Abstract
Bdellovibrio bacteriovorus is an obligate predator of bacteria that grows and divides within the periplasm of its prey. Functions involved in the early steps of predation have been identified and characterized, but mediators of prey invasion are still poorly detailed. By combining omics data available for Bdellovibrio and like organisms (BALO’s), we identified 43 genes expressed in B. bacteriovorus during the early interaction with prey. These included genes in a tight adherence (TAD) operon encoding for two type IVb fimbriae-like pilin proteins (flp1 and flp2), and their processing and export machinery. Two additional flp genes (flp3 and flp4) were computationally identified at other locations along the chromosome, defining the largest and most diverse type IVb complement known in bacteria to date. Only flp1, flp2 and flp4 were expressed; their respective gene knock-outs resulted in a complete loss of the predatory ability without losing the ability to adhere to prey cells. Additionally, we further demonstrate differential regulation of the flp genes as the TAD operon of BALOs with different predatory strategies is controlled by a flagellar sigma factor FliA, while flp4 is not. Finally, we show that FliA, a known flagellar transcriptional regulator in other bacteria, is an essential Bdellovibrio gene.
Introduction
Bdellovibrio and like organisms (BALOs) form a unique group of gram-negative bacteria that thrive by preying on other gram-negative bacteria. They are found in diverse environments including fresh water lakes, oceans, and various terrestrial habitats1–5. BALOs are mainly known in the delta-proteobacteria where they form the families Bdellovibrionaceae and Bacteriovoracaceae. Micavibrio is the only known exception, belonging to the alpha-proteobacteria6, 7 Predation is either epibiotic or periplasmic. In the former, the BALO drains the cytoplasm of its prey while attaching to the prey’s outer membrane1, 6. In the latter, the BALO enters the prey and consumes it while residing in the prey’s periplasm. Both epibiotic and periplasmic BALOs have a life cycle that includes a highly motile small non-replicative attack phase (AP) cell, which adheres to and recognizes suitable prey; a prey envelope-derived signal then triggers a newly described transition phase during which changes in the predator and the detection a prey cytosol-associated cue8 leads to the cell growth phase (GP), and ultimately to cell replication. In B. bacteriovorus, the predator penetrates the prey’s periplasm through a transient rupturing of the prey cell wall, and remodels it into a sphere-shaped cell, the bdelloplast, by release of cell wall modifying enzymes9. The predator consumes the prey’s cytoplasm fueling its growth as a filamentous, multi-nucleoid cell that finally septates to form multiple progeny AP cells9–11.
The early steps of the predator-prey interaction involve many cell cycle-dependent, differentially expressed functions. These include expression of genes specific and indispensable for the predatory process, i.e. genes that are not required for intrinsic survival and growth but specific to the predatory phenotype. These genes are essential in wild type BALOs, but can be deleted in host-independent (HI) mutants11–14. HI mutants are generated by spontaneous mutations in the gene bd0108, enabling B. bacteriovorus to grow both on live prey cells and on rich artificial media in the absence of prey15, 16. Bd0108 may play a role in regulating the production and extrusion/ retraction of type IVa pilus, an appendage shown to be essential for predation in both B. bacteriovorus and in the epibiotic predator, Bdellovibrio exovorus 17, 18. Type IVa pili usually govern twitching motility in other bacteria19, but in B. bacteriovorus they are assumed to facilitate invasion into the prey20. Other predation-essential genes encoding for proteases, regulatory proteins, signal transduction related proteins and flagellar motility, and genes important for cellular structure and organization were identified by transposon mutagenesis performed in HI genetic backgrounds, combined with a screen for alterations in predatory phenotype, or by systematic homology searches12–14, 18.
In this study, we applied computational integrated genome and transcriptome analyses to identify genes essential for predation, based on the rationale that the AP and the earlier steps of prey invasion are unique to obligate predators. A core of 43 AP genes was identified, including genes in an operon coding for type IVb fimbriae-like protein (flp) genes21 that was further characterized. Further protein domain sequence analyses uncovered additional flp B. bacteriovorus genes. Genetic and phenotypic analyses showed that three of these four flp IVb genes are essential for predation and although they are expressed during AP, they are under different transcriptional regulation: flp1 and flp2 are co-regulated with flagellins by the flagella sigma factor FliA, while flp4 is regulated independently by an unknown regulator.
Results
Identification of putative attack phase core genes
Successful initial recognition and attachment to the prey by Bdellovibrio depends on factors expressed during the AP22. In order to identify genes that are involved in the initial step of predation, we searched for known B. bacteriovorus AP genes that are present in other diverse BALOs. Four hundred twenty one B. bacteriovorus HD100 genes that are expressed in AP23 were compared to three fully sequenced Bdellovibrio genomes (Bdellovibrio exovorus JSS24, Bdellovibrio sp. Arhs25 and Bdellovibrio bacteriovorus W (https://www.ncbi.nlm.nih.gov/assembly/GCA_000525675.1), three fully sequenced Bacteriovorax strains (Bacteriovorax SEQ25_V, Bacteriovorax BAL6_X, Bacteriovorax BSW_11IV4 and the fully sequenced Halobacteriovorax marinus SJ26. B. bacteriovorus HD100 and B. bacteriovorus W are periplasmic fresh water BALOs, Bdellovibrio sp. ArHS is a thermophilic BALO, and B. exovorus JSS is a fresh water epibiotic predator within the Bdellovibrionaceae. H. marinus SJ and the three Bacteriovorax strains are marine periplasmic BALOs belonging to the Bacteriovoracaceae1, 4, 25, 26. Forty-three genes that are certain orthologs between the examined species that had high (0.2) BLAST score ratios (BSR) were identified in this computational screen. To further refine our screen, looking for additional evidence for evolutionary conserved genes27, we searched the 43 “AP core” gene set for members that are syntenous in all examined BALOs. Twelve of the 43 genes were found to be syntenous and present in four putative operons (Fig. 1 and Table S1).
Figure 1.
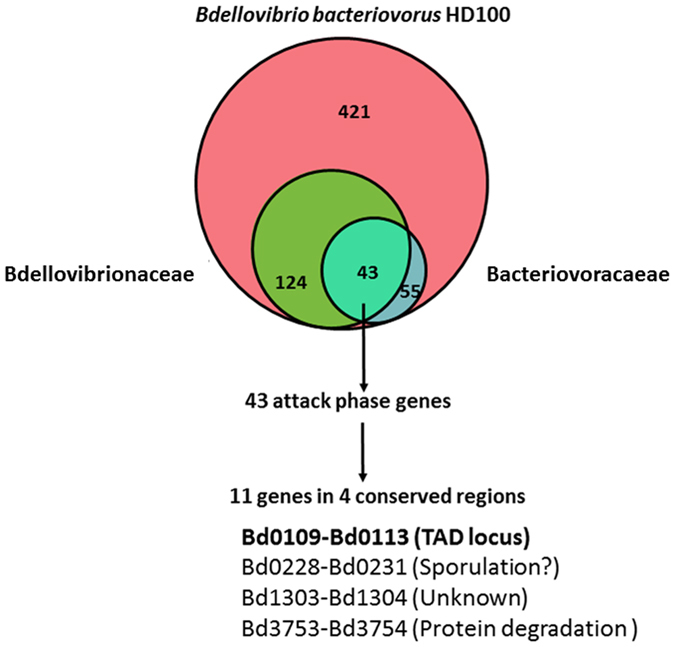
Overlap of attack phase genes between strains of Bdellovibrio and like organisms, identifying putative predation-essential genes. 124 Bdellovibrionaceae and 55 Bacteriovoracaceae genes exhibit high similarity (BSR ≥ 0.2) to 421 B. bacteriovorus HD100 AP-induced genes. The overlap of the three sets depicts 43 AP core genes, 11 of which cluster in four B. bacteriovorus HD100 genomic regions. Each of these regions is a putative operon as indicated by RNAseq expression data (BdelloViewer, http://www.weizmann.ac.il/molgen/Sorek/bdello_browser/viewer/index.php?meta=bdello_combined&jump=bdello,bdello_sRNAs#1 23. The Venn diagram was created using Biovenn69.
