Abstract
Campylobacter is the most common cause of foodborne bacterial illness worldwide. Faecal contamination of meat, especially chicken, during processing represents a key route of transmission to humans. There is a lack of insight into the mechanisms driving C. jejuni growth and survival within hosts and the environment. Here, we report a detailed analysis of C. jejuni fitness across models reflecting stages in its life cycle. Transposon (Tn) gene-inactivation libraries were generated in three C. jejuni strains and the impact on fitness during chicken colonisation, survival in houseflies and under nutrient-rich and –poor conditions at 4 °C and infection of human gut epithelial cells was assessed by Tn-insertion site sequencing (Tn-seq). A total of 331 homologous gene clusters were essential for fitness during in vitro growth in three C. jejuni strains, revealing that a large part of its genome is dedicated to growth. We report novel C. jejuni factors essential throughout its life cycle. Importantly, we identified genes that fulfil important roles across multiple conditions. Our comprehensive screens showed which flagella elements are essential for growth and which are vital to the interaction with host organisms. Future efforts should focus on how to exploit this knowledge to effectively control infections caused by C. jejuni.
Introduction
Infection by Campylobacter is the most common cause of foodborne bacterial diarrhoeal disease worldwide, responsible for ~96 million foodborne illnesses and ~21,000 foodborne deaths in 20101. While most cases are self-limiting, for some, campylobacteriosis is a particularly serious infection, and it is also associated with severe post-infection complications, including irritable bowel and Guillian-Barré syndromes. Consumption of undercooked poultry, unpasteurised dairy products and contaminated water represent the most common sources of human infection2, 3. Campylobacter jejuni has a broad range of environmental reservoirs that include water, birds and other domestic animals3. In addition, flies have been implicated as a transmission vector for C. jejuni for both chicken flocks and possibly for humans4–7.
C. jejuni encounters and has to overcome various stress conditions whilst passing through the gastrointestinal tract of humans and other animals, during processing of food products (e.g. slaughter process of poultry), on/in food (e.g. in milk or poultry meat, generally stored at low temperature) and in the environment (e.g. in surface water, soil, or in flies, the latter representing a transmission vector)4–8. However, compared to other enteric pathogens such as pathogenic Escherichia coli and Salmonella spp, the survival mechanisms used by C. jejuni to cope with these stresses are less well-understood9.
Based on genome analysis, the capacity of C. jejuni to survive outside the host and adapt to environmental stress conditions appears to be limited due to the lack of key stress regulators found in other enteric pathogens10. Colonisation and infection of host organisms by C. jejuni is a multifactorial process with key roles for “swimming” motility, chemotaxis, interaction with gut epithelial cells, toxin production, and oxidative and metabolic stress adaptation11, 12.
In this study, we screened extensive C. jejuni transposon (Tn) gene inactivation mutant libraries to comprehensively assess which genes are required during in vitro growth, chicken colonisation and during exposure to low temperature in nutrient-rich and –poor conditions, and infection of human gut epithelial cells. This study reinforces the importance of flagella for host interactions, and identifies genes required for survival, colonisation and infection in multiple phases of the bacterium’s life cycle.
Results
Identification of genes required for fitness
To assess the genetic basis of C. jejuni growth and survival, genes were randomly inactivated using Tn mutagenesis in three well-characterized C. jejuni strains [M1cam13, 14, NCTC 11168 (hereafter referred to as 11168)10 and 81–17615]. Tn mutant libraries were characterised by Tn insertion site sequencing (Tn-seq16) (Table S1), providing a measure for the relative abundance of each Tn mutant in the library. Genes that are required for growth and survival, hereafter referred to as “fitness” genes, cannot tolerate Tn insertions, or Tn mutants in these genes are severely underrepresented in the libraries. To identify fitness genes, 23,334 unique chromosomal Tn insertions were analysed in M1cam, 15,008 in 11168, and 17,827 in 81–176 (Table S1), reaching near-saturation in terms of the number of genes that could be inactivated (Fig. 1a). In addition to chromosomal Tn insertions, 2,009 and 1,919 unique insertions were in the 81–176 plasmids pVir17 and pTet18, respectively (Table S1). No apparent Tn insertion bias was observed (Fig. S1) and each Tn library predominately yielded unique Tn insertions (Fig. S2).
Figure 1.
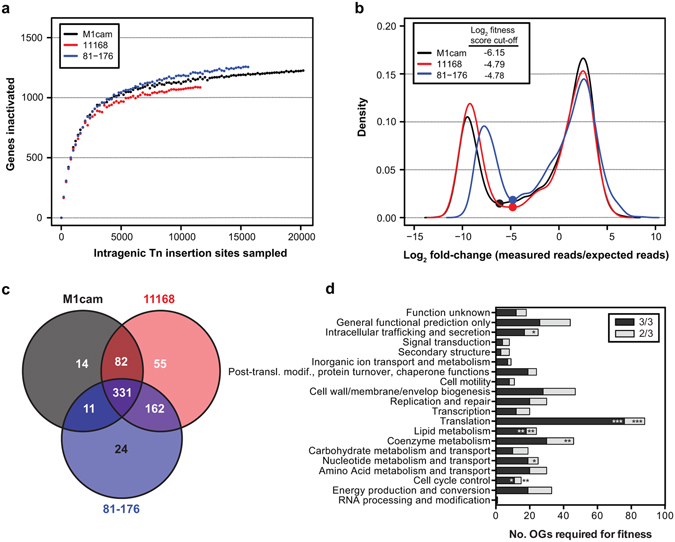
Gene fitness analysis during in vitro growth of C. jejuni M1cam, 11168 and 81–176. (a) Rarefaction analysis of intragenic Tn insertions. (b) Density plot fitness score (Log2 fold-change measured reads/expected reads) per gene. Dots indicate fitness score cut-off values. (c) Overlap of homologous gene (HG) clusters (HGs) required for fitness in C. jejuni M1cam, 11168, and 81–176, representing the “pan-fitness genome”. HG clusters that are required for fitness in one or more strains are displayed. (d) Functional class (COG; Cluster of Orthologous Genes) enrichment analysis of fitness genes. Fisher exact test with Q-value multiple testing correction; *Q < 0.05, **Q < 0.01 and ***Q < 0.001.
Gene fitness score (Log2 fold-change between the observed vs expected sequence reads19) density plots followed a bimodal distribution with the “left” population representing genes required for in vitro growth and survival (Fig. 1b). In total, 445 genes were required for fitness in M1cam, 413 genes in 81–176 and 499 in 11168 (Table S2). Interestingly, cpp13 in the pTet plasmid in strain 81–176 appeared to contribute to fitness. A variant of C. jejuni 81–176 that has lost pTet exists (D. Hendrixson, personal communication), which implies that cpp13 is not obligate essential but may contribute to fitness, or it could be antitoxin of an uncharacterised toxin-antitoxin system. Unexpectedly, Tn insertions were observed in dnaA (1,323 bp) at bp position 352 in M1cam and at position 1,115 in 81–176. Consequently, dnaA did not pass the stringent fitness gene criteria. Disruption of dnaA may be tolerated at the 3′ end of the gene, in the rare event of a secondary site mutation20, or due to the existence of merodiploids21.
To comprehensively assess fitness genes in the species C. jejuni, homologous genes (HG) were compared for the three C. jejuni strains (Table S2). Out of the 1,424 identified HG clusters, M1cam has members in all HG clusters, and 81–176 and 11168 have gene members in 1,423 HG clusters, indicating that in terms of functional capacity the three tested C. jejuni strains are highly related. 331 HG clusters were required for fitness in all three strains tested and 486 HG clusters were required in two or more strains (Fig. 1c), indicating that a large part of the C. jejuni genome is dedicated to growth and survival under the condition tested. We found that 845 HG clusters were not required for fitness in any of the analysed strains. Genes implicated in fitness were relatively dispersed across the C. jejuni genomes, but there were regions that were (almost) devoid of fitness genes, for example, the flagellar glycosylation gene cluster (Fig. S3). The number of fitness HG clusters shared by C. jejuni M1cam, 81–176 and 11168 was substantially larger, at 331, than the 175–233 genes previously reported to be obligatory essential22–25. This may be partially due to the inclusion of genes, in our study, whose inactivation is lethal (obligate essential) as well as genes which when inactivated by a Tn result in severely compromised growth and/or survival. Further, this could be related to C. jejuni strain differences, growth conditions, the Tn element used, the number of Tn mutants analysed and the read-out technology. A systematic review of the literature on C. jejuni 11168 defined gene deletion and Tn mutants revealed that for 38 out of 486 (7.8%) fitness HG clusters (required in two or more strains) identified in this study, mutants have been reported, indicating a low false-positive rate in our study. Although mutants in some of these genes have a growth defect, e.g. as reported for a C. jejuni 11168 pycB mutant26, especially when compared against other ‘more-fit’ mutants or mutants with wild-type fitness, as is the case in the Tn screen.
Genes implicated in translation, lipid metabolism and cell cycle control were overrepresented amongst the genes required for fitness during in vitro growth/survival in all three tested C. jejuni strains. Extending the analysis to fitness genes required in two or more strains also showed overrepresentation of coenzyme metabolism, nucleotide metabolism and intracellular trafficking and secretion genes (Fig. 1d). Required for fitness were, amongst others, genes implicated in replication, transcription, translation (40 out of 49 ribosomal protein genes), purine and pyrimidine metabolism, energy metabolism (ATP and NAD synthase, NADH-quinone oxidoreductase), isoprene biosynthesis, protein secretion (Sec and Tat pathway), as well as genes involved in cofactor biosynthesis (thiamine, folic acid and heme) and oxidative stress (see Table S2 for a complete overview). Further, the complete gluconeogenic pathway was found to be required for fitness whereas the majority of the enzymes of the tricarboxylic acid (TCA) cycle were not, except for aconitase (acnB), probably reflecting flexibility in this part of the bacterium’s metabolism.
