Abstract
Previously developed Asn-Gly-Arg (NGR) peptide-modified multifunctional poly(ethyleneimine)–poly(ethylene glycol) (PEI–PEG)-based nanoparticles (TPIC) have been considered to be promising carriers for the co-delivery of DNA and doxorubicin (DOX). As a continued effort, the aim of the present study was to further evaluate the interaction between TPIC and human umbilical vein endothelial cells (HUVEC) to better understand the cellular entry mechanism. In the present investigation, experiments relevant to co-localization, endocytosis inhibitors and factors influencing the internalization were performed. Without any treatment, there was no co-localization between aminopeptidase N/CD13 (APN/CD13) and caveolin 1 (CAV1). However, co-localization between CD13 and CAV1 was observed when cells were incubated with an anti-CD13 antibody or TPIC. As compared with antibody treatment, TPIC accelerated the speed and enhanced the degree of co-localization. TPIC entered HUVEC not only together with CD13 but also together with CAV1. However, this internalization was not dependent on the enzyme activity of CD13 but could be inhibited by methyl-β-eyclodextfin (MβCD), further identifying the involvement of caveolae-mediated endocytosis (CvME). This conclusion was also verified by endocytosis inhibitor experiments.
KEY WORDS: Asn-Gly-Arg peptide, Aminopeptidase N/CD13, Caveolin 1, Caveolae-mediated endo-cytosis, Endothelial cells, Cellular entry, Co-localization, Doxorubicin
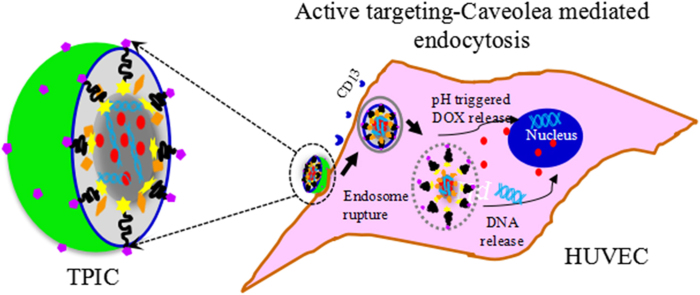
Graphical abstract
Asn-Gly-Arg (NGR)-functioned multifunctional poly(ethyeneimine)–poly(ethyleneglycol) (PEI–PEG)-based nanoparticles (TPIC) could influenced the distribution of CD13 on human umbilical vein endothelial cells through the interaction between CD13 and NGR, causing CD13 clustering, leading to the co-localization of CD13 and caveolin 1, and finally induce the internalization via the caveolea-mediated endocytosis.
1. Introduction
In recent years, gene therapy and drug targeting studies have revealed the importance of identifying intracellular mechanisms of efficient delivery1. Understanding the potential uptake mechanisms involved in the cellular entry of test nanoparticles could be helpful to provide feedback for the rational design of improved vectors2, 3. Accordingly, scientists have been aware of the characteristics of typical trafficking pathways for many targeted therapeutics. Endocytosis pathways other than classical clathrin-mediated endocytosis (CME) have been recently characterized in some details. Such pathways may offer alternative uptake and trafficking pathways for gene delivery vectors4. Caveolae-mediated endocytosis (CvME) has been generally considered to be a non-acidic and non-digestive uptake route, which indicates that it does not sense a drop in pH but travels through pH-neutral caveosomes directly to the Golgi and/or endoplasmic reticulum (ER), from which nuclear entry can take place, thereby avoiding lysosomal degradation5, 6, 7, 8.
CvME is characterized by the evolution of caveolae, which are small, flask-shaped non-clathrin coated invaginations of the hydrophobic membrane subdomains enriched in cholesterol, glycosphingolipids and caveolin protein9. The caveolin protein family has three members: caveolin 1 (CAV1), caveolin 2 (CAV2) and caveolin 3 (CAV 3). Among them, CAV1 is the major structural protein in caveolae possessing the ability to interact with numerous proteins10, 11, 12. Caveolae in vascular endothelial cells were first identified by Paladern13 in 1968. Caveolae exist alone or in a cluster on many types of mammalian cells, particularly on epithelial cells, endothelial cells, fibroblasts, adipocytes and smooth muscle cells14. Caveolae can transport bioactive molecules into cells and participate in the reception and transduction of multiple signals11. In recent years, the cell physiological function of caveolae has drawn increasing attention, especially in signal transduction, cholesterol transport, cell internalization, tumor suppression and muscle cell synthesis15. Additionally, increasing numbers of studies have shown caveolae to be closely related to many diseases, including cancer, arteriosclerosis, muscular dystrophy, early Alzheimer׳s and diabetes16. Because of these characteristics, CvME has attracted tremendous attention in the field of gene delivery research. Among of them, attaching specific ligands to the polymer-based carriers to target CvME has been become a promising approach in gene therapy5, 17, 18.
Aminopeptidase N/CD13 (APN/CD13) is a type II transmembrane protein present in a wide variety of human organs, tissues and cell types (endothelial, epithelial, fibroblast and leukocyte). CD13 has multiple functions related to tumorigenesis, the immune system, and pain19. These functions can facilitate the modulation of bioactive peptide responses, such as pain management and vasopressin release. They can also influence body immune functions and major biological events, such as cell proliferation, secretion, invasion and angiogenesis, thereby providing treatment options for various diseases20. CD13 can be specifically recognized and bound by the specific sequence of Asn-Gly-Arg (NGR) peptide and exhibits high affinity and specificity toward this moiety21. Although CD13 is a ubiquitous enzyme, studies on its expression pattern in normal and neoplastic human tissues suggest that different CD13 forms are expressed in myeloid cells, epithelia and tumor-associated blood vessels22. The CD13 isoform which functions as a vascular receptor for the NGR motif was reported to be selectively overexpressed in tumor vasculature and in some tumor cells21, 23, 24. In fact, many CD13-targeted therapy based on NGR, such as NGR–drug conjugates25, 26, NGR-coated liposomes (http://www.ambrilia.com), NGR-coated PEG-b-PLA polymeric micelles27, NGR-modified PEGylated LPD (liposome-polycation-DNA) nanoparticles28, CNGRC/PEG/PEI/DNA vector for gene therapy29. and so on, are under clinical and late pre-clinical development. Above all, strategy targeting CD13 by NGR for tumor therapy was widely accepted. Characterization of such newly developed formulations is important in the development of vascular-targeted therapies based on the NGR/CD13 system.
Additionally, previous studies have reported that CD13 can participate in receptor-mediated phagocytosis30. Currently, an increasing amount of evidence indicates that the temporal and specific localization of these signaling molecules play critical roles in rapid and efficient CD13-mediated signal transduction events31. Recently, various compounds or particles have been coupled synthetically to NGR peptide targeting CD13 in an attempt to increase its tumor targeting properties32. In our previous studies, NGR peptide was used to modify poly(lactic acid)–poly (ethylene glycol) nanoparticles (PLA–PEG NPs) for targeted gene delivery. It was shown that NGR-modified PLA–PEG NPs (NGR-PLA–PEG NPs) could specifically enhance the in vitro gene transfection efficiency. More importantly, we found that the unique mechanism of CvME was mainly involved in the internalization of NGR-PLA–PEG NPs into HUVEC33. Accordingly, we hypothesized that NGR-modified nanocarriers might induce particular signal transduction events via the CD13 receptor and transport them into CD13 positive cells through CvME. However, detailed work to establish their exact cellular uptake mechanisms is currently lacking. Therefore, it is necessary to gain insight on the cellular entry mechanisms in gene transfection.