Most of the genes in the four putative operons encode for poorly characterized or unknown proteins. Genes bd0228 to bd0231 form a putative operon that encodes a serine protein kinase, an uncharacterized protein and a sporulation protein R, SpoVR. Similar operons were found in many non-sporulating and non-predatory bacteria from the beta-, gamma- and delta-proteobacteria. SpoVR is a non-essential gene that is involved in spore cortex formation in Bacillus subtillis and Myxococcus xanthus 28, 29. Genes bd1103 and bd1104 are in a putative operon and encode two conserved proteins of unknown function, also found in many bacteria. The putative operon bd3753–bd3754 encodes for ClpX and ClpP proteins respectively, forming the ClpXP complex that degrades misfolded proteins and thus assists in maintaining cell proteostasis30. Interestingly, the E. coli ClpXP complex is activated under carbon starvation, the physiological state of the attack phase31, 32.
The syntenous region bd0109–bd0113 is part of a putative operon including genes bd0109–bd0119. It was previously annotated as a tight adherence locus (TAD)33. This operon encodes for proteins that process and export fimbriae like proteins (Flp) to the outer membrane. Flp proteins were found to be crucial for bacteria-host interaction in human, animal, and plant pathogens34, 35.
B. bacteriovorus HD100 genome encodes four different flp pilin genes
The TAD locus operon encodes components essential for the maturation and export of Flp proteins34. The region includes genes coding for the ATPase that drives Flp export, TadA (bd0111); an inner membrane export component, TadB (bd0110); the outer membrane export proteins RcpA (bd0112) and RcpC (bd0113)33, 36. However, TadV, a prepilin peptidase crucial for pilin maturation, is missing from the operon and was not found in the entire B. bacteriovorus genome. Further analysis indicates that TadV is missing from all known Bdellovibrio genomes, but is present in Halobacteriovorax and all of the Bacteriovorax genomes (Fig. 2).
Figure 2.
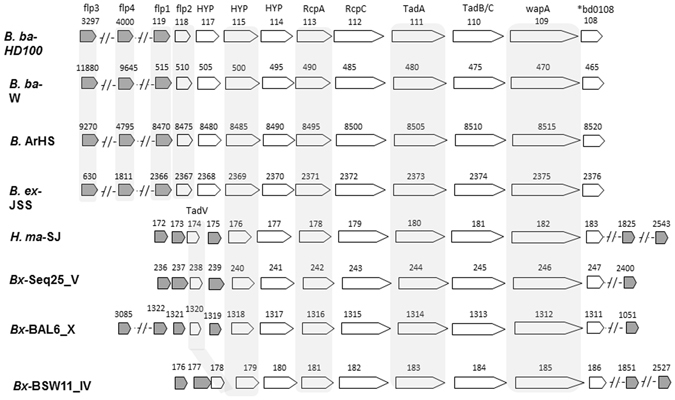
Genomic arrangement of TAD loci in different BALOs. TAD loci from all examined BALOs exhibit synteny. The prepilin peptidase, TadV is missing from all Bdellovibrio genomes but is present in Halobacteriovorax and Bacteriovorax genomes. All BALOs also encode non-TAD associated flp genes. flp2 is a TAD-associated trimmed flp, that is only present in Bdellovibrio TAD loci and is absent from all other BALOs. BALO species designations- B.ba-HD100: Bdellovibrio bacteriovorus HD100; B.ex-JSS: Bdellovibrio exovorus JSS; B.ba-W: Bdellovibrio bacteriovorus W; B-ArHS: Bdellovibrio sp. ArHS; H.ma-SJ: Halobacteriovorax marinus SJ; Bx-SEQ25_V: Bacteriovorax sp. Seq25_v; Bx- BAL6_X: Bacteriovorax sp. BAL6_X; Bx- BSW11_IV: Bacteriovorax sp. BSW11_IV.
Based on their genomic position within the TAD locus, their small size, and the hydrophobic character of the encoded proteins, two B. bacteriovorus flp pilin protein-encoding genes were previously identified (bd0119, flp1, and bd0118, flp2)33. We searched BALOs genomes for additional flp pilin genes, encoded outside the TAD locus. Flp pilins are short and not well conserved37. We thus searched not with specific flp pilin sequences (i.e., by BLAST searches) but with conserved amino acid motifs. We first constructed a position weight matrix (PWM) of conserved motifs from different Flp pilin protein sequences (Experimental procedures, Fig. 3). The PWM corresponded to a known conserved Flp proteins sequence domain, a GXXXXEY followed by a stretch of hydrophobic amino acids37. Searching the B. bacteriovorus genome with the PWM identified three putative flp pilin encoding genes: bd0119 (flp1), bd3297 (flp3) and an un-annotated open reading frame within the bd2719–bd2720 intragenic region that we designated as bd4000.
Figure 3.
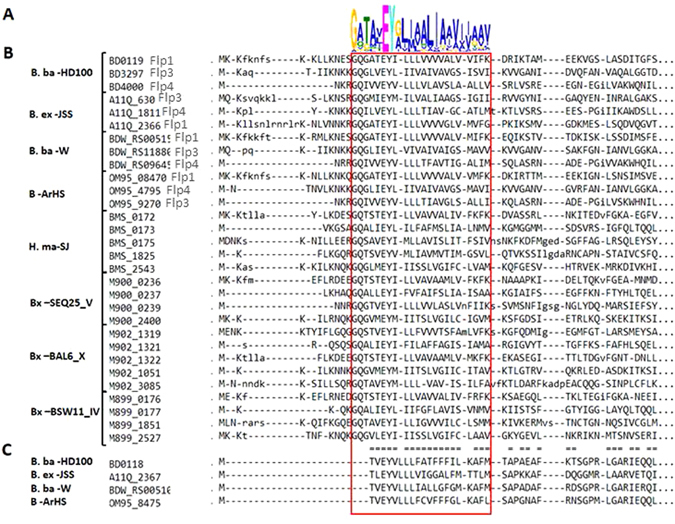
Multiple alignment of predicted flp sequences. Sequence logo representation representation70 of a Flp pilin sequence position weight matrix (PWM), depicting the canonic Flp pilin motif (GXXXXEY and a stretch of hydrophobic amino acids). (A) Multiple alignment of Flp pilin sequences identified by the PWM in various BALO’s with the flp pilin motif indicated by a red outline. (B) Flp2 sequences are truncated forms of Flp pilins, both the N terminus proximal glycine (G) and the amino acid that follows it are missing, creating motif: MXXEY and a stretch of hydrophobic amino acids. The protein logo was created with MEME60. Multiple alignments were calculated using GLAM260, 71. The Flp and the Flp2 multiple alignments were aligned to each other using the Compass program version 2.4572, with the similar regions indicated by the ‘=’ marks in between the multiple alignments. Lower case letters are regions not found to be significantly aligned by GLAM2. BALO designations are as in Fig. 2.