C. jejuni expresses surface structures such as flagella, lipooligosaccharide (LOS) and capsular polysaccharides (CPS). Inactivation of genes in these pathways severely attenuated fitness. This included flagellar basal body rod proteins (encoded by flgAC and fliEL), the MS-ring (fliF) and C-ring (fliG) as well as components of the flagellar type III secretion system (fliQH). Genes required for formation of the LOS lipid A molecule (lpxABCDL), KDO (kdsAB) and the first L-glycero-D-manno-heptose residue (waaC) were required for fitness, whereas the remainder of the genes responsible for the core oligosaccharide were not. Of the capsular biosynthesis gene cluster only the inactivation of the last two genes (kpsDF) resulted in impaired fitness. Genes required for cell envelope generation were also important for fitness including fatty acid biosynthesis genes (accABCD and fabDFGHLZ), peptidoglycan (dapADEF, ddl, murABCDEFG, pbpABC), and the rod-shape determining protein genes (mreBCD). Protein glycosylation is tightly linked to virulence, of which the N-linked protein glycosylation pathway genes pglACD were required for fitness. This is in contrast to flagella glycosylation genes, which had no conserved role in fitness during in vitro growth and survival (Table S1).
Quantitative analysis of genes implicated in the life cycle of C. jejuni
The same extensive Tn library in strain M1cam (Table S1; 9,951 unique Tn insertions with 1,124 genes harbouring Tn insertions) was screened in various in vivo and in vitro models as a proxy for some of the conditions that C. jejuni might encounter during its life cycle, facilitating a comparative analysis across the models. The M1cam library was screened during the colonisation of commercial broiler chickens (natural host), survival in the housefly (transmission vector), survival under nutrient-rich- and nutrient-poor conditions at low temperature, and in models that mimic (stages of) infection of humans, i.e. adhesion and invasion of human gut epithelial cells. For comparative purposes we included data that we obtained in a study analysing the infection of gnotobiotic piglets (de Vries et al., submitted). Tn-seq analysis after exposure to a challenge, compared with the control condition, provided a quantitative measure for the contribution of a gene to fitness in each of the models (Fig. 2 and Tables S3 and S4). The number of Tn mutants recovered from each model confirmed that the complexity of the Tn library was maintained in all models, however library complexity was reduced after colonisation of chickens, i.e. 23% of the input Tns were recovered from chickens (Fig. S4). Therefore, we applied more stringent criteria for selection of candidate genes required in this model (Table S3 and Methods). A detailed analysis for each of the models in this study is provided below.
Figure 2.
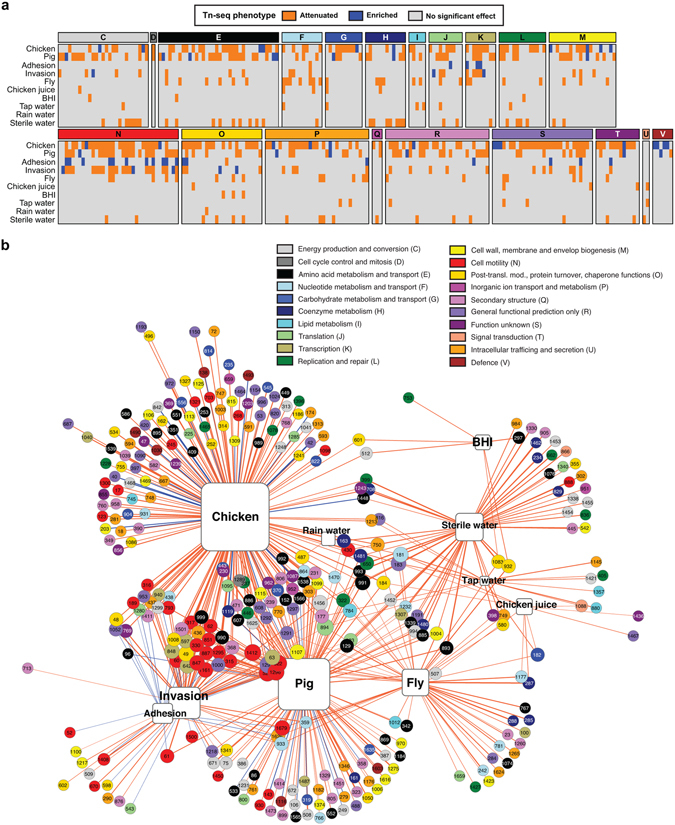
Identification of conditional essential genes in C. jejuni. (a) Effect of Tn insertions on the ability of C. jejuni M1cam to colonize commercial broiler chickens, infect gnotobiotic piglets, adhere and invade gut epithelial tissue culture cells, survive in flies and at 4 °C in various media (chicken juice, BHI, tap water, rain water, and sterile water). Genes of which Tn mutants showed significantly attenuated or enriched fitness in the experimental models are shown and are grouped according to their COG functional classification. Data represented as Log2 fold-change is also presented in Table S4. (b) Gene-model interaction network showing attenuated (orange lines) and enriched (blue lines) Tn-seq scores linked to their respective models; the thickness of the connecting lines corresponds to the Log2 fold-change (input/output). The gene numbers correspond to the C. jejuni M1cam locus-tags13 and are color-coded according to their COG functional class. The size of the genes and the models displayed increases with the number of interactions.
Genes required for chicken colonisation
Broilers generally become colonised with Campylobacter spp. at about 3–4 weeks of age27. We have screened the M1cam Tn library ‘C’ in a relevant chicken colonisation model, i.e. in 3-week-old Ross 308 commercial broiler birds. Four cages, each with 5–7 birds, were inoculated with the library and 6 days post-inoculation (p.i.) colonising Tn mutants were recovered. Due to the coprophagic behaviour of chickens, each individual cage was considered a colonisation unit28. We hypothesised that a group-level approach would improve the robustness of the analysis and reduce any bias introduced by random dropout of Tn mutants as observed in our previous work using a signature-tagged mutagenesis approach29 and using wild-type isogenic-tagged strains (WITS) that have indistinguishable phenotypes in pure culture30. On average, cages harboured 1,641 ± 226 Tn mutants (>10 reads) compared to 7,325 ± 538 Tns in the input (Fig. S4), revealing a population bottleneck, even at the cage level. However, the average read count of the four cages demonstrated recovery of 3,111 Tn mutants (>10 reads) covering 701 genes. This indicated that, with additional stringent filtering steps (see Methods) a large part of the genome could be analysed.
Tn mutants of 172 genes were significantly under-represented in colonised chickens, whereas 24 appeared to enhance fitness during colonisation (Fig. 2 and Tables S3 and S4). Genes linked to the COG class “cell motility” were significantly enriched amongst those required during colonisation (Fig. 2 and Fig. S5), underscoring the prominent role of motility for colonisation of chickens. Previously, Johnson et al., screened 1,155 C. jejuni 81–176 Tn mutants in 1-day-old chicks and identified 130 genes that were required for colonisation and 30 genes that appeared to enhance colonisation23. However, interpretation of the importance of these colonisation genes is complicated due to limited validation; only the importance of mapA was confirmed23. We found a limited overlap of 23 chicken colonisation genes between the two studies, 11 of which were linked to motility and the flagellar system. The datasets had one gene (CJM1_0420; hypothetical protein) in common that was beneficial for colonisation. Differences between identified colonisation genes are likely the result of the model employed, e.g. older birds harbouring a more mature intestinal microbial community and younger birds being more permissive for colonisation by C. jejuni, e.g. as reflected in a lower inoculation dose to establish colonisation31.
For validation, 19 genes were tested individually for their ability to colonise chickens. The importance in chicken colonisation was confirmed for genes involved in chemotaxis (mcp4_1), the flagellar system (pflB, fliD, fliW, and maf3), N-linked protein glycosylation system (capM/pglH), and phosphate transport (pstA) (Fig. 3a). Although motility is considered a driving factor in chicken colonisation, the colonisation deficient maf3, capM and pstA displayed wild-type motility (Fig. S6). In the screen, mutants in eptA (also referred to as eptC 32), glnP, jlpA, fdhA and CJM1cam_1125 showed reduced colonisation. However, deletion mutants in these genes colonised at slightly higher levels compared to the wild-type (Fig. 3a); motility of these gene deletion mutants did not differ significantly from the wild-type (Fig. S6). No difference to the wild-type was found in the colonisation proficiency of deletion mutants in moaA, CJM1cam_0303 (hypothetical protein), CJM1cam_0438 (hypothetical protein), flaG, ilvB, engD and gltA. Identification of these genes in the Tn library screen might be attributed to the competition effect of other Tn mutants in the screen, whilst validation experiments were conducted with single mutant inocula. We opted for validation experiments with single mutant inocula due to the unpredictable outcome of competition experiments, even in the simplest case of two phenotypically indistinguishable WITS30. It is possible that some candidate colonisation genes represent false positives due to the random loss of mutants, i.e. an infection bottleneck that was observed in the chicken model.
Figure 3.
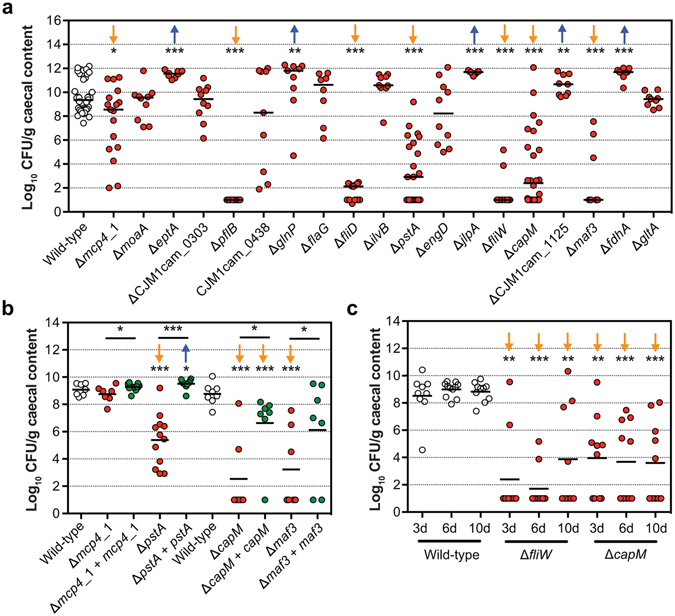
Validation of chicken colonisation Tn mutant library screen. (a) Colonisation levels of C. jejuni M1cam gene deletion mutants 6 d pi. (n ≥ 7) (b) Colonisation of gene deletion mutants and complemented gene deletion mutants (n ≥ 7). (c) Colonisation kinetics of C. jejuni M1cam wild-type and defined mutants in fliW and capM (n ≥ 9). Chickens being colonised vs not colonised were 2/8, 2/8 and 3/6 for the fliW mutant and 4/5, 4/6 and 5/5 for the capM mutant at day 3, 6 and 10 p.i., respectively. Statistical significance was analysed using a Mann-Whitney test with *P < 0.05, **P < 0.01 and ***P < 0.001. The orange arrows represent an attenuated phenotype while the blue arrows represent an enriched chicken colonisation phenotype.