Recently, a NGR-modified multifunctional poly(ethyleneimine)–poly(ethylene glycol) (PEI–PEG)-based nanoparticle (TPIC) has been developed in our group for drug and gene combination therapy, which could enhance the gene transfection efficiency and antitumor activity in vitro34. As a continued effort, the focus of our proceeding studies was to study the exact mechanism involved and clarify whether NGR can interact with CD13 and mediate the subsequent CvME. In the present study, both CD13- and CAV1-positive HUVEC were selected as test cells, and the previously formulated TPIC was the test nanocarrier. Using flow cytometry and confocal laser scanning technology, we presently investigated the co-localization of CD13 and CAV1, the effect of TPIC on this co-localization, the interaction between TPIC and CD13 or TPIC and CAV1, the kinetics of the resulting endocytosis, the effects of endocytosis inhibitors, and factors influencing the internalization of TPIC.
2. Materials and methods
2.1. Materials and cells
The plasmid pCMV-EGFP (pEGFP-N1), which carries the gene of enhanced green fluorescent protein (EGFP) under a cytomegalovirus (CMV) promoter, was propagated in Escherichia coli and purified by an Endo Free Plasmid Maxi Kit (Qiagen, Hilden, Germany). The purity and concentration of pDNA was then measured by a NanoDrop UV-Vis Spectrophotometers (ND-2000C, Thermo, USA). A phycoerythrin (PE)-conjugated anti-human CD13 monoclonal antibody (clone WM15) was purchased from BD Biosciences (USA). A DyLight 488-labelled anti-human caveolin 1 monoclonal antibody (7C8) (NB100-615G) was purchased from Novus Biologicals (USA). Hoechst33342 was purchased from Invitrogen by Life Technologies (USA). Methyl-β-eyclodextfin (MβCD), cholesterol, TritonX-100 and bovine serum albumin (BSA) was purchased from Sigma-Aldrich (USA). All other reagents were of special commercial grade and used without further purification. Deionized water was used throughout the experiment. All other reagents of biochemical and molecular biology grades were obtained from Sigma--Aldrich (China). Doxorubicin (DOX) was purchased from Dalian Meilun Biology Technology Co., Ltd. (Dalian, China).
Human umbilical vein endothelial cells (HUVEC) were purchased from ScienCell Research Laboratories (San Diego, USA) and cultured in the supplied endothelial cell medium (ECM) with 1% endothelial cell growth supplement (ECGS), 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin solution (P/S). All cells were cultured in a 37 °C incubator with 5% CO2.
2.2. The expression of CD13 and CAV1 on HUVEC
HUVEC were cultured in ECM with 1% ECGS, 5% FBS and 1% P/S solution in a 37 °C incubator with 5% CO2. The cells were detached by 0.025% trypsin/EDTA, and the cell density was adjusted to 1×106/100 μL PBS to grow to logarithmic phase. Then, cells were incubated with 2 μL of a PE-conjugated monoclonal antibody specific for CD13 to detect CD13 expression or DyLight 488-labelled anti-human CAV1 monoclonal antibody (1:200 dilution) to detect CAV1 expression for 1 h at 4 °C. After being washed in cold PBS for three times, the cells were resuspended in PBS and analyzed on FACS (BD Biosciences, USA) equipped with a 488-nm argon laser for excitation. For each sample, 10,000 events were collected, and fluorescence was detected. Signals were amplified in the logarithmic mode for fluorescence to determine the positive events by a standard gating technique. The percentage of positive events was calculated as the events within the gate divided by the total number of events, excluding cell debris.
To examine the in suit localization of CD13 on HUVEC, 1×105 of HUVEC were seeded in 35-mm glass bottom dishes in complete medium. The culture media was removed after the cells attached to the glass, and the cells were labelled by a PE-conjugated anti-CD13 antibody (1:100 dilution) for 3 h at 4 °C. Then, cells were washed twice with cold PBS before they were fixed in 4% paraformaldehyde in 0.1 mol/L PBS for 15 min. Finally, the fixed cells were examined under a Zeiss LSM700 laser confocal scanning microscope.
2.3. Co-localization experiment of CD13 and CAV1
To observe the localization of CD13, we treated HUVEC cultured in 35-mm glass bottom dishes (Corning Incorporated, USA) with PE-conjugated anti-CD13 antibody (1:100 dilution) for 3 h at 4 °C, rinsed the cells, and incubated them for 0–3 h at 37 °C. After being fixed with 4% paraformaldehyde in 0.1 mol/L PBS for 15 min, the cells were permeabilized with 0.1% Triton X-100 for 5 min, treated with 3% bovine serum albumin for 20 min, and labelled for CAV1 through incubating HUVEC with DyLight 488-labelled anti-human CAV1 monoclonal antibody (1:200 dilution) at 4 °C overnight. After being washed for three times in PBS, hoechst33342 staining was performed to stain the nucleus as a control. After extensive washing, the glass bottom dishes were examined under a Zeiss LSM700 laser confocal scanning microscope.
2.4. The effect of TPIC on the co-localization between CD13 and CAV1
To examine whether TPIC could change the distribution of CD13 in a manner similar to that of the anti-CD13 antibody and to examine the behavior of CD13 when cells were treated with TPIC, TPIC without DOX was prepared and used to avoid the mutual interference of red fluorescence of DOX and PE. First, HUVEC was incubated with non-DOX-loaded TPIC for 0–4 h at 37 °C with 5% CO2 in an incubator. Then, test nanocarriers were aspirated, and cells were washed for three times with cold PBS. After being fixed with 4% paraformaldehyde in 0.1 mol/L PBS for 15 min, the cells were treated with a PE-conjugated anti-CD13 antibody (1:100 dilution) for 1 h at 4 °C, permeabilized with 0.1% Triton X-100 for 5 min, treated with 3% bovine serum albumin for 20 min, and labelled for CAV1 as described above. After extensive washing, the glass bottom dishes were examined under a Zeiss LSM700 laser confocal scanning microscope.
2.5. The interaction between TPIC and CD13
To avoid the interference of red fluorescence of DOX with the PE-labelled CD13 antibody and to facilitate the observation of the interaction between TPIC and CD13, a green fluorescent labelled and non-DOX-loaded nanocarrier was needed. In this study, 6-carboxyfluorescein (FAM)-NGR was used to prepare green fluorescent-labelled TPIC as described in a previous study34.
To further examine the behavior of CD13 after cells were treated with FAM-TPIC for 4 h at 4 or 37 °C. Then, test nanoparticles were aspirated and cells were washed twice with cold PBS before adding a PE-conjugated anti-CD13 antibody (1:100 dilution). After incubation at 4 °C for 3 h, the cells were fixed and stained for nuclear examination. Then, the glass bottom dishes were examined under a Zeiss LSM700 laser confocal scanning microscope.
2.6. The interaction between TPIC and CAV1
To further study the interaction between TPIC and CAV1, cells were treated with fresh serum free media containing DOX-loaded TPIC for 4 h at 4 or 37 °C. Then, cells were fixed and incubated with a DyLight 488-labelled anti-human CAV1 monoclonal antibody (1:200 dilution) at 4 °C overnight. After extensive washing, the cells were stained by hoechst33342, and the glass bottom dishes were examined under a confocal scanning microscope.
2.7. The effect of CD13 activity on the internalization of TPIC
To determine whether the CD13 activity affected the internalization of TPIC, HUVEC was pre-treated with 100 μg/mL bestatin35, a CD13/APN inhibitor, for 1 h at room temperature in PBS. Then, cells were washed for three times with PBS. TPIC were added into wells and incubated with cells for another 4 h at 37 °C with 5% CO2 in an incubator. Finally, all cells were harvested and washed, and the cell-associated fluorescence was determined by FACS.