Searching other BALO genomes with the PWM revealed that all include similarly located flp pilin open reading frames, i.e. flp sequences in the TAD locus and others that are not TAD associated (Fig. 3, Fig. S1). While only one TAD-associated flp gene was found with the PWM in Bdellovibrio genomes, Bacteriovorax and Halobacteriovorax genomes include three TAD associated flp pilin genes. Interestingly, bd0118, annotated as flp2 in B. bacteriovorus 33 was not identified as a Flp pilin-encoding gene by our PWM. Sequence analysis demonstrated that Flp2 includes a trimmed Flp domain. The Flp2 sequence starts with MXXEY that is followed by a stretch of hydrophobic amino acids (Fig. 3). This domain differs from the consensus Flp domain as it misses four amino acids: the glycine residue that is recognized by the prepilin peptidase and the three residues C-terminal to it that typically are at the N-terminal end of the processed, mature pilin38. Consequently, while the Flp motif is usually 5–20 amino acids from the protein initiator methionine, in Bd0118 it directly follows it (Fig. 3). Genes encoding proteins with such a trimmed Flp motif were also observed within the TAD loci of other Bdellovibrio strains, but not in Bacteriovorax (Figs 2 and 3).
In order to exclude the possibility of mis-annotation of the coding region start position, we added its upstream 21 amino acids to the Bd0118 sequence and compared it to the PWM. No complete flp motif was found in Bd0118 or its orthologs in other BALO genomes. flp3 and flp4 were annotated as Flp proteins in additional Bdellovibrio strains based on the synteny of their surrounding genes with those of the B. bacteriovorus HD 100 flp3, and flp4 genes (Table S2).
A similar screen in four Bacteriovoracaceae genomes identified three TAD-associated and three non-TAD- associated flp pilin genes in H. marinus SJ, Bacteriovorax Seq25_V and Bacteriovorax BAL6_X. Bacteriovorax BSW11_IV was the only exception as it included two flp genes from each kind.
flp1, flp2, and flp4 are expressed during attack phase while flp3 is silent
The expression pattern of the four flp genes were analyzed in AP, and at early (30 min), mid (60 min), and late (180 min) GP. flp1, flp2, and flp4 were only expressed in the AP, while flp3 was silent at all examined time points (Fig. 4). flp gene expression was further examined in the planktonic and biofilm phases of HI mutant cultures. flp1, flp2, and flp4 were expressed in both planktonic and biofilm cultures, while flp3 was silent under both conditions (Fig. 4). Gene bd0199, which is only expressed during GP, served as a control. A protein analysis by mass spectrometry (MS) of wild type B. bacteriovorus cultures in which Bd0110, Bd0111, Bd0112, Bd0113 and the Bd0119 (Flp1) were detected, confirmed that TAD operon-encoded proteins were indeed produced in AP (Table S3).
Figure 4.
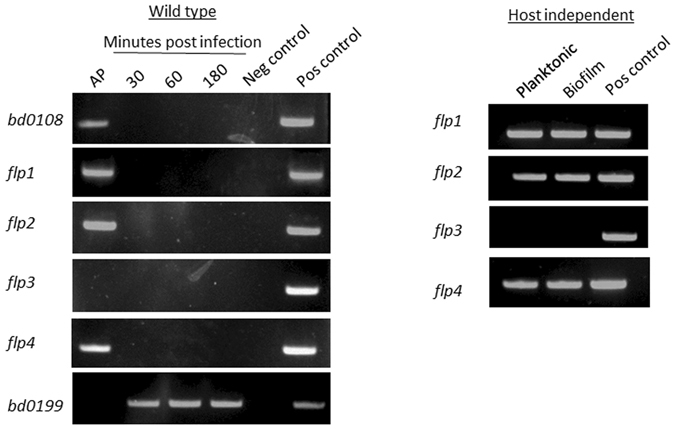
Semi-quantitative RT-PCR (sqRT-PCR) of B. bacteriovorus flp pilin genes. Expression of flp pilin encoding genes during the B. bacteriovorus HD100 life cycle shows that flp1, flp2 and flp4 are expressed during AP, while flp3 is silenced during the whole cycle. The same expression pattern was observed in a host-independent biofilm and in a planktonic culture, with flp1 and flp2 are expressed and flp3 is silent. bd0108 is used as a AP marker and bd0199 as a GP marker. A) Negative control - E. coli DNA. B) positive control - B. bacteriovorus DNA. Gels were acquired with DNR Gel Capture with no further processing.
flp1, flp2, and flp4 pilins knockouts abolish predation
In order to examine the role of Flp pilins-encoding genes in predation, all four flp genes were subjected to in-frame deletion in the background of B. bacteriovorus HD100. While flp3 (which was not expressed under any of the examined conditions) could be deleted in this wild-type background, meaning the mutated strain could grow predatorily, flp1, flp2 and flp4 could not be deleted, as a screen of about 400 plaques of each for putative double recombinants did not yield any. For comparison, the flp3 deletion mutants were identified at a frequency of one per 20 plaques screened with a confirmation rate of 60%. This strongly suggests that both the typical flp1 and flp4 type IVb pilin genes, and the flp2 truncated Type IVb pilin gene, are essential for predation in a wild type background. To examine this hypothesis, these flp genes were deleted separately and also together in the background of HI M11.1, a B. bacteriovorus HD100 spontaneous host-independent type II mutant16. This mutant can grow axenically, in the absence of prey but retains predatory capabilities, albeit with reduced efficiency. This yielded the double mutant HIΔflp1-2, the triple mutant HIΔ flp1-2-4 and the three single mutant strains HIΔflp1, HIΔflp2, and HIΔflp4. These five mutant strains grew axenically, i.e. in the absence of prey on a rich PYE medium as expected from HI strains. However, none was able to prey on living E. coli cells, demonstrating their indispensability in the early stages of the predatory interaction (Fig. 5, Fig. S2). Finally, to confirm that the predation inability of these deletion strains was caused by the introduced deletions, the strains were complemented with the deleted genes under their native promoter by conjugating plasmids pPROBE-NT-flp1-2, pPROBE-NT-flp1, pPROBE-NT-flp2 and pPROBE-NT-flp4 each to its corresponding deletion strain. Complementation resulted in complete restoration of the predatory HI phenotype (Fig. 5, Fig. S2), causing a four-order of magnitude decrease of the prey population. Deletion of flp3 in the HI M11.1 background did not yield any noticeable phenotypic change, confirming that, as in the wild type background and under laboratory conditions, flp3 is not required for predation. To address the possibility of functional complementation between flp genes, plasmid pPROBE::pflp1-flp3, bearing flp3 under the flp1 promoter was introduced in the HIΔflp1and in the HIΔflp4 backgrounds. No complementation was observed, as predation was not restored.