Genetic complementation of pstA, capM and maf3 did significantly increase their colonisation capacity compared to their respective gene deletion mutants (Fig. 3b), confirming their role in chicken colonisation. Attempts to complement fliD (encoding the flagellar cap protein) failed (Fig. S8). When analysing colonisation of the genetically complemented mcp4_1 mutant in chickens, we found that, in contrast to initial validation experiments (Fig. 3a), the mcp4_1 mutant colonised chickens; which could be the result of intrinsically lower colonisation resistance in the batch of chickens used in this experiment. Despite the existence of a colonisation bottleneck and the incomplete reproducibility when comparing defined deletion mutants with the results obtained in the Tn library screen, this work has identified pstA, capM, maf3 and mcp4_1 as novel colonisation factors in 3-week-old broiler chickens.
In validation experiments (Fig. 3a) we observed an ‘all or nothing’ colonisation effect in some chickens infected with particular defined mutants, e.g. pflA, fliD and fliW, whereas for other deletion mutants, such as capM and mcp4_1, colonisation levels varied considerably between the birds. This indicates that there might be differences in the colonisation permissiveness/resistance between birds, within and between experiments. To investigate this in more detail, the colonisation of the wild-type, fliW and capM mutants were measured at different time intervals (3, 6 and 10 days p.i.) (Fig. 3c). Chickens were colonised at high levels from 3 days onwards and neither an increase in the levels of colonisation nor the number of colonised birds was observed. Surprisingly, the level of colonisation of the fliW and capM mutants did not increase between 3 and 10 days p.i. and there was also no obvious increase in the number of colonised chickens (Fig. 3c). We hypothesise that the colonisation responses observed in our validation experiments were potentially confounded by variation in gut microbiota composition or differential inflammatory responses elicited during colonisation33–35.
Genes required for survival in the housefly
As a transmission vector model for C. jejuni 4–7, survival of the M1cam Tn library ‘C’ was examined 4 h after individual inoculation of houseflies. Tn mutants in 48 genes showed reduced survival and no genes were identified for enhanced survival (Fig. 2 and Tables S3 and S4). Genes of the COG class “nucleotide transport and metabolism” were overrepresented amongst the genes linked to survival in the housefly (Fig. 2 and Fig. S5). These were the non-essential genes in the purine (purLMN) and pyrimide (pyrC_1/2 and pyrDF) biosynthesis pathways. Although the Tn library screen identified a number of candidate survival genes, validation experiments with 7 defined gene deletion mutants (inoculated as single mutants) were non-confirmative (Fig. 4a). Our inability to confirm the role of identified candidate genes might be the result of low levels of attenuation, or due to the lack of competition with other mutants. However, the attenuated survival of a capM deletion mutant approached significance (P = 0.0513, two-tailed Mann-Whitney). As the importance of capM in chicken colonisation was confirmed via genetic complementation, we also tested the capM mutant alongside its genetically complemented mutant for survival in the housefly, which confirmed that capM is also involved in survival in the housefly (Fig. 4b).
Figure 4.
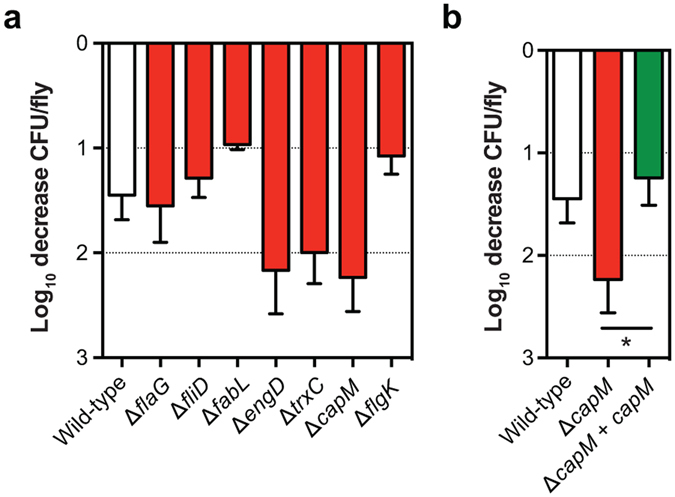
Validation of Tn mutant library screen during survival in the housefly. (a) Survival (Log10 decrease inoculum vs recovered) of defined C. jejuni M1 gene deletion mutant in the housefly after 4 h. (b) Survival of wild-type, capM defined gene deletion mutant and genetically complemented mutant. Data shown are Log10 decrease of CFU per fly relative to the inoculum and plotted as means with SEM (n ≥ 4). Significance was analysed using a Mann-Whitney test with *P < 0.05.
Survival in nutrient-rich and –poor conditions at low temperature
The M1cam Tn library ‘C’ was incubated at 4 °C in “chicken juice” (liquid obtained after thawing chicken carcasses), BHI broth and in water (sterile, tap and rain), with the “water” models being included to assess the survival under more general environmental conditions. Tn mutants that survived after 7 days incubation were compared to the Tn library composition at T = 0 days. Ten genes were implicated in survival in chicken juice and 6 genes in BHI medium. Considerable variation was found for the number of genes implicated in survival at 4 °C in nutrient-poor conditions, with 13 genes identified in tap water and 57 in sterile water. However, only one candidate gene, encoding the heat shock response protein ClpB, was identified as being required for survival in rain water (Fig. 2 and Tables S3 and S4). These variations could arise due to differences in chemical composition, pH and potentially other bacteria present in water samples36, 37. Given the relatively low number of genes associated with survival at low temperature and the relatively mild attenuation, as expressed by Tn-seq fitness score, we hypothesise that survival may be a passive rather than active mechanism38.
At low temperature, an oxidative stress response is induced in C. jejuni 9, 39. In our Tn library screen, the gene encoding the oxidoreductase TrxC was found to be required for survival in sterile water and BHI at low temperature. In addition, the regulator of oxidative stress, PerR, was linked to survival in sterile water. PerR plays a role in controlling oxidative stress resistance and survival under aerobic conditions39, 40. The RacRS two-component system is important for chicken colonisation and is part of a temperature-dependent signalling pathway41. In our study, Tn mutants in racS had reduced survival in chicken juice and tap water at low temperature. Chemotaxis has also been suggested to play a role in survival at low temperature42. In line with this, Tn mutants of mcp4_2 were attenuated for survival in chicken juice and tap water. In addition, Tn mutants in kefB and czcD (antiporters), fabI (fatty acid metabolism) and CJM1cam_0181 to CJM1cam_0183 (purN, nnr, and a gene encoding a hypothetical protein) were linked to survival in both chicken juice and tap water.
For validation, 10 defined gene deletion mutants were assayed under all conditions described above. We found that several deletion mutants were attenuated for survival after 3 days in BHI and water (sterile, tap and rain). However, at 7 days their survival did not differ from the wild-type. This is most likely due to further decreasing levels of the wild-type between day 3 and 7 (Fig. 5a). Confirmatory experiments with deletion mutants highlighted the contribution of hisC (aromatic amino acid aminotransferase) and trxC (thiol-disulphide oxidoreductase) for survival at low temperature (Fig. 5b–e). The hisC mutant showed attenuated survival in BHI (7 days), sterile water (3 days), tap water (3 and 7 days) and rain water (3 days) (Fig. 5b–e). Deletion of trxC, however, resulted in reduced survival of C. jejuni in BHI, sterile water and tap water after 7 days (Fig. 5b–d) and in chicken juice and rain water after 3 days (Fig. 5a,e). Although the deletion of trxC resulted in a prolonged lag-phase during growth in vitro in BHI broth (Fig. S7), genetic complementation of the trxC deletion restored its phenotype under all conditions tested, confirming the importance of trxC in the survival of C. jejuni at 4 °C, both in nutrient-rich and –poor conditions (Fig. 5b). Thus far, the function of TrxC in C. jejuni is unknown. The closest ortholog is TrxC in Helicobacter pylori, which is required for protection against oxidative stress43. The M1cam trxC mutant showed increased sensitivity to hydrogen peroxide, which was restored to near wild type levels in the genetically complemented mutant (Fig. 6), supporting its role in the response to oxidative stress.
Figure 5.
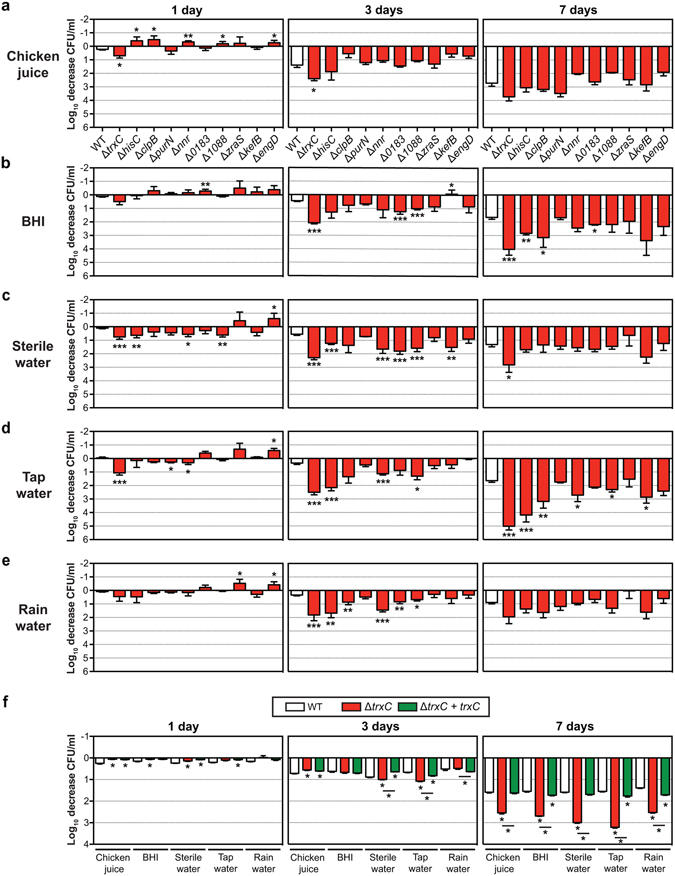
Validation of Tn mutant library screen during survival at low temperature under nutrient-rich and –poor conditions. (a–e) Survival of defined C. jejuni M1cam gene deletion mutant in (a) BHI, (b) chicken juice, (c) sterile water, (d) tap water, (e) rain water. Survival of trxC defined gene deletion mutant and complemented mutant (F), at 4 °C in different media. Data shown are Log10 decrease of CFU/ml relative to the 0 day time-point and plotted as means with SEM (n ≥ 4). Statistical significance was analysed using a Mann-Whitney test with *P < 0.05, **P < 0.01 and ***P < 0.001.