2.8. Inhibition of TPIC intracellular entry by cholesterol depletion
To examine the influence of cholesterol depletion, we treated cells with 2 mmol/L MβCD in ECM for 30 min at 37 °C. After being washed three times with PBS, TPIC were added into wells and incubated with cells for another 4 h at 37 °C with 5% CO2 in an incubator. Then, all cells were harvested and washed, and cell-associated fluorescence was determined by FACS. To exclude the possibility of irreversible damage caused by the above treatment, we further incubated cells with an MβCD-cholesterol complex for 30 min at 37 °C to replenish cholesterol. MβCD-cholesterol was prepared as described previously36. Briefly, cholesterol in methanol-chloroform (1:1) was dried and suspended in MβCD in 0.1 mol/L PBS buffer. The solution was sonicated and rotated at 37 °C overnight. The mixture was then filtered and diluted with ECM. The final concentrations were approximately 1 mmol/L MβCD and 100 μg/mL of cholesterol.
2.9. The effect of cholesterol depletion on the interaction between the TPIC and CAV1
To further study the effect of cholesterol depletion on the interaction between the TPIC and CAV1, cells were pre-treated with MβCD and incubated with TPIC as previously mentioned. After being fixed, cells were incubated with a DyLight 488-labelled anti-human CAV1 monoclonal antibody (1:200 dilution) at 4 °C overnight. After extensive washing, the cells were stained by Hoechst33342, and the glass bottom dishes were examined under a confocal scanning microscope.
2.10. Endocytosis kinetic experiments
To evaluate the endocytosis kinetics of the TPIC in HUVEC, a transfection dose of TPIC was incubated with HUVEC at 4 °C for 0, 30 min, 1, 3 and 5 h, and at 37 °C for 0, 5, 15, 30 min, 1, 3 and 5 h. After incubation, the culture media were removed and the cells were washed for three times with cold PBS. Then, the cells were labelled with a PE-conjugated anti-CD13 antibody (1:100 dilution) for 3 h at 4 °C. Finally, cells were washed twice with cold PBS. The TPIC uptake and surface accessible CD13 after HUVEC incubation with TPIC at 4 or 37 °C for various time periods were determined by FACS.
2.11. Exploring uptake pathways using endocytic inhibitors
To study the effect of endocytic inhibitors on the uptake of TPIC, HUVEC were pre-incubated individually with the following inhibitors at concentrations that were not toxic to the cells33: (1) 10 μg/mL of chlorpromazine for 30 min, (2) 1 μg/mL of genistein for 30 min, (3) 30 mmol/L of cytochalasin D for 30 min. Following the pre-incubation, the cells were further treated with freshly prepared TPIC for 4 h. Subsequently, the cells were washed for three times with PBS, collected according to the methods described above and analyzed by FACS and fluorescence microscopy. In the study, the group without any treatment was used as the background in the FACS analysis, whereas the groups in the presence of TPIC but without inhibitor treatment were used as 100% controls.
2.12. Statistical analysis
All studies were repeated a minimum of three times and measured at least in triplicate. The results are reported as the mean ±standard deviation (SD). Statistical significance was analyzed using Student׳s t-test. Differences between experimental groups were considered significant if the P value was less than 0.05 (P <0.05).
3. Results
3.1. The expression and localization of CD13 on HUVEC
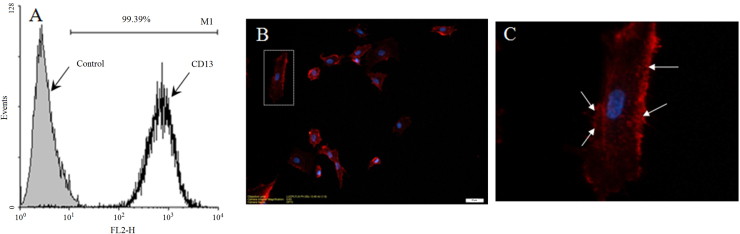
The quantitative results by FACS indicated that the positive percentage of CD13 expression on HUVEC was 99.39%, implying nearly all HUVEC were CD13 positive (Fig. 1A). Localization of CD13 on HUVEC was examined by labelling cells in situ, followed by analysis on a confocal microscope. CD13 was concentrated on sites where the cell membrane began to project into filaments (Fig. 1C), suggesting the role of CD13 in the adhesion and motility of cells. The data are in line with the reported role of CD13 expressed on some tumor cells in cell adhesion37, 38.
Figure 1.
Membrane expression and localization of CD13 on HUVEC. (A) FACS data are presented as histograms. (B) Confocal microscopy of HUVEC labelled in situ using PE anti-CD13 antibody. (C) An enlarged view of (B). (scale bar: 20 μm).
3.2. Both CD13 and CAV1 expressed on HUVEC
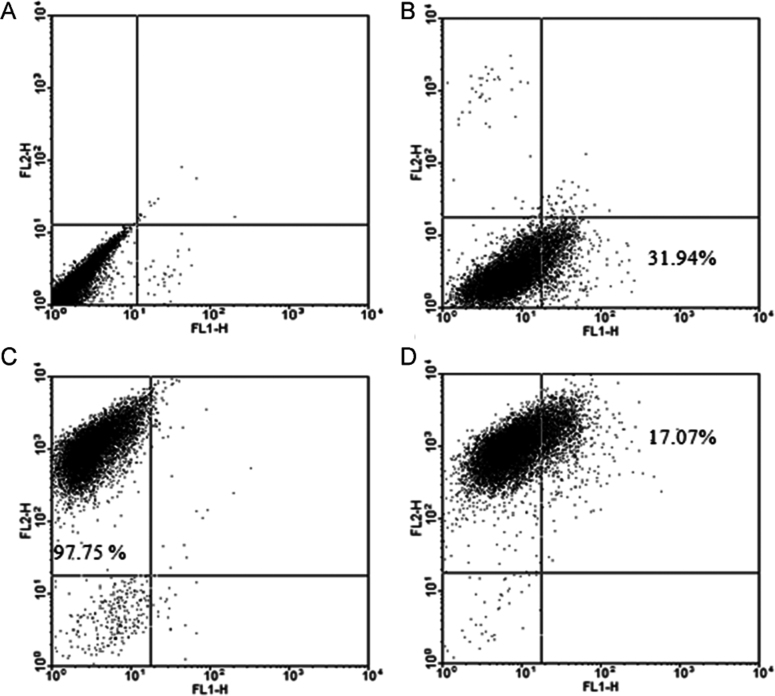
Using an anti-CD13 antibody and anti-CAV1 antibody to label CD13 and CAV1 on HUVEC, respectively, the results of CD13 and CAV1 expression on HUVEC are shown in Fig. 2. A flow cytometric scatter plot was used for the quantitative analysis. Fig. 2B and C present the individual control of PE anti-human CD13 labelled and DyLight 488 anti-human CAV1 antibody labelled cells, indicating the availability of both antibodies. Fig. 2D shows that the cells are both CD13 and CAV1 positive. Those cells expressing CD13 could be labelled by anti-CD13 antibody and exhibited in red fluorescence (FL2-H, PE, upper left corner) and those CAV1 positive could be labelled by anti-CAV1 antibody and exhibited in green fluorescence (FL1-H, FAM, lower right corner). And those cells expressing both CD13 and CAV1 were showed in upper right corner. The quantitative data demonstrated that the positive percentage of CD13 and CAV1 was 97.75% and 31.94%, respectively, and the percentage of HUVEC expressing both CD13 and CAV1 was 17.07%.
Figure 2.
Flow cytometry of CD13 and CAV1 expression on HUVEC. (A) HUVEC cells without any treatment (control); (B) HUVEC labelled by PE anti-human CD13 antibody; (C) HUVEC labelled by DyLight 488 anti-human CAV1 antibody; (D) HUVEC double labelled both by DyLight 488 anti-human CAV1 antibody and PE anti-human CD13.