Figure 5.
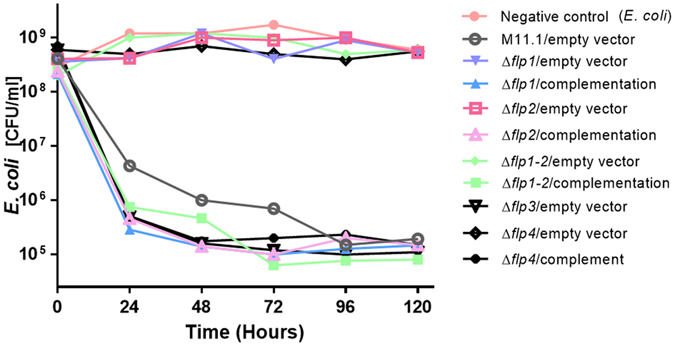
Effects of flp pilin gene deletion on predation. Both TAD-associated flp1, flp2, and non-associated flp4 gene knockouts (Δflp1, Δflp2, and Δflp4, respectively) are unable to prey, while Δflp3 had no effect on predation. All strains regained the predatory phenotype of the parental strain (M11.1) upon complementation (comp.Δflp1, comp.Δflp2 and comp. Δflp4). In order to prevent polar effects, all knockouts were constructed as in-frame deletions, with only small non-functional peptides encoded in the remaining sequence (9–12 amino acids long). Complementation for each mutation was carried out using pPROBE-NT. Control strains carried an empty vector.
flp1, flp2, and flp4 do not mediate binding to biotic or abiotic surfaces
We examined if the non-predatory phenotype of the flp deletions is due to a reduced capacity for surface adhesion, as is the case in other bacteria where flp genes mediate surface attachment39–45. For that purpose, biofilm formation by the parental HI strain, M11.1 and the type IVb pilins triple knockout strain ∆flp1-2-4, was examined. No significant reduction in biofilm formation was observed (Fig. 6).
Figure 6.
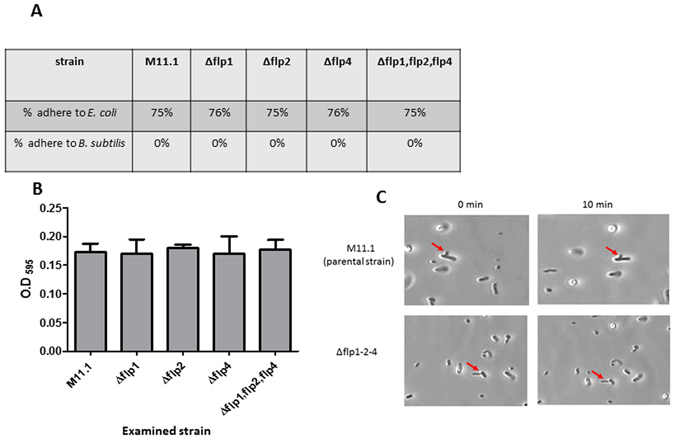
Adherence of flp knockouts to live prey and to abiotic surfaces. (A) HI M11.1 single and triple knockouts of flp1, flp2 and flp4 were examined for their ability to adhere to E. coli cells. No difference was observed between all the knockout strains and the HI M11.1 parental strain. B. subtilis served as a negative control for adhesion. (B) Examination for biofilm formation (adherence to abiotic surfaces), as in section A, no difference was observed between the strains. (C) Phase microscopy illustrating the adhesion assay. As in the parental HI M11.1 strain, the non-predatory mutant Δflp1-2-4 only attached to E. coli through the predatory pole. Similar results were obtained with the individual flp1, flp2 and flp4 mutants (>100 events examined), in suspensions containing 106 or 107 cells.ml−1. In all mutants, the cells did not detach from E. coli during the examination time (10 min). Pictures were taken with a Digital Sight D5 camera (Nikon) and captured with a NIS-element software with no further processing.
The single flp1, flp2, and flp4 mutants as well as the triple flp1-2-4 knockout were then examined for direct binding to E. coli prey cells. No significant difference to the parental strain M11.1 was evident (Fig. 6). Direct observation under phase contrast microscopy showed that the Δflp1-2-4 mutant as well as each single flp mutant can bind to prey cells for significant lengths of time (>10 min).
The B. bacteriovorus HD100 and B. exovorus JSS TAD operons are regulated by the flagellar sigma factor FliA
A transcriptomic analysis of B. bacteriovorus AP and GP23 suggested that the flagellar sigma factor, fliA (bd3318) acts as the main regulator of AP-expressed genes, including those encoded in the TAD operon. To demonstrate FliA control over TAD, we attempted to delete the B. bacteriovorus sigma factor FliA, encoded by gene bd3318. No mutants were obtained after screening 300 plaques of wild type HD100 and 300 colonies of a HI derivative. We then focused our efforts on the HI strain for technical convenience. As FliA is a central regulator of a cell cycle phase, we conjectured that a fliA knockout would be lethal or grow slowly. Accordingly, the knockout procedure was modified (see experimental procedures). No mutants were uncovered under any of the incubation conditions employed: colonies that appeared after 2 to 3 weeks of incubation were all found to be wild type. When low, 2% and 3% sucrose solutions were used for counter-selection, 30 to 50% of the colonies were merodiploid. No full recombinant was obtained. Therefore, the inability to knockout fliA led us to the conclusion that FliA is essential in B. bacteriovorus, unlike what was found in other bacteria46.
The flp1 promoter (i.e., promoter of the TAD operon) (Fig. 7, Fig. S3) was examined in a heterologous E. coli system: fliA coding regions (bd3318 or a11q_0617 for B. bacteriovorus and B. exovorus, respectively) were cloned into pBAD/gIIIA under an arabinose inducible promoter. Since the examined promoters were cloned upstream of a promoter-less β-galactosidase (see experimental procedures), recognition of these promoters by a sigma factor, such as FliA, should result in a measurable β-galactosidase biochemical activity. In other gram-negative bacteria, flagellin-encoding genes are regulated by FliA46, as was also predicted for B. bacteriovorus 11, 23. Therefore, B. bacteriovorus HD100 and B. exovorus JSS flagellin (bd0606 and a11q_1936 respectively) promoters were chosen as positive controls. β-galactosidase under the TAD operon (that includes flp1 and flp2) or under the regulation of flagellin promoters was strongly induced by FliA, as compared to an empty pBBR1-MCS2-LacZ vector. This validated the hypothesis that FliA regulates expression of flp1 and of all the other AP genes present with it in the TAD operon (Fig. 7). In contrast, the flp3 and flp4 promoters from both B. bacteriovorus HD100 and B. exovorus JSS remained silent under these conditions (Fig. 7).
Figure 7.
