved in fatty acid biosynthesis and protein translation38. The up-regulation of these two proteins may result in a change in membrane fatty acid composition, which improves the tolerance of bacteria against environmental stress48.
For CCFM191, significant changes in amino acid metabolism were observed after Cd exposure (Table S3). Two proteins (glyA and gcsH2) involved in glycine biosynthesis and three proteins (cblB, queA and metH) involved sulfur amino acid metabolism were markedly up-regulated. Glycine is one of osmotic protection molecules in bacteria and has been reported to play a role in the response of metal stress37, 49. As Cd binds preferentially to sulfur ligand50, the up-regulation of sulfur amino acid metabolism may be a self-detoxification mechanism of CCFM191 against Cd exposure.
Metal binding and cell wall biosynthesis
In conditions without Cd stress, marked differences were observed in the abundance of extracellular, membrane and cell surface proteins between CCFM8610 and CCFM191 (Table 2). It has been reported that Cd could compete for the binding sites of proteins with other metals such as iron, zinc, and calcium51. The higher abundance of fhuD, an iron chelatin, might improve the Cd-binding capacity of CCFM8610. Another protein with higher abundance in CCFM8610, hydrolase phosphohydrolase (gph1), can release free phosphate from lipids and precipitate Cd as CdHPO4 or other chemical forms onto the cell surface19, 52. Consistent with these analyses, our in vitro adsorption assays in aqueous phase solution showed that CCFM8610 possesses significantly better Cd binding ability and sequesters a higher proportion of Cd in the cellular surface than CCFM191 (Figs S7 and S8). As previously reported, this may be a self-protection of L. plantarum strain to decrease the risk of Cd-induced intracellular toxicity53.
As the cell wall is the first line of defense against environmental stress for bacteria27, the enhancement of cell wall biosynthesis may be a self-protective mechanism of L. plantarum strains in response to Cd exposure. DltC1, a protein that plays a role in lipoteichoic acid (LTA) biosynthesis, showed a 2.56-fold up-regulation in CCFM8610, which was higher than that in CCFM191 (1.71-fold, Table S3). Previous study has demonstrated that LTA can bind metal ions and affect electromechanical characteristics of the cell wall in L. casei 54. This can also partly explain the different Cd-binding abilities of the two L. plantarum strains tested here (Fig. S7).
The observed differences in metal binding and cell wall biosynthesis properties of the two strains were further verified by a significantly higher Cd accumulation in CCFM8610 after Cd exposure compared with CCFM191 (Table S4). The intracellular concentrations of other metals were relatively unaltered after Cd exposure. This may indicate that the Cd binding process in CCFM8610 is selective, which is in correspondence with our earlier in vivo study which showed that essential elements such as Ca, Zn and Mg were unaltered in the tissues of mice after oral administration of CCFM861010. The adsorption of elements on the cell surface have been reported to influence the hydrophobicity and autoaggregation properties, which in turn improves the stress tolerance of the bacteria19, 39. This is consistent with the more significant changes in the surface properties observed in CCFM8610 after Cd exposure (Figs S4, S5 and S6).
The strong Cd tolerance of CCFM8610 might therefore be partly attributed to its inherent Cd-binding ability provided by surface proteins and the effective cell wall biosynthetic ability during Cd exposure, thus blocking the entry of this toxic metal into the cell cytoplasm.
Transporters
Cd exposure inevitably induces osmotic stress in bacterial cells, which in turn leads to further cell damage. The tight regulation of metal import is one of the most basic mechanisms of metal homeostasis20, and ion transporters have been reported to play a role in the Cd stress response in E. coli 12. In the present study, cobalt ABC transporter ATP-binding protein (lp_0100) was observed to be down-regulated by 1.68-fold in CCFM8610 after Cd exposure, which could be regarded as a response of the strain to osmotic stress. The phosphoenolpyruvate-dependent PTS is a major carbohydrate transport system in LAB strains. Two PTS sugar transporters (rafP and pts35B) were down-regulated in CCFM8610 after Cd exposure, indicating the energy-conservation survival mode of this strain during Cd stress.