Figure 6.
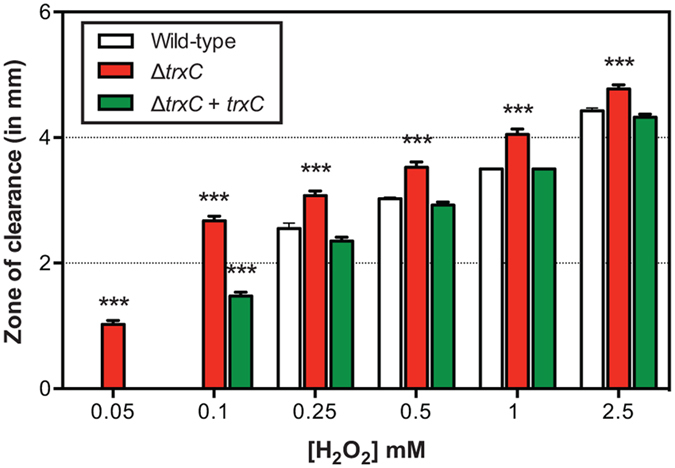
Sensitivity to hydrogen peroxide of C. jejuni M1cam wild-type, trxC defined gene deletion mutant and trxC complemented mutant. Data shown are zones of inhibition and plotted as means with SEM (n ≥ 4). Statistical significance was analysed using a two-way ANOVA test with ***P < 0.001.
Infection of human gut epithelial cells
The M1cam mutant library ‘C’ was used to infect Caco-2 human gut epithelial cells. To identify genes required for adhesion, Tn mutants that adhered to Caco-2 epithelial cells were compared to the non-adherent fraction rather than to the inoculum. This was to restrict the number of false positives due to attenuated survival in the infection medium. Comparing the Tn mutants that invaded the epithelial cells with the non-adherent fraction identified 57 candidate genes involved in cellular invasion, whereas only two genes passed our filtering criteria for adhesion (Fig. 2 and Tables S3 and S4).
In line with previous studies3, 22, 44, 45, genes linked to the COG class “cell motility” were overrepresented amongst the genes required for cellular invasion (Fig. 2 and Fig. S5). Gao et al., screened a Tn mutant library in C. jejuni 81–176 in a Cos-1 monkey kidney fibroblast cell model, which identified 36 invasion genes22, of which 19 genes were also identified in this study using Caco-2 human gut epithelial tissue culture cells. A total of 38 genes were only required for invasion by M1cam and 17 only for 81–17622. Of the M1cam unique invasion genes, 14 were linked to the flagellar system, including fliK, flaG, fliD, fliW, and maf3. The differences in invasion requirements are likely to be caused by the different cell-types or related to the C. jejuni strains.
Validation was performed with 15 defined gene deletion mutants, this included genes not previously reported to be linked to motility or the flagellar system (Fig. 7a,b). This confirmed the role of 13 out of 15 selected invasion genes (Fig. 7b). Tn-seq only identified two genes involved in adhesion, however 14 out of 15 tested deletion mutants displayed attenuated adhesion (Fig. 7a). This apparent discrepancy might be due to differences in the experimental set-up between the Tn library screen and the validation experiments. It is well recognized that between Tn mutants is a confounding factor in Tn library screens23, 45, in contrast to the validation experiments presented here, in which defined gene deletion mutants were allowed to interact with Caco-2 cells as single mutant inocula. Further interactions between adhesion-deficient and –proficient Tn mutants may compensate for adhesion of otherwise adhesion-deficient Tn mutants. Amongst the genes that were confirmed to play a role in interacting with Caco-2 cells were livM (amino acid metabolism), fabL (fatty acid metabolism) and engD (GTP-dependent nucleic acid-binding protein) that have no known link to the flagellar system. However, deletion of livM resulted in ~50% reduced motility compared to the wild-type (Fig. S6). No genomic variations in the livM mutant linked to motility were identified (Table S5). The role of engD in colonisation of chickens was not confirmed in validation experiments with a defined deletion mutant (Fig. 3). However, we confirmed the contribution of engD in adhesion and invasion of Caco-2 tissue culture cells (Fig. 7). The maf3 gene deletion mutant was motile (Fig. S6) but had a reduced capacity to colonise chickens (Fig. 3a,b) and also lacked the ability to adhere to and invade Caco-2 cells (Fig. 7a–d).
Figure 7.
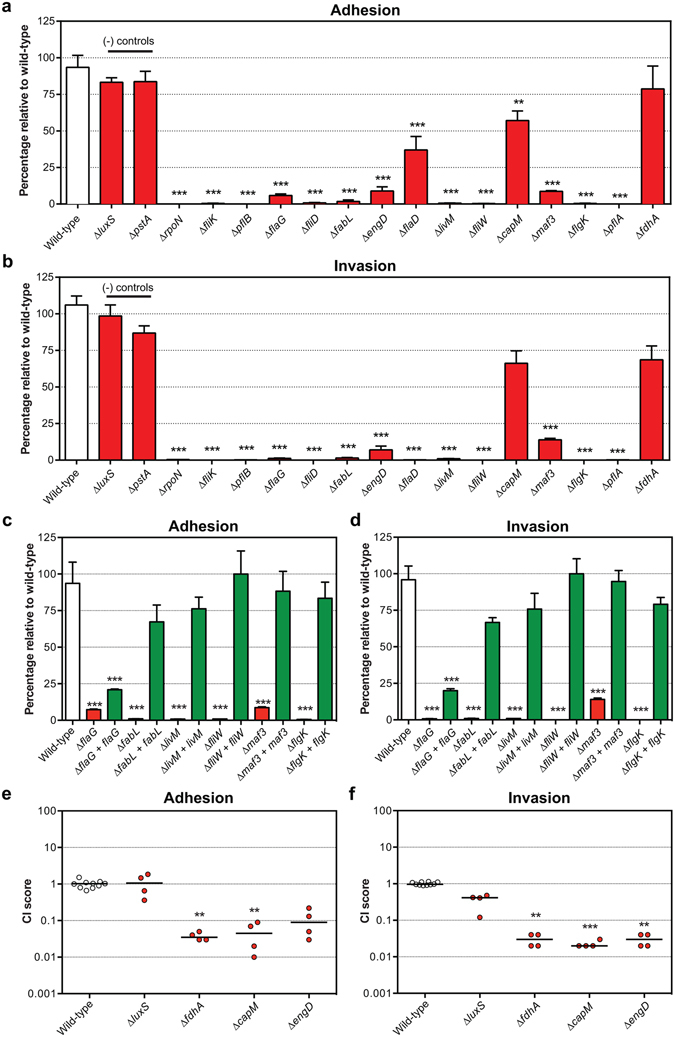
Validation of Tn mutant library screen during adhesion and invasion of human gut epithelial tissue culture cells. Adhesion to (a), and invasion (b) of, Caco-2 cells by C. jejuni M1cam defined gene deletion mutants. Mutants in luxS and pstA, which were not identified in our Tn-seq screen with Caco-2 cells, were included as negative controls and rpoN, which was previously shown to be required for adhesion and invasion81, 82, served as a positive control. Caco-2 adhesion (c), and invasion (d), of gene deletion and genetically complemented mutants. Data is represented as percentage of wild-type (n ≥ 3) and plotted as means and SEM. The competitive index (CI) was calculated by dividing the ratio of mutant to wild-type bacteria recovered upon (e) adhesion to, and (f) invasion of, Caco-2 cells by the ratio of mutant to wild-type bacteria that were used in the inoculum. Statistical significance was calculated using a Mann-Whitney test where *P < 0.05, **P < 0.01 and ***P < 0.001.
Genetic complementation of fabL, livM, fliW, maf3 and flgK mutants restored their adhesion and invasion capacity to wild type-levels, confirming their role in the interaction with human gut epithelial cells (Fig. 7c,d). Genetic complementation of the flaG mutant partially restored its phenotype, which may be due to deregulated fliD or fliS expression as we observed for the fliD mutant (Fig. S8). Complementation of the engD mutant was unsuccessful due to the lack of expression of engD (Fig. S8). Further, the capM and fdhA Tn-seq phenotypes were confirmed when mutants were assayed in competition with the wild-type (Fig. 7e,f), however in mono-infection no attenuation was observed (Fig. 7a–d). The luxS negative control mutant was not attenuated for adhesion and invasion in competition with the wild-type (Fig. 7e,f).
Discussion
Here we present a comprehensive analyses of gene fitness in C. jejuni. Our Tn-seq data identified 486 genes that are essential for fitness in C. jejuni. When compared with the literature, till date, mutants in 38 out of 486 genes have been reported. Although some of these mutants had a severe growth defect, such discrepancies could arise due to the differences in culture conditions or culture medium while generating transposon libraries, or the presence of competition with other Tn mutants (which is not the case during the generation of defined gene deletion mutants). Potential drawbacks of various experimental designs and how to improve Tn-seq based methods have been discussed in great detail in a recent review by Chao et al.46. Profiling the genes required for in vitro growth of three C. jejuni strains underscored that a large part of the genome (~27%) is vital to fitness (Table S2). Most likely as a consequence of a low redundancy in its relatively minimal genome and transcriptional coupling of genes47, 48. This implies that there could be opportunities for targeting some of these genes for novel intervention strategies, e.g. as previously reported by Mobegi et al.49.