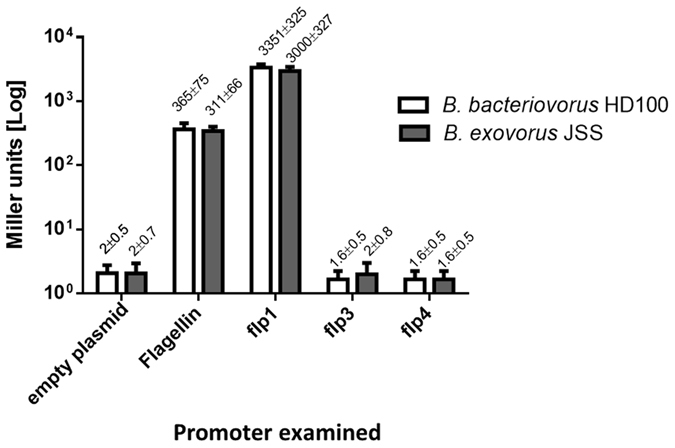
Examination of B. bacteriovorus and B. exovorus FliA-dependent promoter activity in an E. coli heterologous system. B. bacteriovorus HD100 and B. exovorus JSS FliA (bd3318 and a11q_617, respectively) activity was examined in an E. coli heterologous system. Flagellin promoters (Bd0606, A11q_1936) and an empty vector were used as positive and negative controls, respectively. The promoters of flp1 genes (bd0119 and A11q_2366) responded strongly to FliA induction in comparison to the controls. In contrast, promoters of the flp3 (bd3297 and a11q_630) and flp4 (bd4000 and a11q_1811) genes did not respond to FliA induction.
Discussion
Genes essential for predation by B. bacteriovorus have been previously identified by direct random transposon mutagenesis, screening for mutants defective in predation12, 13, or by targeting B. bacteriovorus genes whose orthologs in other bacteria encode functions that could potentially affect predation14. In this work, we aimed to identify genes that are essential for predation by examining early predation stages of the B. bacteriovorus life cycle. Genomic data have so far been used to differentiate between predatory and non-predatory bacteria, between predators exhibiting an epibiotic versus a periplasmic strategy, and to delineate the core genes of periplasmic predators4, 24, 26, 47. By integrating BALOs genomic and transcriptomic data we identified a set of 43 genes designated as “attack phase core genes”.
We focused on the TAD operon (bd0109-0119), that encodes flp (type IVb) pilin-encoding genes and their processing and export machinery. Our analyses showed that type IVb pilin genes, while found in various gram negative bacteria, are more numerous in BALOs than in other known bacteria and that they are very diverse in sequence, in their association with the TAD loci, and in their regulation (Figs 2, 3 and 7). The only other known example of non-TAD associated flp genes is one of the two Agrobacterium tumefaciens flp pili45.
The B. bacteriovorus TAD-encoding flp pilins (flp1, flp2) and flp4 are expressed during AP in wild type cultures as well as in planktonic and in biofilm HI cultures (possibly in the AP cells of these unsynchronized cultures). Flp1 (Bd0119), along with other TAD locus proteins Bd0110, Bd0111, Bd0112, Bd0113, was also detected by proteome analyses, but Flp2 and Flp4 were not detected, possibly because their hydrophobicity and small mass make detection of Flp pilins very difficult33. flp3 was silent under all various conditions we tested and as such is the first documented case of a flp pilin gene in gram-negative bacteria that was not found to be expressed. flp3 does not appear to be a pseudogene and it may be expressed under other environmental conditions.
While a type IVa pilus gene had been shown to be essential for B. bacteriovorus predation14, 17, no functional analysis of type IVb pilin genes have been performed in B. bacteriovorus 33. Since type IVb pilins are crucial for physical interactions with the eukaryotic host cell in pathogenic bacteria48–50 we posited they might also play an important role in BALO predation.
Attempts at deleting flp1, flp2 and flp4 in a wild type background were unsuccessful. In contrast, a flp3 wild type knockout was viable, in accordance with its lack of expression in both AP and GP. In an HI background, triple flp1-flp2-flp4, double flp1-flp2, as well as all individual flp deletions yielded viable strains that could be grown axenically on a rich medium but were unable to prey. The flp genes were not interchangeable as their mutants were not complemented by each other or by the flp3 gene. Thus, each of flp1, flp2 and flp4 is required for predation, and each may have a specific role but none is crucial to the cell cycle, as mutants in an HI background were viable.
B. bacteriovorus, and all other BALO, include both type IVa and type IVb pilin genes. These pilin-encoding genes differ in their sequence characteristics and in the systems that assemble, process and export them35, and different functions have been attributed to each. While in most bacteria PilA pilins are responsible for surface-dependent motility and attachment, the Flp pilins were found to be involved only in surface attachment38–44. In some pathogenic bacteria (such as Pseudomonas aeruginosa, Aggregatibacter actinomycetemcomitans, Haemophilus ducreyi and Agrobacterium tumefaciens) disturbance of flp and TAD components attenuated virulence as well as decreased non-specific surface adhesion that is required for the development of biofilms38, 45, 51, 52. In other pathogens, flp deletion reduced virulence or microcolony formation without affecting biofilm formation53, or bacteria-cell host interaction49. In B. bacteriovorus, deletion of the three active flp genes had no effect on attachment/adhesion and HI biofilm formation. It is remarkable that both Bdellovibrio type IVa and the type IVb pilin –encoding genes, while essential for predation, are unnecessary for adhering or binding to prey cells, suggesting that other adhesion proteins expressed in the AP may be responsible for that step (58).
We provide the first experimental evidence that expression of the TAD-associated flp1 and flp2 gene is regulated by FliA, a flagellar sigma factor, both in the periplasmic predator, B. bacteriovorus HD100, and in the epibiotic predator, B. exovorus JSS. Further analyses of promoter sequences suggested that TAD associated flp genes are also regulated by FliA in other BALOs. Notably, flp4, while co-expressed with the TAD-encoded flp1 and flp2 is not similarly regulated by FliA. In many bacteria FliA regulates flagellar and chemotaxis-related genes46, 54. In several pathogens, FliA has expanded to directly control virulence related genes, such as a cytotoxic protein from Campylobacter jejuni, or indirectly through cylic-di-GMP modulation as with the E. coli fimbriae or Vibrio cholerae cytotoxin55–57. These FliA-regulated traits are unlinked to the cell cycle. In contrast, we previously found that in B. bacteriovorus FliA expanded to become the “AP master regulator” controlling about 77% of the AP genes23. Accordingly, in B. bacteriovorus FliA has turned into an essential gene: B. bacteriovorus FliA knockout was lethal both in wild type and HI mutants, indicating that FliA has expanded to regulate vital functions, a situation not known in any other bacterium. FliA regulation thus appears to be an indispensable component of obligate predators and a central regulator of the Bdellovibrio cell cycle.