3.3. Co-localization experiment of CD13 and CAV1
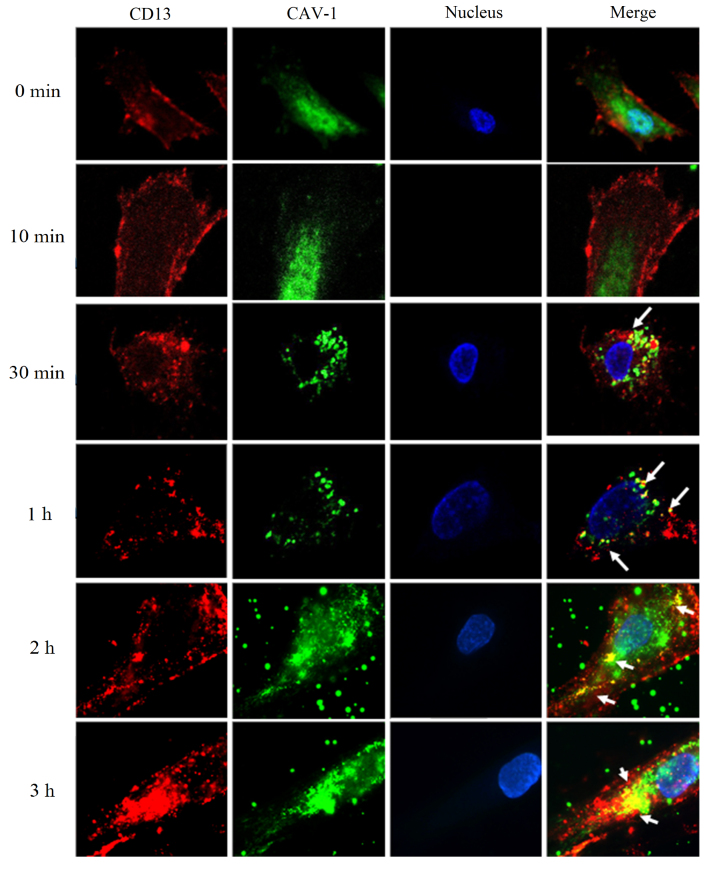
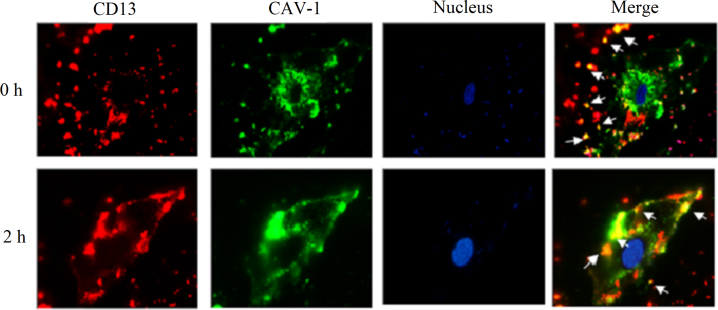
As shown in Fig. 3, when HUVEC were incubated with an anti-CD13 antibody and fixed without warming (0 min), red fluorescence was observed evenly on the cell surfaces. In the same cells, labelling for CAV1 was observed in green. No co-localization occurred between CD13 and CAV1. After cells bound with the antibodies were incubated for 10 min at 37 °C, the labelling of CD13 and CAV1 on the HUVEC surface showed a uniform punctate distribution, and no co-localization between CD13 and CAV1 was observed. When the incubation time was 30 min at 37 °C, the labelling of CD13 and CAV1 showed in spots, and a small extent of co-localization between CD13 and CAV1 was observed (white arrows). With an extended incubation time at 37 °C, co-localization of CD13 with CAV1 became more frequent after 60 min (white arrows). When the incubation time extended to 2 and 3 h, CD13 and CAV1 gathered into clusters and co-localized extensively (white arrows). These results demonstrated that the antibody against CD13 bound to the cell surface evenly when incubated on ice, but the bound antibodies became sequestered to CAV1-positive patches when the temperature increased to 37 °C. Additionally, the results indicate that binding of anti-CD13 antibody and incubation at 37 °C could cause clustering of CD13 and its co-localization with CAV1. These results are exciting and imply that targeting CD13 could initiate a subsequent interaction with CAV1, although there was no co-localization between CD13 and CAV1 in the absence of antibody against CD13. The accumulation of cross-linked molecules in CAV1-positive areas was similar to the previously reported behavior of glycosylphosphatidylinositol (GPI)-anchored proteins and glycosphingolipids22, 39, 40.
Figure 3.
Immunofluorescence microscopy (1000×). HUVEC were treated with a PE-labelled mouse anti-human CD13 antibody at 4 °C firstly and incubated at 37 °C for 0, 10, 30, 60 min, 2 or 3 h. White arrows indicated the co-localization of CD13 and CAV1.
3.4. TPIC can accelerate the speed and enhance the extent of co-localization between CD13 and CAV1
In this section, we tried to examine whether TPIC could change the distribution of CD13 in a manner similar to anti-CD13 antibody and to examine the behavior of CD13 after cells treated by TPIC. HUVEC were incubated with TPIC using the same procedure with anti-CD13 antibody for 0–4 h at 37 °C. As shown in Fig. 4, we found the distribution of CD13 on the HUVEC surface was no longer uniform punctuate but was randomly distributed dots at 0 h, which was different with that after treating with antibody. Moreover, when HUVEC were incubated with TPIC and fixed without warming (0 h), the co-localization of CD13 and CAV1 in small clusters had been already observed (Fig. 4, the top row, white arrows), which was 30 min earlier than that treating with antibody. With the extension of the incubation time at 37 °C, the distribution of CD13 on the HUVEC surface gathered into clusters and extensive co-localization of CD13 and CAV1 occurred at 2 h. However, compared with the co-localization result after anti-CD13 antibody treatment at 2 h, the co-localization result at 2 h after TPIC treatment was much more extensive. These results indicate that TPIC could change the distribution of CD13 in a different manner to antibody, because TPIC could accelerate the speed and enhance the extent of co-localization compared with the anti-CD13 antibody. Additionally, the co-localization also appeared in the cytoplasm, suggesting that after TPIC treatment, CD13 could enter into cell from membrane and co-localize with CAV1 in the cytoplasm for a period of time.
Figure 4.
Immunofluorescence microscopy (1000×). HUVEC were treated with TPIC at 4 °C firstly and incubated at 37 °C for 0 min and 2 h. White arrows indicated the co-localization of CD13 and CAV1.
3.5. TPIC can bind to CD13 and enter into HUVEC together with CD13
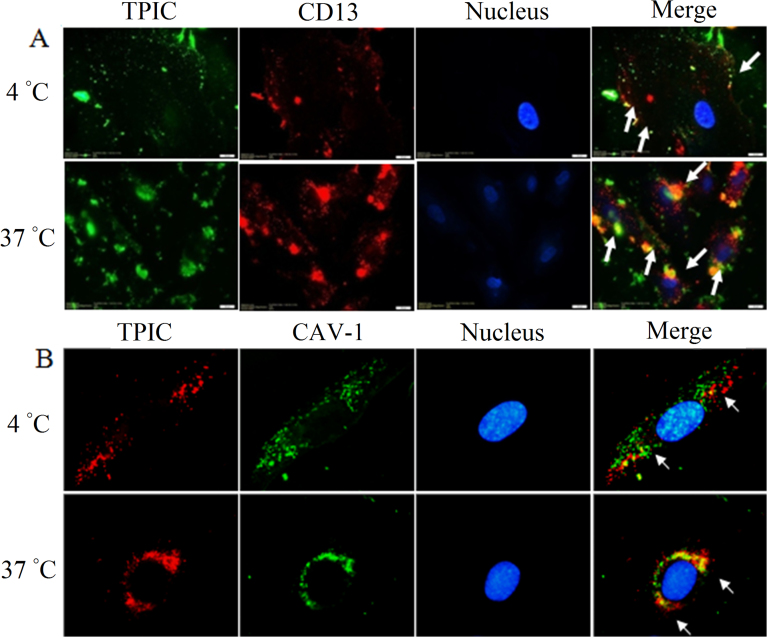
In this section, we tried to examine the interaction between TPIC and CD13. In our previous study, it has been already indicated that NGR-mediated nanocarriers entry into HUVEC was an energy dependent active process33. In the present study, the uptake was significantly inhibited at 4 °C and TPIC mainly stayed at cell surface but not entered cells. At 4 °C, although TPIC was scattered on the cell membrane as a few green spots, and co-localized with red CD13 to yellow (Fig. 5A), indicating that TPIC could bind to CD13 on the cell surface. While, at 37 °C, more green fluorescence was observed inside the cell indicating that TPIC was taken up by cells extensively. Under these conditions, co-localization of red CD13 was observed with green TPIC (yellow and orange dots) accumulated in the perinuclear area, indicating that TPIC and CD13 can interact with each other after entering cells (Fig. 5A). The results indicates that TPIC can first bind to CD13 on the cell surface, and then CD13 enters into HUVEC with the internalization of TPIC, suggesting CD13 might participate in the cellular uptake of TPIC.