Elements of the flagellar system, i.e. the flagellar base and periplasmic rod structure and T3SS (Fig. 8), which when inactivated by a Tn have a severe impact on fitness, provide a potential focus for intervention along with genes required for the LOS lipid A and KDO moieties. We also found that inclusion of the first L-glycero-D-manno-heptose residue (catalyzed by WaaC) was required for fitness. These structural elements have a critical role during host interaction and therefore represent promising targets for developing intervention strategies44, 50. Gene fitness analysis revealed that components of the gluconeogenesis pathway required for biosynthesis of glucose-(derivatives) that serve as building blocks for both LOS and CPS as well as the N- and O-linked protein glycosylation pathways51, were essential for the growth of C. jejuni. In addition, the type II fatty acid synthesis pathway (FASII) genes fabDFGHZ were required for fitness. Interestingly, there are several classes of FASII pathway inhibitors with potent antimicrobial properties and consequently fatty acid synthesis is also considered a lucrative target for antibiotics49, 52.
Figure 8.
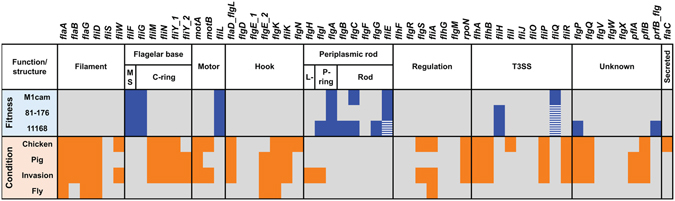
Overview of flagellar system genes required for fitness during in vitro growth and in model representing host interaction. Genes involved in the flagellar system that were required for fitness during in vitro growth of C. jejuni strains M1cam, 81–176 and 11168 are indicated in blue. Genes that passed the fitness score cut-off but did not pass the 0.95 probability for Tn inactivation are indicated with blue dashed boxes. Genes which in the Tn library screens were shown to be required for chicken colonisation, gnotobiotic piglet infection, invasion of human gut epithelial tissue culture cells or survival in houseflies are indicated in orange.
The same C. jejuni M1cam Tn library was screened in all of the experimental models presented in this study and compared with data derived from screening the same Tn library during infection of gnotobiotic piglets (De Vries et al., submitted). Twenty-eight genes were found to be required in all three host interaction models (chicken, pig, and cellular invasion), of which 21 genes belong to the flagellar system. Amongst the genes that were required across three host models and that did not belong to the flagellar system were engD, livM and capM (Figs 3 and 7). The EngD ortholog in E. coli (YchF) belongs to the GTPase family and is a negative regulator of the oxidative stress response53. YchF proteins have been implicated in pathogenesis of other bacterial species53, 54. With branched-chain amino acids (BCAA) being linked to chemotaxis55, livM, part of the ABC-type BCAA transport system (livM, livH, livK, and livJ), may be implicated in within host chemotaxis. CapM/PglH plays a role in the glycan assembly process and adds the final N-acetlygalactosamine residues during surface decoration56. Previous studies demonstrated a role for the N-linked protein glycosylation gene capM/pglH in chicken colonisation57, 58, along with other members of the surface protein glycosylation locus (pglBEF)59. Our chicken colonization Tn-seq screen showed that Tn mutants in pglB, pglF and pglI had reduced colonisation (Table S3), whereas pglACD were required for fitness during in vitro growth (Table S2).
Ninety-six genes were only identified in the chicken colonisation screen and not during infection of gnotobiotic piglets, including phosphate metabolism genes (pstABS and phoR). PstA appears to be unrelated to motility (Fig. S6), deletion of pstA did not affect adhesion and invasion of human gut epithelial cells (Fig. 7), but was required for C. jejuni to colonise chickens (Fig. 3a,b). The phosphate regulon in C. jejuni 81–176 is increased in 1-day-old chicks, which suggests that phosphate levels might be low in chickens, consequently activating the phosphate regulon35. The phosphate regulon might operate in a second messenger system that (in)directly regulates expression of chicken colonisation genes60. However, it remains to be investigated which C. jejuni genes are under control of this system. Tn mutants in genes involved in chemotaxis, mcp4_1 and cheV, were uniquely attenuated during chicken colonisation, whereas Tn mutants in mcp4_3 and mcp4_4 were uniquely attenuated during gnotobiotic piglet infection, suggesting the existence of host- or substrate-specific chemotaxis. A large number of genes involved in the utilisation of amino acids and organic acids such as lactate, pyruvate, acetate and tricarboxylic acid (TCA) cycle intermediates only appear to be required during gnotobiotic piglet infection. These host specific requirements are most likely related to dietary differences between chickens and piglets. Our Tn library screens indicated that pyruvate kinase (pyk) is important in both chickens and piglets. Pyk is part of the Embden-Meyerhof-Parnas pathway, and shifting the metabolic flux at the level of Pyk has previously been suggested as a potential antimicrobial strategy61.
A detailed analysis of the flagellar system across experimental models indicated that the extracellular part of the flagellum, the filament and hook structure, plays a vital role in C. jejuni (Fig. 8). This included the filament chaperone gene fliW 13, 62 and the flagellar cap fliD. A FliD subunit vaccine induced a transient but significant reduction (~2 log) of C. jejuni in the chicken caecum63, demonstrating its potential as a target for intervention. Linked to the flagellar system is also the motility accessory factor 3 (maf3). We found that a maf3 deletion mutant was motile, but displayed attenuated chicken colonisation and adhesion and invasion of Caco-2 cells. Although maf3 is located in the flagellar glycosylation locus64, deletion of maf3 in C. jejuni 81–176 did not result in an altered glycosylation of the flagellar filament65.
Experimental models of C. jejuni survival and transmission, i.e. in the housefly and under low temperature conditions, were less stringent than the in vivo and in vitro host-pathogen models, reflected by subtler fold-changes compared to the host interaction models. This results in a lower number of candidate genes being identified and a lower conformation in validation experiments with defined gene deletion mutants (Figs 4 and 5). Interestingly, this led to a novel potential target for intervention not linked to host interactions. The trxC gene was required for survival at low temperature, both in nutrient-rich and –poor conditions (Fig. 5) and for resistance against peroxide (Fig. 6), indicating a role in the oxidative stress response.
C. jejuni is considered the leading cause of bacterial gastroenteritis, however our understanding of its biology is limited. The development of future intervention strategies might best be aided by a thorough understanding of the biology of C. jejuni in its life cycle. Our work indicates that many genes/pathways make indispensable contributions to the ability of C. jejuni to thrive in the host and environment. We anticipate that the findings from this study and the use of the Tn mutant libraries in future studies will provide a continued insight into the mechanisms required for growth as well as survival within and outside host organisms. Most importantly, our comprehensive screening approach has again clearly shown that the flagella drive C. jejuni interaction with its hosts. Therefore, future efforts should focus on how to exploit this to effectively control infections caused by C. jejuni.
Methods
Bacterial strains and growth conditions
Wild-type strains, defined gene deletion mutants, genetically complemented mutants and plasmids are summarised in Table S6. C. jejuni strains M1cam (derivative of M1 used in our laboratory)13, 14, 1116810, and 81–17615 were cultured in Brain Heart Infusion (BHI) broth or on BHI agar supplemented with 5% (v/v) defibrinated horse blood in the presence of 5 µg/ml trimethoprim (TrM). Tn mutant libraries and defined gene deletion mutants were grown in the presence of 10 µg/ml chloramphenicol (Cm) and 50 µg/ml kanamycin (Km) was added for culturing genetically complemented mutants. C. jejuni were grown under microaerophilic conditions (5% O2, 10% CO2, 85% N2) in a MACS VA500 Variable Atmosphere Work Station (Don Whitley, Shipley, United Kingdom). Liquid cultures were grown with agitation (200 rpm). For use in the experimental models, C. jejuni M1cam wild-type, defined gene deletion mutants and genetically complemented mutants were cultured for ~48 h, re-plated on fresh plates and grown for another 16 h. Tn mutant libraries were grown from freezer stocks on 9 × 90 mm BHI blood agar plates (200 μl per plate) and grown for 16 h. E. coli NEB 5α or 10-β (New England Biolabs) were used for cloning and were cultured in Luria Bertani (LB) medium with appropriate antibiotics, at 37 °C.
Construction of defined gene deletion mutants and genetically complemented mutants
C. jejuni M1cam gene deletion mutants were constructed by allelic replacement of the gene with a chloramphenicol (cat) resistance cassette as described in de Vries et al.13. Gene deletion mutants were subjected to phenotypic and genotypic characterisation, including motility (Fig. S6) and in vitro growth in liquid culture (Fig. S7). Whole genome sequencing (WGS) based variant analysis (single nucleotide polymorphisms [SNP] and insertion/deletions [INDELS]) of defined deletion mutants was used to screen for second-site mutations that might have affected the phenotypes under investigation. Variants were detected at 129 positions relative to the C. jejuni M1cam reference genome13 (Table S5). The variant database was cross-referenced with data obtained in Tn mutant library screens to assess whether the gene affected by the variant had a potential impact on the phenotype under investigation. In addition, phenotypes of deletion mutants sharing a variant were compared to predict possible confounding effects of the second-site mutation. This in-depth analysis did not identify confounding effects of second-site mutations.