Although functional analyses show that type IVa and type IVb pilins mutants are phenotypically undistinguishable from each other, genetic and biochemical analyses suggest that each may have a different role in signaling prey cells to the predator. We propose to integrate our data into previously proposed models8, 18, 20. BALOs reversely bind the prey outer membrane (with a yet unknown component), followed by irreversible anchoring to a prey envelope-associated structure8. Type IV pili with their varied pilins, bring about the sensing, transduction, and an initial output of the interaction in the form of tightening of the predator-prey complex. According to a recent study, transition from AP to GP defines a third phase in B. bacteriovorus’ life cycle – the transition phase (TP)8. The TP expresses a specific transcription profile in response to signals triggered by the prey. Characteristically, response to the prey envelope results in bd0108 shutdown, and in pilA and fliA down-regulation as the first step resulting from attachment to the prey. We suggest that type IVa and type IVb pili generate differentiated signaling outputs: type IVa pilus sensing leads to its retraction through Bd0108–Bd010918 (AP genes not regulated by FliA) initiating a signal sensed at the invasive pole of the predatory cell20. This signal could be further processed through cyclic-di-GMP regulation20, leading to bd0108 and other non-fliA regulated genes (pilA) downregulation; the signal generated by interacting type IVb pilins during attachment and further, during penetration and establishment in the periplasm, brings about alterations in Bdellovibrio FliA-regulated genes, including flp locus genes and pilin genes. This two-pronged signaling scheme, which still has to be validated, would ensure a rapid response based on enzymatic activation (c-di-GMP signaling) followed by a somewhat slower change at the transcription level.
Experimental Procedures
Bacterial strains and growth conditions
B. bacteriovorus HD100 wild type (Table S4) was grown with 109 cells.ml−1 E. coli prey cells in diluted nutrient broth (DNB) at pH 7.4, supplemented with 2 mM CaCl2 and 3 mM MgCl2 58. HI strains were grown in peptone-yeast extract medium (PYE) similarly supplemented with CaCl2 and MgCl2 58. Unless mentioned otherwise, kanamycin and streptomycin were added to final concentrations of 25 µg.ml−1 and 100 µg.ml−1, respectively.
Predation assay of the mutated and complemented strains
The tested strains were grown axenically to OD600 0.7, centrifuged, washed twice in DNB, and suspended in 5 ml of OD600 1 of E. coli in DNB supplemented with the appropriate antibiotic. E. coli were counted by serial dilution on LB agar every 24 hours for 120 h. Plates were incubated overnight at 28 °C.
Orthologs identification and synteny analysis
Genuine orthologs were identified using the BLAST Score Ratio (BSR) approach59 using a ratio cutoff of 0.2. BSR similarity measure is the ratio between the blast score of one protein to another with the blast score of the protein to itself.
flp pilins identification
A position weight matrix (PWM) was generated with the MEME program60 using flp pilins sequences from the Pfam database (entry PF04964)61, Fig. 3. The PWM was searched against selected BALO genomes using the MAST program61. Only high scoring hits (expected values < 0.1) within short (<100 amino acids) proteins or ORFs were further considered.
Biofilm formation assay
B. bacterivorous HI M11.1 was grown from an initial 100 μl OD600 0.7 in 96 well plates at 28 °C. Biofilm formation was measured every 24 hours using the standard crystal violet staining procedure13.
Sample collection and RNA extraction
RNA was extracted using a MasterPure kit (epicenter) according to the manufacturer’s instructions. This was followed by an additional DNAse treatment using TurboDNASE kit (ambion). RNA integrity was validated by agarose gel electrophoresis. All the RNA samples were tested for DNA contamination by PCR using 16 S rRNA gene primers (8 f and 1492 R in Table S5). RNA extraction from biofilms was performed in glass test tubes: a turbid culture of the host independent strain M11.1 was diluted to OD600 0.7 into 1 ml fresh PYE for 4 days. Next, the planktonic phase (AP) cells were harvested for RNA extraction. The biofilm was washed three times with HEPES buffer then immersed in RNALatter (Ambion), extracted from the test tube by scraping the flanks, centrifuged and kept at −80 °C until RNA was extracted. Samples were collected as described previously8: 1 ml of each time point was centrifuged and immersed in RNAlatter and then kept in −80 °C until RNA was extracted.
Semi-quantitive RT-PCR (sqRTPCR)
100 to 200 ng of RNA were used for cDNA synthesis using Improm II kit (Promega) according to the manufacturer’s protocol. 50 µl of RNAse free water were added to the cDNA mixture to a final volume of 80 µl. sqRTPCR was performed using 1 μl of a diluted cDNA mixture, primers as listed in Table S5 and a Readymix polymerase (Lambda biotech) according to manufacturer’s instructions. Gels were recorded using a DNR MiniBIsPro BioImager with the DNR Gel Capture software with no further processing (DNR Bio-Imaging Systems, Israel).
Proteome analyses
Analysis of the HD100 proteome was performed by MS. SDS-PAGE separation and digestion was performed as previously described62. The resulting peptide mixtures were fractioned by with strong cation exchange and analyzed by subsequent high-performance liquid chromatography coupled to an Orbitrap XL. MS data (survey and fragmentations scans) was analyzed using Thermo Proteome Discoverer™ (Thermo Fisher Scientific, Version 1.4.1.12) through SEQUEST HT search engine with a UniProt database (5th November, 2015) containing the whole HD100 proteome63. Proteins were considered, if they were identified with at least two unique peptides and a minimum score of 20.
Plasmid construction
All plasmids were constructed by the “Restriction Free” (RF) method as described by64. PCR was performed using Q5 polymerase (New England Biolabs) according to manufacturer instructions. Plasmids backbones and primers are listed in Tables S4 and S5, respectively.
For the purpose of in-frame deletions, 500–1000 bp long DNA fragments were amplified from both sides of the target gene and fused together by the RF method to the suicide plasmid pSSK1065. Knockout primers were designed to leave a 16–17 amino acid remaining to avoid upstream polar effects.
Complementation was carried by pPROBE-NT, which carried the examined gene with its promoter (usually 200–400 bp upstream to the translation start site).
Examined fliA orthologs were cloned to replace the gIII secretion signal peptide, downstream to the arabinose-induced promoter in pBAD/gIII (Invitrogen). Examined promoters were cloned upstream to a promoter-less full-length β-galactosidase (lacZ) in pBBR1-MCS2-LacZ66.
Conjugation in B. bacteriovorus
E. coli donor strains were grown with the appropriate antibiotics (listed in Table S4) to an OD600 of 0.4, then washed twice with DNB and concentrated 10-fold. A 100 µl volume of each donor was mixed with 100 µl of B. bacteriovorus HD100 or B. bacteriovorus M11.1 from a turbid culture or with an E. coli-B. bacteriovorus HD100 overnight- dual culture. The mixture was then spread over a sterile membrane that was placed upon a DNB agar plate. Following overnight incubation at 28 °C, the membrane was washed with 1 ml of DNB, diluted and plated on E. coli double layer agar or PYE agar plates supplemented with 50 µg.ml−1 streptomycin and 25 µg.ml−1 kanamycin. When E. coli MFDpir was used, diaminopimelic acid (DAP) was added to a final concentration of 100 µg.ml−1.