Figure 5.
(A) Immunofluorescence microscopy of co-localization of TPIC and CD13 at 4 and 37 °C, respectively (scale bar: 20 μm). The arrows indicate the co-localization of the PE anti-CD13 antibody and TPIC. (B) Immunofluorescence microscopy (1000×) of co-localization of TPIC and CAV1 at 4 and 37 °C, respectively. The arrows indicate the co-localization of the DyLight 488 anti-human CAV1 antibody and TPIC.
3.6. TPIC can co-localize with CAV1 and enter HUVEC together with CAV1
The results are shown in Fig. 5B. At 4 °C, red TPIC and green CAV1 overlapped into yellow stains, which demonstrated that TPIC could co-localize with CAV1. At 37 °C, the extent of co-localization increased and concentrated around the perinuclear area, suggesting that TPIC entered HUVEC together with CAV1 and that CAV1 participated in the internalization of TPIC. The co-localization behavior of TPIC and CAV1 was similar to that produced by the combination of TPIC and CD13. The difference was TPIC could not cause clustering of CAV1.
3.7. TPIC entry into HUVEC is not dependent on the enzyme activity of CD13
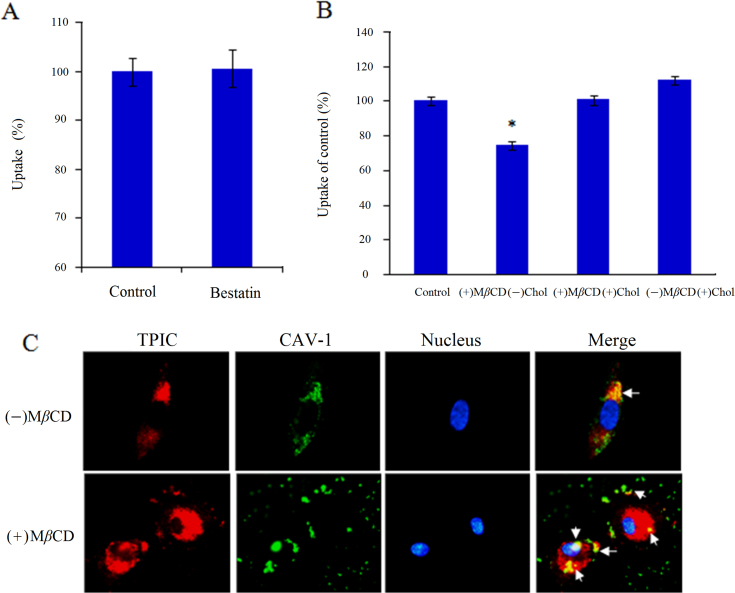
Bestatin is a potent CD13 inhibitor that can inhibit the activity of APN/CD13 and exert anti-tumor activity40. The concentration of 100 μg/mL of bestatin was chosen since it was shown to effectively inhibit enzyme activity of CD1335 but non-toxic to cells. The results of the inhibition study of CD13 enzyme activity are shown in Fig. 6A. Without bestatin treatment, TPIC could be efficiently internalized by HUVEC, and the positive percentage was over 99%. After bestatin treatment, TPIC was also efficiently internalized by HUVEC but not be significantly suppressed (P>0.05), suggesting the internalization of TPIC by HUVEC was not dependent on the enzyme activity of CD13.
Figure 6.
Effect of (A) bestatin and (B) MβCD on the internalization of TPIC in HUVEC. (C) Immunofluorescence microscopy (1000×) of the co-localization of TPIC and CAV1 with or without MβCD.
3.8. The cellular entry of TPIC was inhibited by cholesterol depletion
Caveolae are invaginated membrane structures and are enriched in cholesterol, sphingomyelin, and caveolin protein10. The integrity of these sphingolipid- and cholesterol-rich microdomains depends on cholesterol. To examine whether the behavior of TPIC changed if plasmalemmal cholesterol was depleted, MβCD was used to deplete cholesterol from the cell membrane, thereby selectively destroying the formation of caveolae. According to FACS analysis (Fig. 6B), the amount of internalized TPIC was significantly inhibited by cholesterol depletion (P<0.05). The replenishment of cholesterol by MβCD-cholesterol did not affect the amount of TPIC internalized because the exogenous cholesterol significantly reversed MβCD-induced effects41. Furthermore, addition of exogenous cholesterol alone did not change the amount of internalized TPIC. The amount of TPIC internalized decreased significantly in cholesterol-depleted cells but was recovered to the control level in cholesterol-depleted and -replenished cells, demonstrating that cholesterol depletion can inhibit the intracellular entry of TPIC.
3.9. The co-localization of TPIC and CAV1 can be affected by cholesterol depletion
After MβCD treatment, the co-localization of TPIC and CAV1 was also evaluated to further determine the important role of caveolae in TPIC internalization. As shown in Fig. 6C, the co-localization of TPIC and CAV1 decreased significantly by pretreatment with MβCD. Without MβCD, CAV1 and TPIC co-localized at the perinuclear area to a large extent. However, after MβCD treatment, cell cholesterol was depleted and the integrity of the caveolae was destroyed. Thereby, the distribution of CAV1 became discontinuous on the cell surface and gathered into a cluster. Consequently, the extent of the co-localization of TPIC and CAV1 was greatly reduced, and only a small number of overlapping spots appeared, suggesting the important effect of caveolae on the internalization of TPIC.
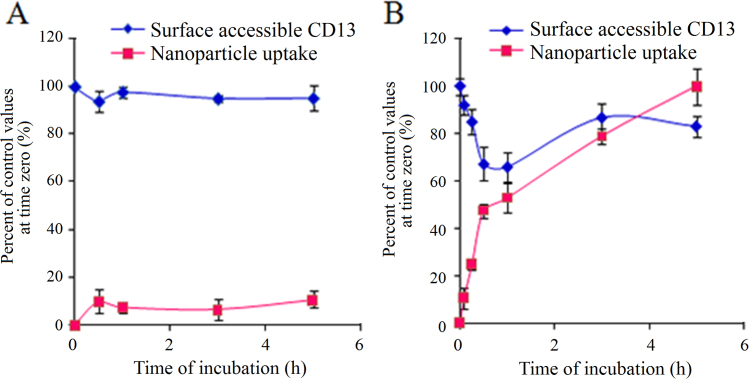
3.10. Endocytosis kinetic experiments
The results of the endocytosis kinetic experiments are shown in Fig. 7. As shown in Fig. 7, TPIC uptake was enhanced as the incubation time increased at 37 °C, whereas the internalization of TPIC at 4 °C was inhibited, which demonstrated that endocytic uptake of TPIC was an energy-dependent mechanism. Additionally, surface accessible CD13 was also affected by temperature. At 4 °C, there was approximately less than 10% TPIC internalized, resulting in the decrease in surface accessible CD13. At 37 °C, rapid internalization of TPIC was observed and the uptake of TPIC increased with the incubation time. Interestingly, with TPIC uptake increased, the amount of cell surface CD13 appeared to decrease first and then increase. Over the following 1 h, the internalization of TPIC increased rapidly and reached 53% and there was a sharp decline of surface CD13, which reached the lowest value of 66%. This finding implies that CD13 was involved in the endocytosis process of TPIC. After 1 h, the TPIC endocytosis process continued, but the endocytosis speed reduced because of the lower amount of CD13 on the cell surface. With the increase of the extension of time, the cell CD13 could return to the cell surface through the receptor cycle. With the endocytosis process, the amount of CD13 on the surface was dynamically changed. However, at any time point, the recycled CD13 continued to participate in the uptake process of TPIC so that internalized TPIC gradually increased.