Genetic complementation of mcp4_1, fliK, flaG, ΔfliD, pstA, fabL, engD, livM, trxC, capM, maf3 and flgK mutants was performed using the pSV009 plasmid as described in de Vries et al.13 (Fig. S8). Predicted promoter region(s), including a ribosome-binding site (RBS) were derived from the Dugar et al. dataset47. For genes arranged in an operon, the promoter region was mostly identified upstream of the 5′ end of the transcript. These promoter regions were synthesised as a DNA string (GeneArt, Life Technologies, UK) and flanked with appropriate restriction sites. The fliK, fliD, pstA, engD and maf3 open reading frames (ORFs) were cloned into XbaI and BamHI sites in pSV009 and promoter regions were inserted in the XhoI and XbaI sites, resulting in the promoter-gene fusions. For mcp4_1, flaG, fabL, trxC, capM and flgK, a promoter consensus region was identified upstream of the start codon. A fragment containing the ORF plus ~200 bp upstream was cloned in the XhoI and BamHI sites of the plasmid pSV009. As no obvious promoter was identified for livM, its transcription was placed under the control of a cat promoter by cloning the ORF into XbaI and BamHI sites of pSV009. Genetic complementation fragments were amplified from the pSV009-derived plasmids using primers pSV009_GCampl _FW1/RV1. The genetic complementation fragments were introduced into respective M1cam defined gene deletion mutants by electroporation as described in de Vries et al.13. Oligonucleotide sequences are listed in Table S7.
Gene expression analysis in genetically complemented gene deletion mutants
Total RNA was isolated as previously described in de Vries et al.66. DNA-free total RNA (500 ng) was reverse transcribed into cDNA using the QuantiTect reverse transcription kit (Qiagen). Real-time quantitative PCR was performed using the SensiFast SYBR No-ROX Kit (Bioline) on a Rotor-Gene Q machine (Qiagen). Gene expression fold-changes in genetically complemented mutants were calculated relative to wild-type using the ΔΔCt method67 with gyrA as a reference (two biological replicates with two technical replicates).
Growth kinetics
C. jejuni wild-type and gene deletion mutants were harvested from BHI blood agar plates in BHI broth. Culture suspensions were diluted to OD600nm ~ 0.2 and used to inoculate 5 ml pre-warmed BHI broth at an OD600nm of ~0.005. Growth was monitored after 4, 8 and 24 h incubation at 200 rpm at 42 °C under microaerophilic conditions (n ≥ 3), by optical density and by plating ten-fold serial dilutions.
Analysis of motility
Motility assays (n = 3) were performed in semi-solid agar as described in de Vries et al.13.
Genome sequencing of defined gene deletion mutants
Sequencing libraries were prepared using the NEBNext Ultra or Ultra II DNA library prep kit (New England Biolabs) and sequenced on the MiSeq platform as described in de Vries et al.13. Reads were mapped to the M1cam reference genome (accession no. CP01214913) using Stampy68, variants were identified with Samtools69 and the effect at the protein level was predicted using SnpEff70. Deletion mutants in pflA and flaD were analysed previously13.
Construction of Tn mutant libraries in C. jejuni
A Tn donor plasmid suitable for Tn-seq16 was constructed by amplifying the mariner Tn encoding the Cm-resistance cassette from pAJG3929 using a single 5′-phosphorylated primer PBGSF20, introducing MmeI restriction sites within the inverted repeats of the Tn element. The Tn element was sub-cloned into pJET1.2 (Thermo Scientific) and the resulting plasmid pSV006, was used for in vitro Tn mutagenesis as described in Holt et al.71 with minor adjustments. Briefly, 2 μg of C. jejuni DNA was incubated for 5 h at 30 °C with 1 µg of pSV006 and ~250 ng Himar1-C9 transposase, which was purified as described in Akerley et al.72. After end-repair with T4 DNA polymerase and E. coli DNA ligase the mutagenesised DNA was transferred to C. jejuni by natural transformation13. Tn transformants were harvested from plates and pooled. The pooled library was used to inoculate 100–200 ml BHI-TrM-Cm broth to an OD600nm of ~0.1 and grown overnight), yielding “working stocks”. Genomic DNA was isolated with Genomic-tip columns (Qiagen).
Gene fitness analysis
Tn insertion sites were identified using Tn-seq16, as described in Burghout et al.73 with minor adjustments. Briefly, Tn mutant library DNA was fragmented by MmeI restriction digestion, adapters containing inline barcodes were ligated to the MmeI fragments, and amplified using the primers PBGSF29 and PBGSF30 with NEBNext high fidelity polymerase (New England Biolabs). Tn insertion sites were sequenced using single-end 40–50 bp sequencing on the Illumina HiSeq 2500. Sequence reads were demultiplexed using the FastX toolkit barcode splitter and analysed further with the ESSENTIALS pipeline19. Sequence reads were aligned to the C. jejuni genomes13, 15, 74 with a match of ≥16 nt. Kernel density plots were generated in R to distinguish “true” Tn insertions from “noise” sequencing reads, yielding a read count cut-off per Tn library. Insertion “hot spotting” was analysed by plotting the Log2 read count per chromosomal position using an in-house Perl script. As a measure of gene fitness, the Log2 fold-change of observed vs expected reads was calculated per gene, with Kernel density plots allowing accurate delineation of fitness (required for in vitro growth) and non-fitness genes19. Additional criteria were: a Benjamini & Hochberg adjusted P < 0.05 and a probability that the gene was inactivated by a Tn insertion of >0.95, as calculated using a derivative of Poisson’s law; 1 − eN x Ln(1−f), with N = number of unique Tn insertion mutants and f being the gene size divided by the size of the genome. In addition, genes for which no sequence reads were detected and the probability of inactivation was >0.95 were considered to be required for fitness. For functional class enrichment analysis COGs were assigned to M1cam, 11168 and 81–176 proteins and consensus COGs were assigned to homologous groups (HGs; see next section). The overrepresentation of COG classes was assessed using a Fisher exact test with Q-value multiple testing correction75.
Identification of homologs in C. jejuni strains
Protein sequences of C. jejuni M1cam, 11168 and 81–176 were clustered into putative HGs with OrthAgogue76. For this, a collective database was generated and proteins of each strain were queried against this database. The reciprocal best-hit protein pairs were identified by applying an e-value filter cut-off of 1e-5 to the “all against all” BLAST output (only proteins >50 amino acids in length were included). The putative HGs were identified by clustering with MCL with an inflation parameter of 2.677.
Chicken colonisation experiments
All chicken infection work was conducted in accordance with UK legislation governing experimental animals under project licence 40/3652 and was approved by the University of Liverpool ethical review process prior to the award of the licence. One-day-old Ross 308 broiler chicks were obtained from a commercial hatchery. Chicks were housed in the University of Liverpool, High Biosecurity Poultry unit. Chicks were individually tagged with leg rings or wing bands and maintained in floor pens at UK legislation recommended stocking levels allowing a floor space of 2,000 cm2 per bird at 25 °C on wood-shavings litter that was changed weekly prior to inoculation, and were given ad libitum access to water and a pelleted laboratory grade vegetable protein-based diet (SDS). Prior to experimental infection, all birds were confirmed as Campylobacter-free by taking cloacal swabs, which were streaked onto selective blood-free agar (mCCDA) (Lab M) supplemented with Campylobacter Enrichment Supplement (SV59) and grown for 48 h at 42 °C under microaerophilic conditions.
At 21 days of age, birds were inoculated by oral gavage with ~1.8 × 108 CFU C. jejuni M1cam Tn library ‘C’. The birds were split into five cages (n = 6, 7, 7, 6, 6; group 1–5, respectively). Six days post-inoculation (p.i.), birds were killed by cervical dislocation. At necroscopy the caeca were removed aseptically and caecal content was collected. Next, the caecal contents were diluted 5- and 50-fold with Maximum Recovery Diluent (MRD) and 0.5 ml was plated per mCCDA-Cm (10 plates in total per dilution) for recovery of Tn mutants that successfully colonised the birds. Ten-fold dilution series were plated on mCCDA-Cm for enumeration. After 2 days growth, C. jejuni Tn mutants were recovered from the plates by scraping in 2 ml MDR, pelleted by centrifugation and stored at −80 °C. Genomic DNA was isolated from plate harvest pellets with Genomic-tip columns (Qiagen).
The composition of the input Tn mutant library (n = 3) and the library recovered per cage of birds (n = 4, cage 1–4), a cage was considered as a single unit of colonisation, was analysed by Tn-seq16. For this, 1 μg of DNA was pooled per group of birds (n = 6, 7, 7, 5 for groups 1 to 4, respectively). One bird from group 4 and 2 birds from group 5 could not be analysed due to heavy contamination on the recovery plates. As a result of this group 5 was eliminated from further analysis.
For validation with C. jejuni M1cam defined gene deletion mutants and genetically complemented mutants, birds were inoculated with ~1.8 × 108 CFU, at 6 days p.i. chickens were killed by cervical dislocation, the caeca were removed aseptically and the caecal contents plates onto mCCDA plates for enumeration.
Infection of houseflies
Newly pupated female houseflies (Musca domestica) were inoculated with 1 μl C. jejuni suspension via their proboscis78. For screening of Tn mutant survival, 5 groups of 10 flies were inoculated with ~106 CFU M1cam Tn library ‘C’ on four different days. The flies were incubated for 4 h at 20 °C (in the dark) after which flies were homogenized using a Drigalsky spatula and 5 ml BHI was added. Large debris was removed through a low speed (700 × g) spin and the supernatant was adjusted to 5 ml with BHI before plating 0.5 ml per mCCDA-Cm (10 plates in total). Ten-fold dilution series were plated on mCCDA-Cm for enumeration. After 24 h growth, Tn mutants were recovered from plates by scraping in 2 ml BHI medium, centrifuged and pellets were stored at −80 °C for DNA isolation with Genomic-tip columns (Qiagen). Chromosomal DNA from the 5 groups of 10 flies per day was pooled; resulting in a single pooled sample for each 4 replicate. The composition of the input Tn mutant library (n = 4) and the library recovered per group of flies (n = 4) was analysed by Tn-seq16.
For validation experiments with C. jejuni M1cam gene deletion mutants (n ≥ 4) and genetically complemented mutants (n = 6), groups of 5 flies were inoculated with ~106 CFU, killed 4 h p.i., and the bacterial load was quantified from pools of five flies or five flies individually (only for 2 replicates with gene deletion mutants), in both cases the average CFU per fly was used to calculate the Log10 decrease of CFU per fly relative to the inoculum.