B. bacteriovorus knockouts
After conjugation with the appropriate pSSK10 derived construct, plaques or colonies were picked up and checked for merodiploidicity by PCR (Table S4). Selected plaques or colony were transferred into 5 ml of OD600 1 of an E. coli suspension or to PYE, respectively, supplemented with streptomycin and grown for several days. An aliquot was transferred to OD600 1 of an E. coli suspension or PYE both supplemented with streptomycin and 5% sucrose and grown for 24 and 72 hours, respectively, then diluted and plated on E. coli double layer agar or PYE agar plate both supplemented with 50 µg.ml−1 streptomycin. Plaques and colonies appeared after a week of incubation. Plaques were transferred to a 96 wells plate, each well containing 150 to 200 μl of a OD600 1 E. coli suspension. HI colonies were transferred to a grid PYE agar plate (53 colonies per plate), then after appropriate incubation, samples were examined for allelic exchange by PCR. Parental strains (WT HD100 or HI) and the appropriate merodiploid strain were used in each PCR reaction as a control.
Selection for fliA knockout strain was performed with the following modifications: Recombinants were screened as above in counter-selection tubes containing PYE and 5% sucrose. After 2, 4 and 5 days (instead of after 3 days in the standard procedure), suspensions were plated on PYE agar and incubated for a week. Colonies were examined for allelic exchange by PCR. Thereafter, plates were incubated for an additional 2 to 3 weeks under humid conditions, to enable the development of slowly growing colonies, typically 10 to 20 which were examined for allelic exchange by PCR. This experiment was performed three times.
E. coli fliA knockout
E. coli fliA was knocked out using a Lambda-red recombination system67. Briefly, E. coli DH5α/pKD46 was induced for lambda red recombinase by adding 0.05% arabinose. A linear DNA fragment containing a chloramphenicol resistance cassette flanking 30 bp with the corresponding upstream and downstream to E. coli fliA was created by PCR using primers: fliAlambdared-F and fliAlambdared-R and pKD3 as a template. Next, the liner fragment was electroporated into the induced lambda red E. coli DH5α/pKD46. This resulted in chloramphenicol resistant E. coli colonies that were isolated and validated for fliA knockout using primers Colifliadel-R and Colifliadel-F.
E. coli - Bdellovibrio attachment assay
Attachment assays were performed according to Milner et al.20 with slight modifications. HI and HI flp mutants were grown in PYE medium to an OD600 of 0.5, washed twice in DNB, and mixed with E. coli. The experiments were carried out at the final cell concentrations of 106 and of 107 ml−1. The Bdellovibrio - E. coli co-culture was incubated for recovery of 30 min in 28 °C, then concentrated by low speed centrifugation (4000 rpm for 10 min), the pellet was re-suspended in fresh DNB and re-incubated in 28 °C. 10 µl were taken at different time points, placed on positively charged polylysine-coated adhesive microscope slide (X-tra slides, Leica) and examined under phase contrast microscopy. As a negative control, E. coli was replaced with Bacillus subtilis, a gram-positive bacterium that Bdellovibrio does not attach to.
Examination of B. bacteriovorus and B. exovorus flp pilin gene promoters response to FliA in an E. coli heterologous system
Examined promoters were cloned upstream to β-Galactosidase (lacZ) in pBBR1-MCS2-LacZ. The resulting constructs were introduced into E. coli DH5α ∆fliA together with FliA cloned under arabinose-induced promoter in pBAD. The endogenous E. coli fliA was knocked out in order to reduce the background signal. Each E. coli strain carrying the two plasmids was grown overnight, then diluted into fresh LB to OD600 0.05, supplemented with either 0.5% glucose or 0.5% arabinose. When cultures reached mid-log (OD600 0.4–0.6), bacteria were harvested and measured for LacZ activity following a standard protocol68. Final β-Galactosidase activity was calculated by the following formula:
In this manner any background signals stemming from E. coli were eliminated.
Electronic supplementary material
Acknowledgements
This research was supported by the Israel Science Foundation (Grant 1583/12 to Edouard Jurkevitch) and by the German-Israel Foundation for Scientific Research and Development (Grant I-1217-342.13/2012 to Edouard Jurkevitch, Shmuel Pietrokovski and Michael W. Linscheid). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors thank Or Rotem for insightful discussions, Yoav Peleg and Rena Gorovits for advanced technical help, and the Proteome Factory AG for technical support.
Author Contributions
O.A. planned and performed most of the experimental work and most of the writing. M.P. performed the attachment experiments. R.B., S.B. and M.L. were responsible for the proteomics analysis. S.P. and E.J. planned and supervised experiments, and participated in the writing of the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00951-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shmuel Pietrokovski, Email: Shmuel.Pietrokovski@weizmann.ac.il.
Edouard Jurkevitch, Email: edouard.jurkevitch@mail.huji.ac.il.
References
- 1.Koval SF, et al. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int. J. Syst. Evol. Microbiol. 2013;63:146–151. doi: 10.1099/ijs.0.039701-0. [DOI] [PubMed] [Google Scholar]
- 2.Hobley L, et al. Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics. 2012;13:670. doi: 10.1186/1471-2164-13-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pineiro SA, et al. Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environ. Microbiol. 2007;9:2441–2450. doi: 10.1111/j.1462-2920.2007.01362.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, et al. Draft genome sequences for the obligate bacterial predators Bacteriovorax spp. of four phylogenetic clusters. Stand. Genomic Sci. 2015;10:11. doi: 10.1186/1944-3277-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurkevitch E, Minz D, Ramati B, Barel G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 2000;66:2365–2371. doi: 10.1128/AEM.66.6.2365-2371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Kadouri DE, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC genomics. 2011;12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidov Y, Jurkevitch E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2004;54:1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- 8.Rotem O, et al. Cell-cycle progress in obligate predatory bacteria is dependent upon sequential sensing of prey recognition and prey quality cues. Proc. Natl. Acad. Sci. USA. 2015;112:E6028–E6037. doi: 10.1073/pnas.1515749112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerner TR, et al. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Path. 2012;8:e1002524. doi: 10.1371/journal.ppat.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomashow MF, Rittenberg SC. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J. Bacteriol. 1978;135:998–1007. doi: 10.1128/jb.135.3.998-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert C, et al. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 2006;60:274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tudor JJ, Davis JJ, Panichella M, Zwolak A. Isolation of predation-deficient mutants of Bdellovibrio bacteriovorus by using transposon mutagenesis. Appl. Environ. Microbiol. 2008;74:5436–5443. doi: 10.1128/AEM.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina AA, Shanks RM, Kadouri DE. Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol. 2008;8:33. doi: 10.1186/1471-2180-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J. Bacteriol. 2007;189:4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotter TW, Thomashow MF. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J. Bacteriol. 1992;174:6018–6024. doi: 10.1128/jb.174.19.6018-6024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J. Bacteriol. 2011;193:1745–1756. doi: 10.1128/JB.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahmoud KK, Koval SF. Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology. 2010;156:1040–1051. doi: 10.1099/mic.0.036137-0. [DOI] [PubMed] [Google Scholar]