Figure 7.
The TPIC uptake and surface accessible CD13 after HUVEC incubated with TPIC at (A) 4 °C or (B) 37 °C at various time points.
3.11. Exploring uptake pathways using endocytic inhibitors
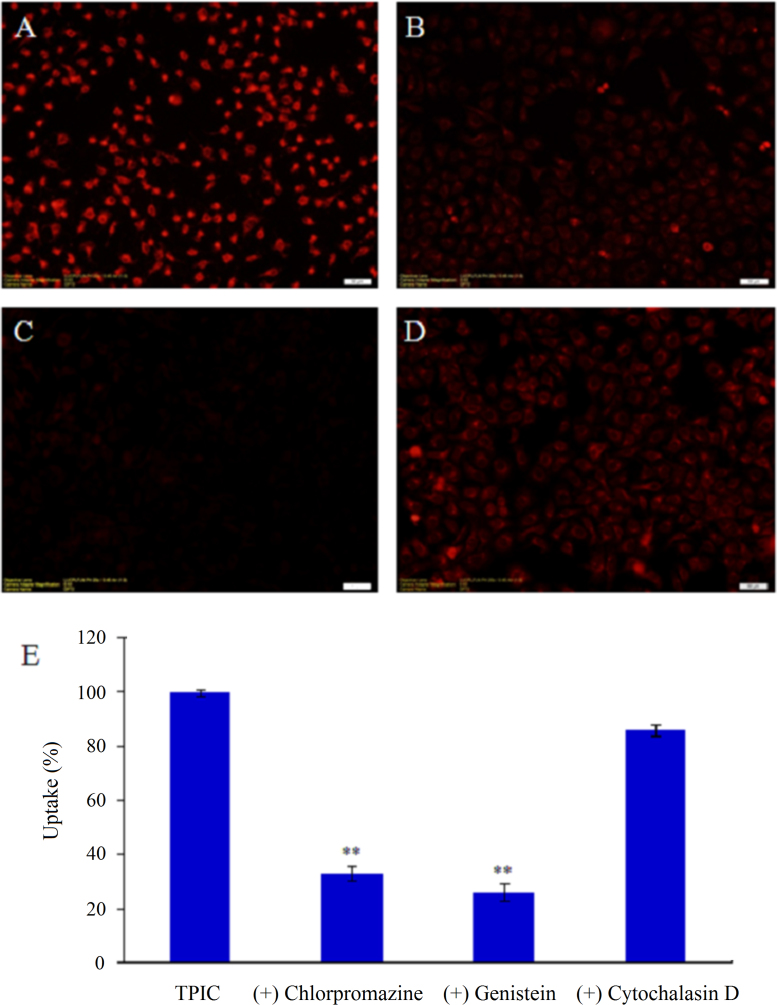
Nanoparticles enter cells through endocytic pathways, and selective inhibition of these various pathways was initiated to identify the relevant intracellular track adopted by TPIC for cell internalization. Hence, HUVEC were treated separately with each inhibitor against different internalization routes including CME, CvME, and macropinocytosis before cell incubation with the TPIC. Chlorpromazine is effective to inhibit CME. Treatment with chlorpromazine (10 μg/mL) for 30 min resulted in a 67.19% inhibition of TPIC uptake into HUVEC (as shown in Fig. 8), which indicated that CME was involved. Although the involvement of CME was previously mentioned34, other types of transcellular mechanism were also implicated in the previously reported intracellular traffic experiment of TPIC in HUVEC. Inhibition of the CvME was tested using genistein, a tyrosine kinase inhibitor, which was reported to inhibit the CvME in some virus42. After treated by genistein (1 μg/mL) for 30 min, the cell uptake of TPIC was significantly decreased by 74.2% (Fig. 8), suggesting the internalization of TPIC was mainly through CvME. Cytochalasin D was reported to inhibit actin polymerization and membrane ruffling involved in macropinocytosis43. The cell uptake of TPIC into HUVEC was reduced by 13.86% (Fig. 8) after cytochalasin D treatment, implying that macropinocytosis was not a major cell uptake mechanism for TPIC under normal condition. Taken together, the endocytic inhibition study presented herein indicated that the internalization of TPIC in HUVEC was a combined process of CME and CvME. The relative ratio between CME and CvME was about 1:1.1. Nevertheless, the unique mechanism of caveolae-mediated endocytosis was indeed mainly involved in the internalization which was consistent with our original hypothesis.
Figure 8.
Fluorescent micrographs of TPIC uptake into HUVEC pre-incubated with different inhibitors (100×). (A) Control; (B) chlorpromazine; (C) genistein; (D) cytochalasin D. (E) The quantitative results of (A), (B), (C) and (D). Compared with (A), **P<0.01. The groups in the presence of TPIC but without inhibitor treatment as controls and their fluorescence intensities were expressed as 100%. The decreased fluorescence of other groups compared with control suggested the potential mechanism of internalization.
4. Discussion
In our previous studies34, TPIC has been developed and extensively characterized. The average particle size and zeta potential of TPIC were 199.8±9.2 nm and 4.390±0.83 mV, respectively. TPIC was shown to be a promising carrier for the co-delivery of DNA and DOX leading to the efficiency of gene transfection and anti-tumor activity in vitro. As an extension of the intracellular study of TPIC for drug/gene co-delivery, the aim of the present study was to determine how NGR-functionalized TPIC interacts with targeted cells and promotes intracellular delivery process.
NGR peptide showed highly specific recognition of CD13. It was also reported that aggregated labelling of CD13 co-localized with CAV1 in most cells. Based on these findings and the previously obtained conclusion, we hypothesized that NGR might be able to mediate the TPIC into CD13 positive cells via the CvME. In the present study, it was verified that there was no co-localization between CD13 and CAV1 without treatment (Fig. 3, 0 min). However, when cells were incubated with an anti-CD13 antibody, co-localization between CD13 and CAV1 was observed (Fig. 3, 10 min to 3 h). These exciting results imply that targeting CD13 could initiate a subsequent interaction with CAV1. When HUVEC were treated with TPIC, a greater degree of co-localization of CD13 and CAV1 was found. It was shown that TPIC could accelerate the speed and enhance the degree of the co-localization of CD13 and CAV1 to a greater degree than anti-CD13 antibody. This difference could be attributed to the differences in the cross-linking degree and size between ligands and antibodies. Compared with antibodies, the NGR-modified nanocarrier was supposed to be a polyvalent ligand for CD13. Therefore, after treating with TPIC, both the speed and the extent of co-localization were enhanced compared with antibody. Notably, although the pattern was less conspicuous, cross-linked CD13 molecules after TPIC treatment showed a streaky distribution along longitudinal lines (Figure 3, Figure 2h; Figure 3, Figure 3h). This phenomenon was probably because the cross-linking of CD13 by TPIC would induce binding to actin filaments, either directly or indirectly, and further cause the longitudinal alignment. This similar distribution was also reported for cross-linked β2-microglobulin44 and HCoV-229E45 and both were eventually internalized by the CvME.