Cold survival assay
A suspension was prepared from C. jejuni M1cam Tn mutant library ‘C’ plate harvests, to an OD600nm ~ 0.5 (~5 × 108 CFU/ml). The Tn mutant library suspension was harvested by centrifugation and resuspended in either BHI or sterile tissue-culture grade water (experiment 1) or chicken juice, tap water or rain water (experiment 2). The chicken juice was prepared as follows: 10 frozen whole chickens were purchased from a commercial supplier and were allowed to defrost within the packaging for at least 16 h, as described by Brown et al.79. Concentrated liquid was recovered (>200 ml), sterilised through a 0.2 μM filter and stored at −20 °C. Tap water was obtained from a mains-fed tap in our laboratory that was allowed to run for at least 2 min prior to collecting the water used in experiments. To collect rain water, a large tub was left outside our laboratory overnight on a rainy evening. The rain water and tap water were passed through a 0.22 µM filter unit and stored at 4 °C. Chicken juice, tap water and rain water were screened for the presence of Campylobacter spp. by plating on mCCDA plates. The Tn library samples were incubated at 4 °C and aliquots were taken at 0 h, 6 h, 1 and 7 days for experiment 1 (n = 3) or at 1, 3, and 7 days for experiment 2 (n = 4) and plated for recovery of Tn mutants. Ten-fold dilution series were plated on BHI-TrM-Cm for enumeration. After 2 days incubation, Tn mutants were recovered from plates by scraping in 2 ml BHI per plate, centrifuged and pellets were stored at −80 °C for DNA isolation with Genomic-tip columns (Qiagen). The Tn library composition at time = 0 h was compared to the library recovered after incubation at 4 °C under the above mentioned conditions by Tn-seq16.
For validation experiments, the survival of gene deletion mutants and genetically complemented mutants was assayed as described above (n = 4).
Infection of human gut epithelial cells
Caco-2 cells (ATCC CC-L244 HTB-37), were cultured in DMEM (Life Technologies) supplemented with 10% (v/v) heat inactivated FBS (Gibco), and 1% (v/v) non-essential amino acids (Sigma Aldrich), at 37 °C with 5% CO2. The C. jejuni M1cam Tn mutant library ‘C’ was used to infect seven 143 cm2 dishes per replicate (n = 4) containing a monolayer of Caco-2 cells at a multiplicity of infection (MOI) of 100 in low phosphate HEPES buffer (10 mM HEPES, 5.4 mM KCl, 145 mM NaCl, 5 mM glucose, 1 mM MgCl2, 1 mM CaCl2, 2 mM phosphate buffer pH 7.4)80. Cells were incubated for 2 h after which the non-adherent fraction from two dishes was recovered onto BHI blood agar plates. For adherence, two dishes were washed three times in Dulbecco’s PBS (D-PBS), Caco-2 cells were lysed in 10% (v/v) Triton-X100 solution in D-PBS, and bacteria were recovered on BHI blood agar plates. The remaining five dishes were washed three times in D-PBS and incubated for an additional 2 h in DMEM with 250 µg/ml gentamycin. Tn mutants that invaded Caco-2 cells were recovered after washing three times in D-PBS and Caco-2 cell lysis in 10% (v/v) Triton-X100 in D-PBS. After two days of growth on plates, Tn mutants were recovered from the plates, centrifuged and pellets were stored at −80 °C. DNA was isolated from harvested pellets using Genomic-tip columns (Qiagen).
Validation of the screen was performed using 24-well plates, for which Caco-2 cells were infected with C. jejuni M1cam defined gene deletion mutants and genetically complemented strains (n ≥ 4) as described above. Bacteria recovered from different fractions were serially diluted and plated on BHI agar plates containing appropriate antibiotics. Adhesion and invasion of Caco-2 cells by various C. jejuni strains was calculated relative to the matched wild-type (n ≥ 3). To test the effect of competition, Caco-2 cells were infected at an MOI of 100 with a mix of the wild-type strain to a defined gene deletion mutant at a ratio of 100:1. A competitive index (CI) score was calculated by dividing the ratio of mutant to wild-type recovered from adherent and invaded fractions by the ratio of mutant to wild-type bacteria in the inoculum (n ≥ 4).
Conditionally essential gene analyses
Tn-seq data from the different experimental models (conditionally essential genes screens) was processed as described in “gene fitness analysis”. To identify genes of which Tn mutants were attenuated or enriched in the tested models, read counts were collected per gene and compared between output (recovered) and the input or control conditions (as defined above). Only genes covered by >100 reads in the control condition were considered, allowing assessment of 809 ± 58 (67 ± 5%) non-fitness genes (define ‘non-fitness genes’) in the selected models. The following filter steps were applied: a Log2 fold-change (FC) below the attenuated cut-off value or higher than the enriched cut-off value, Benjamini & Hochberg false discovery rate <0.05, and two or more Tn mutants showing a Log2 fold-change below the attenuated cut-off value or higher than the enriched cut-off value (analysed using a custom Python script). The Log2 fold-change cut-offs were selected based on MA-plots. In addition, 514 genes that were obligate essential or required for fitness (defined above) were eliminated from the analysis, see “gene fitness analysis”. COG functional class enrichment was analysed using a Fisher exact test with Q-value multiple testing correction75.
Hydrogen peroxide sensitivity assay
C. jejuni strains were added to pre-cooled BHI agar (~45 °C) to a calculated OD600nm ~0.005 and 25 ml of this media-bacterial suspension was poured into 90 mm petri dishes. Blank filter paper discs, 6 mm, were loaded with 10 µl of 0.05, 0.1, 0.25, 0.5, 1, 2.5, 5 or 7.5 M H2O2 solution. The discs were allowed to air dry and were then placed in the center of the solidified agar plates. The plates were incubated for 24 h, after which time the inhibition zone diameter was measured using a ruler (n = 4).
Nucleotide sequence accession numbers
Tn-seq and genome sequencing data has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) and are available via study accession numbers PRJEB18797 and PRJEB18765, respectively.
Statistical analysis
Statistical analysis was performed in GraphPad Prism v6.
Electronic supplementary material
Acknowledgements
This work was funded by Biotechnology and Biological Sciences Research Council (http://www.bbsrc.ac.uk) grant BB/K004514/1. D.P.W. was funded by a Wellcome Trust (https://wellcome.ac.uk) Infection and Immunity PhD rotation studentship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interests exist. We thank Gemma Murray from the Department of Genetics, University of Cambridge for providing the R script used for COG assignment and Diane Newell for providing the C. jejuni M1 strain. We thank David Lampe for providing the pMALC9 plasmid and Roy Chaudhuri for sharing the read count mapping Perl script.
Author Contributions
Conceived and designed the experiments S.P.d.V., S.G., A.N.J., N.W., P.W., T.H., D.J.M., A.J.G.; Performed the experiments S.P.d.V., S.G., E.W., A.W., A.N.J., L.L.L., S.H., H.S., K.M., E.P., D.P.W., J.L’H., D.G.E.S., P.E., N.W., P.W.; Analysed the data S.P.W.d.V., S.G., A.B., A.N.J., F.M.M., A.Z., A.J.G.; Wrote the paper: S.P.W.d.V., S.G., A.G.; All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Stefan P. de Vries and Srishti Gupta contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01133-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Havelaar AH, et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphrey T. Are happy chickens safer chickens? Poultry welfare and disease susceptibility. Br. Poult. Sci. 2006;47:379–391. doi: 10.1080/00071660600829084. [DOI] [PubMed] [Google Scholar]
- 3.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 4.Bahrndorff S, Gill C, Lowenberger C, Skovgard H, Hald B. The effects of temperature and innate immunity on transmission of Campylobacter jejuni (Campylobacterales: Campylobacteraceae) between life stages of Musca domestica (Diptera: Muscidae) J. Med. Entomol. 2014;51:670–677. doi: 10.1603/ME13220. [DOI] [PubMed] [Google Scholar]
- 5.Evers EG, Blaak H, Hamidjaja RA, de Jonge R, Schets FM. A QMRA for the transmission of ESBL-producing Escherichia coli and Campylobacter from poultry farms to humans through flies. Risk Anal. 2016;36:215–227. doi: 10.1111/risa.12433. [DOI] [PubMed] [Google Scholar]
- 6.Gill, C., Bahrndorff, S. & Lowenberger, C. Campylobacter jejuni in Musca domestica: An examination of survival and transmission potential in light of the innate immune responses of the house flies. Insect Sci., doi:10.1111/1744-7917.12353 (2016). [DOI] [PubMed]
- 7.Hald B, Sommer HM, Skovgard H. Use of fly screens to reduce Campylobacter spp. introduction in broiler houses. Emerg. Infect. Dis. 2007;13:1951–1953. doi: 10.3201/eid1312.070488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronowski C, James CE, Winstanley C. Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 2014;356:8–19. doi: 10.1111/1574-6968.12488. [DOI] [PubMed] [Google Scholar]
- 9.Murphy C, Carroll C, Jordan KN. Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 2006;100:623–632. doi: 10.1111/j.1365-2672.2006.02903.x. [DOI] [PubMed] [Google Scholar]
- 10.Parkhill J, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 11.Haddad N, et al. Adhesion ability of Campylobacter jejuni to Ht-29 cells increases with the augmentation of oxidant agent concentration. Curr. Microbiol. 2010;61:500–505. doi: 10.1007/s00284-010-9644-z. [DOI] [PubMed] [Google Scholar]
- 12.Zilbauer M, Dorrell N, Wren BW, Bajaj-Elliott M. Campylobacter jejuni-mediated disease pathogenesis: an update. Trans. R. Soc. Trop. Med. Hyg. 2008;102:123–129. doi: 10.1016/j.trstmh.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 13.de Vries SP, et al. Motility defects in Campylobacter jejuni defined gene deletion mutants caused by second-site mutations. Microbiology. 2015;161:2316–2327. doi: 10.1099/mic.0.000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friis C, et al. Genomic characterization of Campylobacter jejuni strain M1. PLoS One. 2010;5:e12253. doi: 10.1371/journal.pone.0012253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofreuter D, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacon DJ, et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81–176. Infect. Immun. 2002;70:6242–6250. doi: 10.1128/IAI.70.11.6242-6250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology. 2004;150:3507–3517. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- 19.Zomer A, Burghout P, Bootsma HJ, Hermans PWM, van Hijum SA. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One. 2012;7:e43012–29. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogoma T, von Meyenburg K. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 1983;2:463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston C, et al. Natural genetic transformation generates a population of merodiploids in Streptococcus pneumoniae. PLoS Gen. 2013;9:e1003819. doi: 10.1371/journal.pgen.1003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao B, Lara-Tejero M, Lefebre M, Goodman AL, Gálan JE. Novel components of the flagellar system in epsilonproteobacteria. mBio. 2014;5:e01349–01314. doi: 10.1128/mBio.01349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JG, Livny J, Dirita VJ. High-throughput sequencing of Campylobacter jejuni insertion mutant libraries reveals mapA as a fitness factor for chicken colonization. J. Bacteriol. 2014;196:1958–1967. doi: 10.1128/JB.01395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metris A, Reuter M, Gaskin DJ, Baranyi J, van Vliet AH. In vivo and in silico determination of essential genes of Campylobacter jejuni. BMC Genomics. 2011;12:535. doi: 10.1186/1471-2164-12-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahl M, Stintzi A. Identification of essential genes in C. jejuni genome highlights hyper-variable plasticity regions. Funct. Integr. Genomics. 2011;11:241–257. doi: 10.1007/s10142-011-0214-7. [DOI] [PubMed] [Google Scholar]