Compared with CCFM8610, CCFM191 showed extra changes in three carbohydrate transporters (pts29C, pts6C and malT), five ABC transporters (lp_3018, lp_2974, lp_2893, lp_2543 and lp_2541), one branched-chain amino acid transporter (livA) and two glycerol uptake facilitators (glpF4 and glpF3) after Cd exposure. These changes might indicate that in order to survive the Cd stress, the sensitive strain CCFM191 needs to shut down more carbohydrate transporters, optimize ABC transporters and increase the proportion of hydrophobic amino acids.
Potential key proteins
In the non-treated conditions, the abundance of prophage P2b protein 18 (lp_2463) in CCFM8610 was 6.88-fold higher than that in CCFM191, which was the second-highest fold change among all up-regulated proteins between the two strains. In addition, this prophage protein was observed to be the most significantly up-regulated protein (4.45-fold) in CCFM8610 after Cd exposure. In the Cd-sensitive strain CCFM191, prophage P2b protein 18 remained unaffected (Table S3). In pathogenic bacteria, phage genes are related to genetic islands encoding virulence and colonization factors and play a role in the environmental adaption of the bacteria55, 56. Prophage-dependent thermo-resistance via plasmid integration has been reported in Staphylococcus aureus 57. The phage genes have also been observed to be up-regulated in E. coli upon Cd stress12. These analyses highlighted the prophage P2b protein 18 as a potential key determinant in L. plantarum strains for the response to Cd stress. The acquisition of this stress resistance-related protein by CCFM8610 is a clear benefit to the strain.
Cd-tolerance-associated proteins, including CadA, mntA and lp_3327, are believed to be Cd transporters in the cell membranes of L. plantarum strains and some well-studied Cd-tolerant microorganisms25, 58. Since the information of these membrane proteins is easily to be lost during proteomic analysis due to their low abundance and the insufficient extraction, the expressions of their genes were quantified by RT-qPCR (Fig. 5). The expression of CadA, a P-type ATPase which catalyzes the active efflux of Cd2+, was markedly up-regulated (fold change >17) after Cd exposure in CCFM8610, while in CCFM191 it was only marginally down-regulated (0.85 fold). The Cd-resistant function of CadA is well established in S. aureus, L. monocytogenes and L. lactis 23, 59, 60, the significantly enhanced efflux of Cd by the up-regulation of CadA might be a detoxification strategy of CCFM8610. The expression of mntA, a Cd2+-Mn2+ shared transporter, and lp_3327, a Cd-/zinc-/cobalt-transporting ATPase, were up-regulated 6.06-fold and 4.57-fold in CCFM8610 after Cd exposure, respectively. However, the expressions of these two proteins slightly decreased in CCFM191. Besides their metal transporting ability, mntA and lp_3327 have been reported to harbor multiple transmembrane domains that bind metal. Therefore, these proteins may also prevent Cd-induced cytotoxicity by Cd sequestration. This may also explain the difference of Cd tolerance between these two strains.