In evaluating the interaction between TPIC and CD13, we found that TPIC could bind to CD13 (Fig. 5A, 4 °C) and enter HUVEC together with CD13 at 37 °C (Fig. 5A, 37 °C). However, TPIC changed the distribution of CD13 on HUVEC, causing CD13 clustering. The cluster of CD13 was possibly because of the accumulation after interacting with NGR on the TPIC surface. This aggregation of receptors was a ubiquitous phenomenon in the process of receptor-mediated endocytosis, which further demonstrated that the uptake of TPIC in HUVEC was via CD13-mediated active targeting endocytosis. Moreover, CD13 and TPIC were not completely co-localized, and some separately entered the cells, which might be because CD13 needed to separate from the CD13-NGR complex and recycle to the cell surface to be used again. This type of receptor cycle was verified in the following endocytosis kinetic experiments (Fig. 7).
Subsequently, TPIC was demonstrated to co-localize with CAV1 and enter into HUVEC together with CAV1 (Fig. 5B). However, compared with their interaction with CD13, TPIC did not cause CAV1 clustering, suggesting that the co-localization of TPIC and CAV1 did not result from the receptor–ligand interaction of CD13 and NGR. Moreover, some TPIC were internalized independent of CAV1 (red dots), suggesting that there might be other mechanisms independent of CAV1, such as CME. We then verified that CME was indeed involved by the following endocytosis inhibitor experiments, which revealed results consistent with our hypothesis from previous studies34.
It was reported the different functions of CD13 were independent of each other. For example, the antibodies that target coronavirus-binding sites on CD13 can potentially block coronavirus infections, without interfering with the physiological functions of CD13. It was demonstrated that coronaviruses had evolved a mechanism to use CD13 as their cell entry receptor without interfering with the physiological functions of this important host enzyme46. In this study, the results further verified that the internalization of TPIC by HUVEC was not dependent on the enzyme activity of CD13, implying that TPIC might also use CD13 as their cell entry receptor without interfering with the physiological functions in the same way as coronaviruses do.
Then, MβCD was used to deplete cholesterol from the cell membrane and thereby damage caveolae, leading to the blocked internalization of TPIC. Additionally, the co-localization of TPIC and CAV1 was also inhibited after MβCD treatment. Obviously, cholesterol depletion can destroy the integrity of caveolae, affect the interaction between TPIC and CAV1 and inhibit TPIC entry into the cells. The above studies supported the important role of caveolae in TPIC entry into HUVEC. The subsequent inhibition studies also suggested that the internalization of TPIC in HUVEC was a process combining CME and CvME. The existence of CME was inferred through our above and previously reported results. Nevertheless, the dominant intracellular mechanism of TPIC entry into HUVEC was the CvME. This result was consistent with our original hypothesis.
5. Conclusions
Based on the previously constructed TPIC, we further systematically studied the interaction between the carriers and targeted cells and their intracellular delivery process to determine whether NGR used the CvME to transport TPIC into CD13 positive cells. The results indicated that TPIC influenced the distribution of CD13 on HUVEC through the interaction between CD13 and NGR, causing CD13 clustering, leading to the co-localization of CD13 and CAV1 and internalization via the CvME. Additionally, this internalization was not dependent on the enzyme activity of CD13 but was inhibited by cholesterol depletion. In this paper, we elucidated the intracellular mechanisms of multifunctional self-assembled nanocarriers and first proposed that NGR can mediate nanocarriers entering the cell through CvME. The elucidation of the mechanism was crucial for understanding the structure-bioactivity relationships of gene-loaded nanoparticles and was important to make a contribution to direct the design of novel drug delivery systems.
Acknowledgments
This work was supported by the National Natural Science Foundation (No. 81402867).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Medina-Kauwe L.K., Xie J., Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- 2.Peng S.F., Tseng M.T., Ho Y.C., Wei M.C., Liao Z.X., Sung H.W. Mechanisms of cellular uptake and intracellular trafficking with chitosan/DNA/poly(γ-glutamic acid) complexes as a gene delivery vector. Biomaterials. 2011;32:239–248. doi: 10.1016/j.biomaterials.2010.08.081. [DOI] [PubMed] [Google Scholar]
- 3.Gu J., Hao J., Fang X., Sha X. Factors influencing the transfection efficiency and cellular uptake mechanisms of Pluronic P123-modified polypropyleneimine/pDNA polyplexes in multidrug resistant breast cancer cells. Colloids Surf B Biointerfaces. 2016;140:83–93. doi: 10.1016/j.colsurfb.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 4.Singh J., Michel D., Chitanda J.M., Verrall R.E., Badea I. Evaluation of cellular uptake and intracellular trafficking as determining factors of gene expression for amino acid-substituted gemini surfactant-based DNA nanoparticles. J Nanobiotechnol. 2012;10:7. doi: 10.1186/1477-3155-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vercauteren D., Rejman J., Martens T.F., Demeester J., De Smedt S.C., Braeckmans K. On the cellular processing of non-viral nanomedicines for nucleic acid delivery: mechanisms and methods. J Control Release. 2012;161:566–581. doi: 10.1016/j.jconrel.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Douglas K.L. Toward development of artificial viruses for gene therapy: a comparative evaluation of viral and non-viral transfection. Biotechnol Prog. 2008;24:871–883. doi: 10.1021/bp070319o. [DOI] [PubMed] [Google Scholar]
- 7.Medina-Kauwe L.K. "Alternative" endocytic mechanisms exploited by pathogens: new avenues for therapeutic delivery? Adv Drug Deliv Rev. 2007;59:798–809. doi: 10.1016/j.addr.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rewatkar P.V., Parekh H.S., Parat M.O. Molecular determinants of the cellular entry of asymmetric peptide dendrimers and role of caveolae. PLoS One. 2016;11:e0147491. doi: 10.1371/journal.pone.0147491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alex S.M., Sharma C.P. Enhanced intracellular uptake and endocytic pathway selection mediated by hemocompatible ornithine grafted chitosan polycation for gene delivery. Colloids Surf B Biointerfaces. 2014;122:792–800. doi: 10.1016/j.colsurfb.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Podar K., Anderson K.C. Caveolin-1 as a potential new therapeutic target in multiple myeloma. Cancer Lett. 2006;233:10–15. doi: 10.1016/j.canlet.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 11.Liu C., Zhang N. Pharmaceutical strategies enhancing cell penetration efficiencies of non-viral gene delivery systems. Curr Gene Ther. 2009;9:267–290. doi: 10.2174/156652309788921053. [DOI] [PubMed] [Google Scholar]
- 12.Xiang S., Tong H., Shi Q., Fernandes J.C., Jin T., Dai K. Uptake mechanisms of non-viral gene delivery. J Control Release. 2012;158:371–378. doi: 10.1016/j.jconrel.2011.09.093. [DOI] [PubMed] [Google Scholar]