- 18.Capeness MJ, et al. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links Type IVa pilus extrusion/retraction status to prey-independent growth signalling. PloS one. 2013;8:e79759. doi: 10.1371/journal.pone.0079759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry JL, Pelicic V. Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 2015;39:134–154. doi: 10.1093/femsre/fuu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milner DS, et al. Ras GTPase-like protein MglA, a controller of bacterial social-motility in Myxobacteria, has evolved to control bacterial predation by Bdellovibrio. PLoS Gen. 2014;10:e1004253. doi: 10.1371/journal.pgen.1004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rendulic S, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 22.Varon M, Shilo M. Interaction of Bdellovibrio bacteriovorus and host bacteria. I. Kinetic studies of attachment and invasion of Escherichia coli B by Bdellovibrio bacteriovorus. J. Bacteriol. 1968;95:744–753. doi: 10.1128/jb.95.3.744-753.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PloS one. 2013;8:e61850. doi: 10.1371/journal.pone.0061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasternak Z, et al. In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J. 2013;8:625–635. doi: 10.1038/ismej.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sangwan N, et al. Arsenic rich Himalayan hot spring metagenomics reveal genetically novel predator-prey genotypes. Environ. Microbiol. Rep. 2015;7:812–823. doi: 10.1111/1758-2229.12297. [DOI] [PubMed] [Google Scholar]
- 26.Crossman LC, et al. A small predatory core genome in the divergent marine Bacteriovorax marinus SJ and the terrestrial Bdellovibrio bacteriovorus. ISME J. 2013;7:148–160. doi: 10.1038/ismej.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denielou YP, Sagot MF, Boyer F, Viari A. Bacterial syntenies: an exact approach with gene quorum. BMC Bioinformatics. 2011;12:193. doi: 10.1186/1471-2105-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beall B, Moran CP., Jr. Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J. Bacteriol. 1994;176:2003–2012. doi: 10.1128/jb.176.7.2003-2012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tengra FK, et al. CbgA, a protein involved in cortex formation and stress resistance in Myxococcus xanthus spores. J. Bacteriol. 2006;188:8299–8302. doi: 10.1128/JB.00578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerth U, et al. Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 2008;190:321–331. doi: 10.1128/JB.01233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalik S, et al. Life and death of proteins: a case study of glucose-starved Staphylococcus aureus. Mol. Cel. Proteomics. 2012;11:558–570. doi: 10.1074/mcp.M112.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwudke D, et al. Transcriptional activity of the host-interaction locus and a putative pilin gene of Bdellovibrio bacteriovorus in the predatory life cycle. Curr. Microbiol. 2005;51:310–316. doi: 10.1007/s00284-005-0030-1. [DOI] [PubMed] [Google Scholar]
- 34.Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nature Rev. Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 35.Roux N, Spagnolo J, de Bentzmann S. Neglected but amazingly diverse type IVb pili. Res. Microbiol. 2012;163:659–673. doi: 10.1016/j.resmic.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Lambert C, et al. A predatory patchwork: membrane and surface structures of Bdellovibrio bacteriovorus. Adv. Microb. Physiol. 2009;54:313–361. doi: 10.1016/S0065-2911(08)00005-2. [DOI] [PubMed] [Google Scholar]
- 37.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 2001;9:429–437. doi: 10.1016/S0966-842X(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 38.de Bentzmann S, Aurouze M, Ball G, Filloux A. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 2006;188:4851–4860. doi: 10.1128/JB.00345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Ann. Rev. Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 40.Conrad JC, et al. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys J. 2011;100:1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunger G, Guzzo CR, Andrade MO, Jones JB, Farah CS. Xanthomonas citri subsp. citri type IV pilus is required for twitching motility, biofilm development, and adherence. Mol. Plant Microb. Interact. 2014;27:1132–1147. doi: 10.1094/MPMI-06-14-0184-R. [DOI] [PubMed] [Google Scholar]
- 42.Gibiansky ML, et al. Bacteria use type IV pili to walk upright and detach from surfaces. Science. 2010;330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- 43.Keilberg D, Sogaard-Andersen L. Regulation of bacterial cell polarity by small GTPases. Biochemistry. 2014;53:1899–1907. doi: 10.1021/bi500141f. [DOI] [PubMed] [Google Scholar]
- 44.Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Haitjema CH, Fuqua C. The Ctp Type IVb Pilus Locus of Agrobacterium tumefaciens Directs Formation of the Common Pili and Contributes to Reversible Surface Attachment. J. Bacteriol. 2014;196:2979–2988. doi: 10.1128/JB.01670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson JK, Smith TG, Hoover TR. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 2010;18:30–37. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasternak Z, et al. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 2013;7:756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernard CS, Bordi C, Termine E, Filloux A, de Bentzmann S. Organization and PprB-dependent control of the Pseudomonas aeruginosa tad Locus, involved in Flp pilus biology. J. Bacteriol. 2009;191:1961–1973. doi: 10.1128/JB.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schilling J, et al. Transcriptional activation of the tad type IVb pilus operon by PypB in Yersinia enterocolitica. J. Bacteriol. 2010;192:3809–3821. doi: 10.1128/JB.01672-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nika JR, et al. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect. Immun. 2002;70:2965–2975. doi: 10.1128/IAI.70.6.2965-2975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreiner HC, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janowicz DM, et al. Expression of the Flp proteins by Haemophilus ducreyi is necessary for virulence in human volunteers. BMC Microbiol. 2011;11:208. doi: 10.1186/1471-2180-11-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nykyri J, et al. Role and regulation of the Flp/Tad pilus in the virulence of Pectobacterium atrosepticum SCRI1043 and Pectobacterium wasabiae SCC3193. PloS one. 2013;8:e73718. doi: 10.1371/journal.pone.0073718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fitzgerald DM, Bonocora RP, Wade JT. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 2014;10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poly F, et al. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Claret L, et al. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 2007;82:33275–33283. doi: 10.1074/jbc.M702800200. [DOI] [PubMed] [Google Scholar]
- 57.Tsou AM, Frey EM, Hsiao A, Liu Z, Zhu J. Coordinated regulation of virulence by quorum sensing and motility pathways during the initial stages of Vibrio cholerae infection. Commun. Integr. Biol. 2008;1:42–44. doi: 10.4161/cib.1.1.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 2008;74:7152–7162. doi: 10.1128/AEM.01736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finn RD, et al. Pfam: the protein families database. Nucleic acids Res. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalho RN, Lettieri T. Proteomic analysis of the marine diatom Thalassiosira pseudonana upon exposure to benzo(a)pyrene. BMC Genomics. 2011;12:159. doi: 10.1186/1471-2164-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Becker R, Schwarz G, Beck S, Linscheid MW. Software assisted data analysis for relative quantification of differentially metal labeled proteins based on HPLC/ESI-MS and -MS/MS experiments. J. Mass. Spectrom. 2015;50:1120–1123. doi: 10.1002/jms.3627. [DOI] [PubMed] [Google Scholar]
- 64.Peleg Y, Unger T. Application of the Restriction-free (RF) cloning for multicomponents assembly. Methods Mol. Biol. 2014;1116:73–87. doi: 10.1007/978-1-62703-764-8_6. [DOI] [PubMed] [Google Scholar]
- 65.Steyert SR, Pineiro SA. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 2007;73:4717–4724. doi: 10.1128/AEM.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fried L, Lassak J, Jung K. A comprehensive toolbox for the rapid construction of lacZ fusion reporters. J. Microbiol. Methods. 2012;91:537–543. doi: 10.1016/j.mimet.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 67.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 69.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frith MC, Saunders NF, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sadreyev R, Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/S0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