Conclusion
In this study, we investigated the mechanism of Cd stress response of L. plantarum strains using comparative and functional proteomic analysis of L. plantarum CCFM8610 (strongly resistant to Cd) and L. plantarum CCFM191 (sensitive to Cd). The proteomic profiles in non-stimulating conditions and the altered proteomic profiles after Cd stress in both strains were explored. Of the total 1415 identified proteins, 206 were differentially expressed for the comparison of natural proteomic profiles of CCFM8610 and CCFM191, 27 were differently regulated in CCFM8610 after Cd exposure, and 111 were changed in CCFM191 in response to Cd stress. Both strains showed physiological alterations in energy metabolism, purine and pyrimidine metabolism, global stress responses, lipid and amino acid metabolism, metal binding properties, cell wall biosynthesis and transporters in response to Cd exposure. The underlying mechanism of the intra-species distinctions between CCFM8610 and CCFM191 on Cd tolerance can be attributed to the following aspects. (a) CCFM8610 possesses a specific energy-conservation survival mode, which can be regarded as an inherent “poised” physiological status primed for Cd exposure. (b) CCFM8610 can cope with environmental stress with mild induction of the cellular defense and repair system, which enables the strain to survive Cd exposure without drastic physiological response. (c) CCFM8610 induces the biosynthesis of hydrophobic amino acids that enhance surface hydrophobicity of the cell and prevent Cd-induced protein damage. (d) CCFM8610 has inherent superior Cd binding ability and effective cell wall structures, which promotes Cd sequestration on the surface of the cell, preventing the uptake of this toxic metal into the cytoplasm. (e) CCFM8610 exhibits a tight regulation on ion transport to withstand the Cd-induced osmotic stress. (f) Several key proteins, including prophage P2b protein 18, CadA, mntA and lp_3327, also play a potential role in Cd tolerance in CCFM8610. This study provides an overview of the Cd stress response network (Fig. 6) of L. plantarum CCFM8610 that enables this strain to be strongly resistant to Cd.
Experimental Procedures
Cadmium chloride (CdCl2) and other chemicals were purchased from Shanghai Sinopharm Chemical Reagent Company (China). Trizol reagent was obtained from Ambion Life Technologies (USA). TAKARA RR047A kits were purchased from TAKARA BIO INC (China). iTaqTM Universal SYBR® Green Supermix was purchased from Bio-Rad (USA). iTRAQ reagents were purchased from Applied Biosystems (USA).
Bacterial Strains and Growth Conditions
Twenty L. plantarum strains, including CCFM11, CCFM166, CCFM191, CCFM198, CCFM231, CCFM232, CCFM239, CCFM240, CCFM241, CCFM308, CCFM309, CCFM405, CCFM436, CCFM578, CCFM579, CCFM595, CCFM602, CCFM605, CCFM8610, and CCFM8661, were all obtained from the Culture Collections of Food Microbiology, Jiangnan University (Wuxi, China). All strains were cultured in de Man, Rogosa and Sharpe (MRS) broth (Hopebio Company, Qingdao, China) at 37 °C for 18 h routinely.
Cd tolerance assay
As it is normally to incubate the bacteria in liquid medium for proteomic analysis33, 61, twenty L. plantarum strains were inoculated (2% v/v) into MRS broth containing Cd2+ ranging from 0 mg/L to 50 mg/L and then incubated at 37 °C. The OD600 values were measured at different time points until the stationary phase of the strains. The relative growth rate of each strain was expressed as a percentage of that of the control culture (without Cd exposure) which was assigned a value of 100%33, 61. The MIC value of each strain was determined as the lowest Cd concentration that completely inhibited the growth rate of the strains.
Proteomics analysis
Whole cell protein extraction
Based on the data of Cd tolerance assay, two strains with the widest difference in Cd tolerance values were selected for proteomics analysis. The strains were grown in the Cd containing MRS broth at 37 °C and the Cd concentration was set as the 1/2 MIC value of the sensitive strain (5 mg/L) as that was established previously33, 34. The strains were also incubated in Cd-free media as a control.
At the early stationary phase (OD600 = 6.0)38, 61, the cells were harvested and washed twice with ice-cooled phosphate-buffered saline (PBS) solution and pelleted. The collected pellets were immediately stored at −80 °C and used for protein extraction. The cell protein was extracted as previously described with minor modifications27, 62. The detailed procedure of protein extraction could be found in supplementary materials.
Protein sample preparation and iTRAQ labeling
The detailed description of protein sample preparation and iTRAQ labeling can be seen in the supplementary materials. The workflow of the iTRAQ experiment with 9 replicates (3 biological replicates × 3 mechanical replicates in mass spectrometry runs) in each of four conditions is shown in Fig. 1.
Liquid chromatography tandem mass spectrometry (LC/LC−MS/MS) Analysis
The detailed procedures can be seen in the supplementary materials.