- 13.Paladern G.E. Structure modullation of plasmalemmal vesicle. J Cell Biol. 1968;37:633–649. doi: 10.1083/jcb.37.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Tiruppathi C., Minshall R.D., Malik A.B. Size and dynamics of caveolae studied using nanoparticles in living endothelial cells. ACS Nano. 2009;3:4110–4116. doi: 10.1021/nn9012274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penacho N., Rosa M., Lindman B., Miguel M.G., Simões S., de Lima M.C. Physicochemical properties of transferrin-associated lipopolyplexes and their role in biological activity. Colloids Surf B Biointerfaces. 2010;76:207–214. doi: 10.1016/j.colsurfb.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 16.López C.A., de Vries A.H., Marrink S.J. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLoS Comput Biol. 2011;7:e1002020. doi: 10.1371/journal.pcbi.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahay G., Alakhova D.Y., Kabanov A.V. Endocytosis of nanomedicines. J Control Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabrielson N.P., Pack D.W. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J Control Release. 2009;136:54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Hajitou A., Pasqualini R., Arap W. Vascular targeting: recent advances and therapeutic perspectives. Trends Cardiovasc Med. 2006;16:80–88. doi: 10.1016/j.tcm.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan Y., Xu W. The structure and main functions of aminopeptidase N. Curr Med Chem. 2007;14:639–647. doi: 10.2174/092986707780059571. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z., Hou L., Feng L., Huang S., Luo M., Shao S. An antimicrobial peptide containing NGR motif has potent antitumor activity against CD13+ and CD13− tumor cells. Tumour Biol. 2015;36:8167–8175. doi: 10.1007/s13277-015-3402-6. [DOI] [PubMed] [Google Scholar]
- 22.Wickström M., Larsson R., Nygren P., Gullbo J. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011;102:501–508. doi: 10.1111/j.1349-7006.2010.01826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soudy R., Ahmed S., Kaur K. NGR peptide ligands for targeting CD13/APN identified through peptide array screening resemble fibronectin sequences. ACS Comb Sci. 2012;14:590–599. doi: 10.1021/co300055s. [DOI] [PubMed] [Google Scholar]
- 24.Porcellini S., Asperti C., Valentinis B., Tiziano E., Mangia P., Bordignon C. The tumor vessel targeting agent NGR-TNF controls the different stages of the tumorigenic process in transgenic mice by distinct mechanisms. Oncoimmunology. 2015;4:e1041700. doi: 10.1080/2162402X.2015.1041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Hensbergen Y., Broxterman H.J., Elderkamp Y.W., Lankelma J., Beers J.C., Heijn M. A doxorubicin–CNGRC-peptide conjugate with prodrug properties. Biochem Pharmacol. 2002;63:897–908. doi: 10.1016/s0006-2952(01)00928-5. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S., Barnés C.M., Haskel A., Short S.M., Barnes K.R., Lippard S.J. Conjugated platinum (IV)–peptide complexes for targeting angiogenic tumor vasculature. Bioconjug Chem. 2008;19:39–49. doi: 10.1021/bc070031k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Wang Y., Chen X., Wang J., Zhang X., Zhang Q. NGR-modified micelles enhance their interaction with CD13-overexpressing tumor and endothelial cells. J Control Release. 2009;139:56–62. doi: 10.1016/j.jconrel.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Wu J.J., Huang L. Nanoparticles targeted with NGR motif deliver c-myc siRNA and doxorubicin for anticancer therapy. Mol Ther. 2010;18:828–834. doi: 10.1038/mt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffatt S., Wiehle S., Cristiano R.J. Tumor-specific gene delivery mediated by a novel peptide–polyethylenimine–DNA polyplex targeting aminopeptidase N/CD13. Hum Gene Ther. 2005;16:57–67. doi: 10.1089/hum.2005.16.57. [DOI] [PubMed] [Google Scholar]
- 30.Tokuda N., Levy R.B. 1,25-Dihydroxyvitamin D3 stimulates phagocytosis but suppresses HLA-DR and CD13 antigen expression in human mononuclear phagocytes. Proc Soc Exp Biol Med. 1996;211:244–250. doi: 10.3181/00379727-211-43967. [DOI] [PubMed] [Google Scholar]
- 31.Smith N.J., Luttrell L.M. Signal switching, crosstalk, and arrestin scaffolds: novel G protein-coupled receptor signaling in cardiovascular disease. Hypertension. 2006;48:173–179. doi: 10.1161/01.HYP.0000232641.84521.92. [DOI] [PubMed] [Google Scholar]
- 32.Corti A., Curnis F., Arap W., Pasqualini R. The neovasculature homing motif NGR: more than meets the eye. Blood. 2008;112:2628–2635. doi: 10.1182/blood-2008-04-150862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C., Yu W., Chen Z., Zhang J., Zhang N. Enhanced gene transfection efficiency in CD13-positive vascular endothelial cells with targeted poly(lactic acid)–poly(ethylene glycol) nanoparticles through caveolae-mediated endocytosis. J Control Release. 2011;151:162–175. doi: 10.1016/j.jconrel.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 34.Liu C., Liu F., Feng L., Li M., Zhang J., Zhang N. The targeted co-delivery of DNA and doxorubicin to tumor cells via multifunctional PEI–PEG based nanoparticles. Biomaterials. 2013;34:2547–2564. doi: 10.1016/j.biomaterials.2012.12.038. [DOI] [PubMed] [Google Scholar]
- 35.Menrad A., Speicher D., Wacker J., Herlyn M. Biochemical and functional characterization of aminopeptidase N expressed by human melanoma cells. Cancer Res. 1993;53:1450–1455. [PubMed] [Google Scholar]
- 36.Mina-Osorio P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med. 2008;14:361–371. doi: 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujimoto T. GPI-anchored proteins, glycosphingolipids, and sphingomyelin are sequestered to caveolae only after crosslinking. J Histochem Cytochem. 1996;44:929–941. doi: 10.1177/44.8.8756764. [DOI] [PubMed] [Google Scholar]
- 38.Mayor S., Rothberg K.G., Maxfield F.R. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 1994;264:1948–1951. doi: 10.1126/science.7516582. [DOI] [PubMed] [Google Scholar]
- 39.Gabrilovac J., Cupić B., Zivković E., Horvat L., Majhen D. Expression, regulation and functional activities of aminopeptidase N (EC 3.4.11.2; APN; CD13) on murine macrophage J774 cell line. Immunobiology. 2011;216:132–144. doi: 10.1016/j.imbio.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 40.Winiarska M., Bil J., Wilczek E., Wilczynski G.M., Lekka M., Engelberts P.J. Statins impair antitumor effects of rituximab by inducing conformational changes of CD20. PLoS Med. 2008;5:e64. doi: 10.1371/journal.pmed.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai S.K., Hida K., Man S.T., Chen C., Machamer C., Schroer T.A. Privileged delivery of polymer nanoparticles to the perinuclear region of live cells via a non-clathrin, non-degradative pathway. Biomaterials. 2007;28:2876–2884. doi: 10.1016/j.biomaterials.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Raghu H., Sharma-Walia N., Veettil M.V., Sadagopan S., Chandran B. Kaposi׳s sarcoma-associated herpesvirus utilizes an actin polymerization-dependent macropinocytic pathway to enter human dermal microvascular endothelial and human umbilical vein endothelial cells. J Virol. 2009;83:4895–4911. doi: 10.1128/JVI.02498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huet C., Ash J.F., Singer S.J. The antibody-induced clustering and endocytosis of HLA antigens on cultured human fibroblasts. Cell. 1980;21:429–438. doi: 10.1016/0092-8674(80)90479-1. [DOI] [PubMed] [Google Scholar]
- 44.Han J., Hajjar D.P., Tauras J.M., Nicholson A.C. Cellular cholesterol regulates expression of the macrophage type B scavenger receptor, CD36. J Lipid Res. 1999;40:830–838. [PubMed] [Google Scholar]
- 45.Nomura R., Kiyota A., Suzaki E., Kataoka K., Ohe Y., Miyamoto K. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J Virol. 2004;78:8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L., Lin Y.L., Peng G., Li F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc Natl Acad Sci U S A. 2012;109:17966–17971. doi: 10.1073/pnas.1210123109. [DOI] [PMC free article] [PubMed] [Google Scholar]