- 26.Velayudhan J, Kelly DJ. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology. 2002;148:685–694. doi: 10.1099/00221287-148-3-685. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs-Reitsma WF, van de Giessen AW, Bolder NM, Mulder RW. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol. Infect. 1995;114:413–421. doi: 10.1017/S0950268800052122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conlan AJ, Coward C, Grant AJ, Maskell DJ, Gog JR. Campylobacter jejuni colonization and transmission in broiler chickens: a modelling perspective. J. R. Soc. Interface. 2007;4:819–829. doi: 10.1098/rsif.2007.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant AJ, et al. Signature-tagged transposon mutagenesis studies demonstrate the dynamic nature of cecal colonization of 2-week-old chickens by Campylobacter jejuni. Appl. Environ. Microbiol. 2005;71:8031–8041. doi: 10.1128/AEM.71.12.8031-8041.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coward C, et al. Competing isogenic Campylobacter strains exhibit variable population structures in vivo. Appl. Environ. Microbiol. 2008;74:3857–3867. doi: 10.1128/AEM.02835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringoir DD, Szylo D, Korolik V. Comparison of 2-day-old and 14-day-old chicken colonization models for Campylobacter jejuni. FEMS Immunol. Med. Microbiol. 2007;49:155–158. doi: 10.1111/j.1574-695X.2006.00181.x. [DOI] [PubMed] [Google Scholar]
- 32.Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. USA. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey S, et al. Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio. 2014;5:e01364-14–e01364-14. doi: 10.1128/mBio.01364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringoir DD, Korolik V. Colonisation phenotype and colonisation potential differences in Campylobacter jejuni strains in chickens before and after passage in vivo. Vet. Microbiol. 2003;92:225–235. doi: 10.1016/S0378-1135(02)00378-4. [DOI] [PubMed] [Google Scholar]
- 35.Taveirne ME, Theriot CM, Livny J, DiRita VJ. The complete Campylobacter jejuni transcriptome during colonization of a natural host determined by RNAseq. PLoS One. 2013;8:e73586. doi: 10.1371/journal.pone.0073586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kay D, et al. The microbiological quality of seven large commercial private water supplies in the United Kingdom. Journal of water and health. 2007;5:523–538. doi: 10.2166/wh.2007.042. [DOI] [PubMed] [Google Scholar]
- 37.Kulthanan K, Nuchkull P, Varothai S. The pH of water from various sources: an overview for recommendation for patients with atopic dermatitis. Asia Pacific allergy. 2013;3:155–160. doi: 10.5415/apallergy.2013.3.3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beales N. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Comp. Rev. Food Sci. Food Safety. 2004;3:1–20. doi: 10.1111/j.1541-4337.2004.tb00057.x. [DOI] [PubMed] [Google Scholar]
- 39.Stintzi A. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 2003;185:2009–2016. doi: 10.1128/JB.185.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handley RA, et al. PerR controls oxidative stress defence and aerotolerance but not motility-associated phenotypes of Campylobacter jejuni. Microbiology. 2015;161:1524–1536. doi: 10.1099/mic.0.000109. [DOI] [PubMed] [Google Scholar]
- 41.Bras AM, Chatterjee S, Wren BW, Newell DG, Ketley JM. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J. Bacteriol. 1999;181:3298–3302. doi: 10.1128/jb.181.10.3298-3302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stintzi A, Whitworth L. Investigation of the Campylobacter jejuni cold-shock response by global transcript profiling. Genome Letters. 2003;2(10):18–27. [Google Scholar]
- 43.Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15:456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Novik V, Hofreuter D, Galan JE. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 2010;78:3540–3553. doi: 10.1128/IAI.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao MC, Abel S, Davis BM, Waldor MK. The design and analysis of transposon insertion sequencing experiments. Nat. Rev. Microbiol. 2016;14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dugar G, et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Gen. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porcelli I, Reuter M, Pearson BM, Wilhelm T, van Vliet AH. Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria. BMC Genomics. 2013;14:616. doi: 10.1186/1471-2164-14-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mobegi FM, et al. From microbial gene essentiality to novel antimicrobial drug targets. BMC Genomics. 2014;15:958. doi: 10.1186/1471-2164-15-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwata T, et al. Effects of lipooligosaccharide inner core truncation on bile resistance and chick colonization by Campylobacter jejuni. PLoS One. 2013;8:e56900. doi: 10.1371/journal.pone.0056900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofreuter D. Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2014;4:137. doi: 10.3389/fcimb.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons JB, Rock CO. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol. 2011;14:544–549. doi: 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenk M, et al. A universally conserved ATPase regulates the oxidative stress response in Escherichia coli. J. Biol. Chem. 2012;287:43585–43598. doi: 10.1074/jbc.M112.413070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Vries SP, et al. Deciphering the genetic basis of Moraxella catarrhalis complement resistance: a critical role for the disulphide bond formation system. Mol. Microbiol. 2014;91:522–537. doi: 10.1111/mmi.12475. [DOI] [PubMed] [Google Scholar]
- 55.Rahman H, et al. Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog. 2014;10:e1003822. doi: 10.1371/journal.ppat.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glover KJ, Weerapana E, Imperiali B. In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. USA. 2005;102:14255–14259. doi: 10.1073/pnas.0507311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones MA, et al. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 2004;72:3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karlyshev AV, et al. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology. 2004;150:1957–1964. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- 59.Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 60.D’Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 61.Murima P, McKinney JD, Pethe K. Targeting bacterial central metabolism for drug development. Chem. Biol. 2014;21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 62.Radomska KA, Ordonez SR, Wosten MM, Wagenaar JA, van Putten JP. Feedback control of Campylobacter jejuni flagellin levels through reciprocal binding of FliW to flagellin and the global regulator CsrA. Mol. Microbiol. 2016;102:207–220. doi: 10.1111/mmi.13455. [DOI] [PubMed] [Google Scholar]
- 63.Chintoan-Uta C, Cassady-Cain RL, Stevens MP. Evaluation of flagellum-related proteins FliD and FspA as subunit vaccines against Campylobacter jejuni colonisation in chickens. Vaccine. 2016;34:1739–1743. doi: 10.1016/j.vaccine.2016.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlyshev AV, Linton D, Gregson NA, Wren BW. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology. 2002;148:473–480. doi: 10.1099/00221287-148-2-473. [DOI] [PubMed] [Google Scholar]
- 65.Guerry P, et al. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 2006;60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Vries SP, et al. Genome analysis of Moraxella catarrhalis strain BBH18, a human respiratory tract pathogen. J. Bacteriol. 2010;192:3574–3583. doi: 10.1128/JB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 68.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holt JP, Grant AJ, Coward C, Maskell DJ, Quinlan JJ. Identification of Cj1051c as a major determinant for the restriction barrier of Campylobacter jejuni strain NCTC11168. Appl. Environ. Microbiol. 2012;78:7841–7848. doi: 10.1128/AEM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akerley BJ, Lampe DJ. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 2002;358:100–108. doi: 10.1016/S0076-6879(02)58082-4. [DOI] [PubMed] [Google Scholar]
- 73.Burghout P, et al. Streptococcus pneumoniae folate biosynthesis responds to environmental CO2-levels. J. Bacteriol. 2013;195:1573–1582. doi: 10.1128/JB.01942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gundogdu O, et al. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics. 2007;8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ekseth OK, Kuiper M, Mironov V. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics. 2014;30:734–736. doi: 10.1093/bioinformatics/btt582. [DOI] [PubMed] [Google Scholar]
- 77.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skovgard H, Kristensen K, Hald B. Retention of Campylobacter (Campylobacterales: Campylobacteraceae) in the house fly (Diptera: Muscidae) J. Med. Entomol. 2011;48:1202–1209. doi: 10.1603/ME11061. [DOI] [PubMed] [Google Scholar]
- 79.Brown HL, et al. Chicken juice enhances surface attachment and biofilm formation of Campylobacter jejuni. Appl. Environ. Microbiol. 2014;80:7053–7060. doi: 10.1128/AEM.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Putten JP, Duensing TD, Cole RL. Entry of OpaA+ gonococci into HEp-2 cells requires concerted action of glycosaminoglycans, fibronectin and integrin receptors. Mol. Microbiol. 1998;29:369–379. doi: 10.1046/j.1365-2958.1998.00951.x. [DOI] [PubMed] [Google Scholar]
- 81.Elvers KT, Park SF. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology. 2002;148:1475–1481. doi: 10.1099/00221287-148-5-1475. [DOI] [PubMed] [Google Scholar]
- 82.Fernando U, Biswas D, Allan B, Willson P, Potter AA. Influence of Campylobacter jejuni fliA, rpoN and flgK genes on colonization of the chicken gut. Int. J. Food Microbiol. 2007;118:194–200. doi: 10.1016/j.ijfoodmicro.2007.07.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