Protein identification and screening of differently expressed proteins
The raw data obtained from LC/LC−MS/MS analysis were processed using Thermo Proteome Discoverer software (v1.0 build 43, Thermo Fisher Scientific) and searched with Mascot (Matrix Science, London, UK) at the in-house server to perform database comparisons against L. plantarum WCFS1, based on our preliminary studies revealing that there is a utmost genome similarity between our CCFM8610 and WCFS1. The function of the identified proteins was annotated using Gene Ontology (GO) analysis, and the metabolomics pathway was analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The threshold for differentiated expressed proteins was set as P < 0.05 and fold change >1.5 or <−1.5.
Quantifications of key proteins in proteomics
The total RNA of the strains was extracted using Trizol reagent (Life Technologies). Reverse transcription was performed with TAKARA RR047A kits according to the instruction manual. The alterations of mRNA expressions were evaluated as previously reported63. The primers listed in Table S1 were designed using Primer 5.0 software based on the genome sequence of L. plantarum WCFS164. The L. plantarum 16S rRNA gene was used as an expression control with primers specific for L. plantarum strains, and each reaction was conducted in four duplicates65.
Biological phenomena
Intracellular metal accumulation, Cd binding, bacterial hydrophobicity, autoaggregation, scanning electron microscope (SEM), intracellular reactive oxygen species (ROS) production, cellular components involved in Cd binding, glucose consumption and hydrophobic amino acid production.
The detailed procedures of these assays can be seen in the material supplementary materials.
Statistical analysis
Data were expressed as the mean ± standard error of the mean (SEM) for each group. Differences between groups were analyzed using one-way analysis of variance (ANOVA), followed by the Tukey post-hoc test. A P value of <0.05 was considered to indicate statistical significance.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China Key Program (No. 31530056), National Natural Science Foundation of China (No. 31601452), the Natural Science Foundation of Jiangsu Province (BK20160175), the General Financial Grant from the China Postdoctoral Science Foundation (2016M590412), the General Financial Grant from the Jiangsu Postdoctoral Science Foundation (No. 1601113 C), BBSRC Newton Fund Joint Centre Award.
Author Contributions
Q.Z. and Y.X. carried out the experiment and drafted the manuscript. F.T., J.Z. and H.Z. participated in the RT-qPCR assay, Cd tolerance assay, intracellular metal accumulation and Cd binding, hydrophobicity and autoaggregation assays, and analyzed the data. A.N. and W.C. conceived of the study, managed the project design and helped to revise the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Qixiao Zhai and Yue Xiao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01180-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Uetani M, et al. Tissue cadmium (Cd) concentrations of people living in a Cd polluted area, Japan. Biometals. 2006;19:521–525. doi: 10.1007/s10534-005-5619-0. [DOI] [PubMed] [Google Scholar]
- 2.Hyder O, et al. Cadmium exposure and liver disease among US adults. J. Gastrointest. Surg. 2013;17:1265–1273. doi: 10.1007/s11605-013-2210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satarug S, Garrett SH, Sens MA, Sens DA. Cadmium, Environmental Exposure, and Health Outcomes. Environ. Health Perspect. 2010;118:183. doi: 10.1289/ehp.0901234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.IARC. In Lyon IARC Monographs on the evaluation of carcinogenic risks to humans Vol. 58, 148–161, 206–210 (International Agency for Research on Cancer, 1993).
- 5.Goyer, R. A. & Clarkson, T. W. In Casarett & Doull’s Toxicology: The Basic Science of Poisons (ed. C. D. Klaassen) Ch. 23, 825–826 (McGraw-Hill Health Professions Division, 2001).
- 6.Nordberg, G. F., Nogawa, K., Nordberg, M. & Friberg, L. T. In Handbook on the Toxicology of Metals (eds Gunnar F. Nordberg, Bruce A. Fowler, Monica Nordberg & Lars T. Friberg) vii, 446, 463–470 (Academic Press, 2011).
- 7.Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2001;2:43–53. [PubMed] [Google Scholar]
- 8.Foligné B, Daniel C, Pot B. Probiotics from research to market: the possibilities, risks and challenges. Curr. Opin. Microbiol. 2013;16:284–292. doi: 10.1016/j.mib.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Zhai Q, et al. Protective effects of Lactobacillus plantarum CCFM8610 against acute cadmium toxicity in mice. Appl. Environ. Microbiol. 2013;79:1508–1515. doi: 10.1128/AEM.03417-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai Q, et al. Protective effects of Lactobacillus plantarum CCFM8610 against chronic cadmium toxicity in mice indicate routes of protection besides intestinal sequestration. Appl. Environ. Microbiol. 2014;80:4063–4071. doi: 10.1128/AEM.00762-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhai Q, et al. Oral administration of probiotics inhibits heavy metal cadmium absorption by protecting intestinal barrier. Appl. Environ. Microbiol. 2016;AEM:00695–00616. doi: 10.1128/AEM.00695-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang A, Crowley DE. Global gene expression responses to cadmium toxicity in. Escherichia coli. J. Bacteriol. 2005;187:3259–3266. doi: 10.1128/JB.187.9.3259-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vido K, et al. A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:8469–8474. doi: 10.1074/jbc.M008708200. [DOI] [PubMed] [Google Scholar]
- 14.Banjerdkij P, Vattanaviboon P, Mongkolsuk S. Exposure to cadmium elevates expression of genes in the OxyR and OhrR regulons and induces cross-resistance to peroxide killing treatment in Xanthomonas campestris. Appl. Environ. Microbiol. 2005;71:1843–1849. doi: 10.1128/AEM.71.4.1843-1849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhai Q, et al. Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control. 2015;54:23–30. doi: 10.1016/j.foodcont.2015.01.037. [DOI] [Google Scholar]
- 16.Matyar F, Kaya A, Dinçer S. Antibacterial agents and heavy metal resistance in Gram-negative bacteria isolated from seawater, shrimp and sediment in Iskenderun Bay, Turkey. Sci. Total Environ. 2008;407:279–285. doi: 10.1016/j.scitotenv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Abou-Shanab R, Van Berkum P, Angle J. Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere. 2007;68:360–367. doi: 10.1016/j.chemosphere.2006.12.051. [DOI] [PubMed] [Google Scholar]
- 18.Lee S-W, Glickmann E, Cooksey DA. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microbiol. 2001;67:1437–1444. doi: 10.1128/AEM.67.4.1437-1444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez L, et al. Exploration of intraclonal adaptation mechanisms of Pseudomonas brassicacearum facing cadmium toxicity. Environ. Microbiol. 2007;9:2820–2835. doi: 10.1111/j.1462-2920.2007.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 2005;57:27–40. doi: 10.1111/j.1365-2958.2005.04642.x. [DOI] [PubMed] [Google Scholar]
- 21.Schirawski J, Hagens W, Fitzgerald GF, van Sinderen D. Molecular characterization of cadmium resistance in Streptococcus thermophilus strain 4134: an example of lateral gene transfer. Appl. Environ. Microbiol. 2002;68:5508–5516. doi: 10.1128/AEM.68.11.5508-5516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delorme C. Safety assessment of dairy microorganisms: Streptococcus thermophilus. Int. J. Food Microbiol. 2008;126:274–277. doi: 10.1016/j.ijfoodmicro.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Liu C-Q, et al. Genetic Analysis of Regions Involved in Replication and Cadmium Resistance of the Plasmid pND302 from Lactococcus lactis. Plasmid. 1997;38:79–90. doi: 10.1006/plas.1997.1301. [DOI] [PubMed] [Google Scholar]
- 24.O’Sullivan D, et al. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. Microbiol. 2001;67:929–937. doi: 10.1128/AEM.67.2.929-937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao Z, Chen S, Wilson DB. Cloning, expression, and characterization of cadmium and manganese uptake genes from Lactobacillus plantarum. Appl. Environ. Microbiol. 1999;65:4746–4752. doi: 10.1128/aem.65.11.4746-4752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Kranenburg R, et al. Functional analysis of three plasmids from Lactobacillus plantarum. Appl. Environ. Microbiol. 2005;71:1223–1230. doi: 10.1128/AEM.71.3.1223-1230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JY, Pajarillo EAB, Kim MJ, Chae JP, Kang D-K. Proteomic and transcriptional analysis of Lactobacillus johnsonii PF01 during bile salt exposure by iTRAQ shotgun proteomics and quantitative RT-PCR. J. Proteome Res. 2012;12:432–443. doi: 10.1021/pr300794y. [DOI] [PubMed] [Google Scholar]
- 28.Yun S-H, et al. Proteomic characterization of the Pseudomonas putida KT2440 global response to a monocyclic aromatic compound by iTRAQ analysis and 1DE-MudPIT. J. Proteomics. 2011;74:620–628. doi: 10.1016/j.jprot.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Evans FF, Raftery MJ, Egan S, Kjelleberg S. Profiling the secretome of the marine bacterium Pseudoalteromonas tunicata using amine-specific isobaric tagging (iTRAQ) J. Proteome Res. 2007;6:967–975. doi: 10.1021/pr060416x. [DOI] [PubMed] [Google Scholar]
- 30.Timms JF, Cramer R. Difference gel electrophoresis. Proteomics. 2008;8:4886–4897. doi: 10.1002/pmic.200800298. [DOI] [PubMed] [Google Scholar]
- 31.Wu WW, Wang GH, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J. Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 32.Surowitz KG, Titus JA, Pfister RM. Effects of cadmium accumulation on growth and respiration of a cadmium-sensitive strain of Bacillus subtilis and a selected cadmium resistant mutant. Arch. Microbiol. 1984;140:107–112. doi: 10.1007/BF00454911. [DOI] [Google Scholar]
- 33.Guo Z, et al. Proteomic analysis of the copper resistance of Streptococcus pneumoniae. Metallomics. 2015;7:448–454. doi: 10.1039/C4MT00276H. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y-M, et al. Proteomic analysis of cadmium exposure in cultured lung epithelial cells: evidence for oxidative stress-induced cytotoxicity. Toxicol. Res. 2013;2:280–287. doi: 10.1039/c3tx50014d. [DOI] [Google Scholar]
- 35.Aguilar-Barajas, E., Ramírez-Díaz, M. I., Riveros-Rosas, H. & Cervantes, C. In Pseudomonas 255–282 (Springer, 2010).
- 36.Miller C, et al. Copper and cadmium: responses in Pseudomonas putida KT2440. Lett. Appl. Microbiol. 2009;49:775–783. doi: 10.1111/j.1472-765X.2009.02741.x. [DOI] [PubMed] [Google Scholar]
- 37.Tremaroli V, et al. Metabolomic investigation of the bacterial response to a metal challenge. Appl. Environ. Microbiol. 2009;75:719–728. doi: 10.1128/AEM.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heunis T, Deane S, Smit S, Dicks LM. Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J. Proteome Res. 2014;13:4028–4039. doi: 10.1021/pr500353x. [DOI] [PubMed] [Google Scholar]
- 39.An H, et al. Integrated transcriptomic and proteomic analysis of the bile stress response in a centenarian-originated probiotic Bifidobacterium longum BBMN68. Mol. Cell Proteomics. 2014;13:2558–2572. doi: 10.1074/mcp.M114.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 41.Wall T, et al. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 2007;73:3924–3935. doi: 10.1128/AEM.01502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeiler EA, Azcarate-Peril MA, Klaenhammer TR. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 2007;189:4624–4634. doi: 10.1128/JB.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltramo C, Grandvalet C, Pierre F, Guzzo J. Evidence for multiple levels of regulation of Oenococcus oeni clpP-clpL locus expression in response to stress. J. Bacteriol. 2004;186:2200–2205. doi: 10.1128/JB.186.7.2200-2205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y-M, Rock CO. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 46.Chang YY, Cronan JE. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 1999;33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- 47.Kast P, et al. A strategically positioned cation is crucial for efficient catalysis by chorismate mutase. J. Biol. Chem. 2000;275:36832–36838. doi: 10.1074/jbc.M006351200. [DOI] [PubMed] [Google Scholar]
- 48.Fozo EM, Quivey RG. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 2004;70:929–936. doi: 10.1128/AEM.70.2.929-936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith DS, Bell RA, Kramer JR. Metal speciation in natural waters with emphasis on reduced sulfur groups as strong metal binding sites. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2002;133:65–74. doi: 10.1016/S1532-0456(02)00108-4. [DOI] [PubMed] [Google Scholar]
- 51.Bridges CC, Zalups RK. Molecular and ionic mimicry and the transport of toxic metals. Toxicol. Appl. Pharmacol. 2005;204:274–308. doi: 10.1016/j.taap.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macaskie LE, Dean AC, Cheetham AK, Jakeman RJ, Skarnulis AJ. Cadmium accumulation by a Citrobacter sp.: the chemical nature of the accumulated metal precipitate and its location on the bacterial cells. Microbiology. 1987;133:539–544. doi: 10.1099/00221287-133-3-539. [DOI] [Google Scholar]
- 53.Zhai Q, et al. The cadmium binding characteristics of a lactic acid bacterium in aqueous solutions and its application for removal of cadmium from fruit and vegetable juices. RSC Adv. 2016;6:5990–5998. doi: 10.1039/C5RA24843D. [DOI] [Google Scholar]
- 54.Neuhaus FC, Heaton MP, Debabov DV, Zhang Q. The dlt operon in the biosynthesis of D-alanyl-lipoteichoic acid in Lactobacillus casei. Microb. Drug Resist. 1996;2:77–84. doi: 10.1089/mdr.1996.2.77. [DOI] [PubMed] [Google Scholar]
- 55.Hayashi T, et al. Complete genome sequence of enterohemorrhagic Eschelichia coli O157: H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 56.McLeod SM, Kimsey HH, Davis BM, Waldor MK. CTXϕ and Vibrio cholerae: exploring a newly recognized type of phage–host cell relationship. Mol. Microbiol. 2005;57:347–356. doi: 10.1111/j.1365-2958.2005.04676.x. [DOI] [PubMed] [Google Scholar]
- 57.Schwesinger MD, Novick R. Prophage-dependent plasmid integration in Staphylococcus aureus. J. Bacteriol. 1975;123:724–738. doi: 10.1128/jb.123.2.724-738.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kleerebezem M, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lebrun M, Audurier A, Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J. Bacteriol. 1994;176:3040–3048. doi: 10.1128/jb.176.10.3040-3048.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nucifora G, Chu L, Misra TK, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamon E, et al. Comparative proteomic analysis of Lactobacillus plantarum for the identification of key proteins in bile tolerance. BMC Microbiol. 2011;11:1. doi: 10.1186/1471-2180-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y, Chen WN. iTRAQ-coupled 2-D LC–MS/MS analysis of cytoplasmic protein profile in Escherichia coli incubated with apidaecin IB. J. Proteomics. 2011;75:511–516. doi: 10.1016/j.jprot.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 63.Xiao, Y. et al. Metabolomics analysis reveals heavy metal copper-induced cytotoxicity in HT-29 human colon cancer cells. RSC Adv. 6 (2016).
- 64.Spano G, Chieppa G, Beneduce L, Massa S. Expression analysis of putative arcA, arcB and arcC genes partially cloned from Lactobacillus plantarum isolated from wine. J. Appl. Microbiol. 2004;96:185–193. doi: 10.1046/j.1365-2672.2003.02132.x. [DOI] [PubMed] [Google Scholar]
- 65.Groot MNN, et al. Genome-based in silico detection of putative manganese transport systems in Lactobacillus plantarum and their genetic analysis. Microbiology. 2005;151:1229–1238. doi: 10.1099/mic.0.27375-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


