SUMMARY
mTORC1 inhibitors were first approved for the use in metastatic kidney cancer. However, observed treatment benefit was highly heterogeneous among patients. Through case-based cancer genomic sequencing of therapeutic outliers, we can begin to appreciate the convergent evolution of given cancer pathways/phenotypes beyond genes in kidney cancer, like a braided river.
In this issue of Clinical Cancer Research, Kwiatkowski and colleagues (1) report findings correlating mutations in TSC1, TSC2, and MTOR with response to rapalogs in patients with metastatic renal cell carcinoma (mRCC). The authors analyzed 560 cancer genes from tumor tissues of 79 mRCC patients treated with mTOR inhibitors who displayed distinct clinical outcomes. Response was defined as either partial response, or stable disease with any tumor shrinkage for at least 6 months, and non-response was progressive disease within the first 3 months of therapy. Based on such definitions, 43 responders and 36 non-responders were included. By focusing on somatic mutations of the 5 core mTOR pathway genes (MTOR, TSC1, TSC2, PTEN, and PIK3CA), these authors found that each of these five gene mutations were more frequently detected in responders than non-responders. This study added significant support to the available next generation sequencing (NGS) literature suggesting that TSC1 and TSC2 loss-of-function, or MTOR activating mutations could predict therapeutic benefits to Everolimus or Temsirolimus in various cancer types (2). Intriguingly, this study also demonstrated that most (56%) responders did not and some (22%) non-responders did carry these mutations, respectively, explaining why only marginal statistical significance was observed.
What can we learn from the cancer genomics of these outliers in terms of deciphering individual cancer pathobiology and possibly making treatment decision? We cancer doctors/researchers need to better leverage NSG data with a few caveats in mind. The first remark is “Number Matters”, i.e. sequencing coverage, copy number, and how many regions sequenced are important determinants for the value of individual mutations. High-coverage and multi-regional sequencing better capture the genomic landscape of analyzed tumors (3, 4). The second remark is “Frequency Matters”, i.e. varied allelic frequencies of mutations connote important therapeutic significance. Reported mutation frequencies of a given gene are heavily influenced by stromal contribution and clonal evolution. For example, clear cell renal cell carcinoma (ccRCC) is in principle a VHL mutated disease (5). VHL mutations represent the truncal mutation event. Hence, the allelic frequency of VHL mutation can be used to gauge tumor purity. By assessing allelic frequencies of co-detected mutations within the same tumor sample one could assess the clonal composition within the tumor. The third remark is “Position Matters”, i.e. not all missense mutations are the same. The position of detected missense mutations and corresponding amino acid changes could either represent a real gain-of-function or simply a passenger mutation. For example, missense mutations clustered within the FAT or Kinase domains are likely activating mutations whereas those scattered thorough the HEAT domains are likely passenger mutations (5–7). The fourth remark is “Site Matters”, i.e. sequencing obtaining from primary or metastatic tumors of the same patient carries different therapeutic significance. For example, the primary tumors of ccRCC tend to be fairly large, and encompass tens/hundreds or more subclones (3). Which clone(s) eventually metastasize and take life of the afflicted patient is likely miss- or under-represented if only a small piece of the primary tumors was sequenced. The fifth remark is “Time Matters”, i.e. the chronology of samples acquired for sequencing is important. For example, cancer genomics before treatment could offer prognostic/predictive values, whereas genomics after treatment likely offer clues concerning adaptive resistance mechanism. Incorporating these NGS “Matters” with corresponding therapeutic outcomes, we can now attempt reconciling these seemingly contradictory results about mTOR pathway mutations detected in both responders and non-responders.
One key lesson learned from studying targeted therapeutic outliers of a given cancer type is the recurrent theme about convergent evolution on select sets of oncogenic pathways (2, 6). More importantly, such pathway or phenotype convergences take places within the same tumor, among tumors of the same patient, and probably shared by the same histopathological subtype of given cancer types despite intra- or inter-tumor heterogeneity (2). For example, mTOR pathway activation due to either MTOR activation mutations or TSC1 loss-of-function mutations occurred at high frequencies in ccRCC where VHL mutation serves as the universal tumor-initiating event. This could explain why the two main categories of targeted therapeutic agents approved for the treatment of metastatic ccRCC are inhibitors of vascular endothelial growth factor (VEGF) or mTOR signaling pathways (8). It also supports the notion that cancer metabolism plays key roles in ccRCC pathogenesis (9). As VHL-loss and the resulting HIF hyperactivation are nearly universal in the pathogenesis of ccRCC, it would predict VEGF inhibitors such as Sunitinib, Pazopanib, and Axitinib to be more efficacious than mTOR inhibitors such as Everolimus and Temsirolimus in ccRCC (10). This was indeed supported by multiple randomized clinical trials comparing these two kinds of agents in mRCC. For example, Record-3 (11) a randomized trial comparing Sunitinib with Everolimus in previously untreated mRCC patients demonstrated median progression-free survival (PFS) with Sunitinib at 10.7 months and Everolimus at 7.9 months.
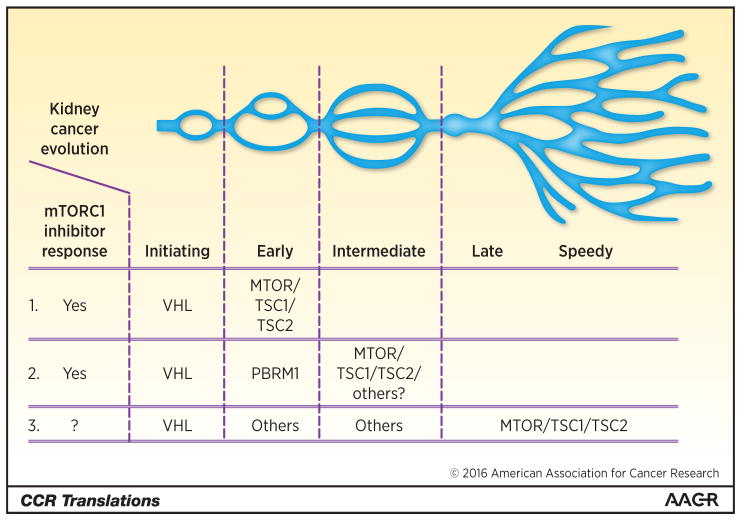
On the other hand, some RCC patients experienced markedly longer survival on mTOR inhibitors. To help improve depiction of cancer evolution and advise therapeutic selection of targeted agents, we proposed a novel “braided river model” (2). This model illustrates parallel and convergent events occurring throughout tumorigenesis. Starting from initiating driver mutations, it depicts the stepwise acquisition of different driver mutations (early, intermediate, late and speedy drivers) during cancer evolution. In ccRCC, PBRM1, a chromatin remodeling SWI/SNF complex gene, is the second most commonly mutated gene (~45%) (5). It was reported that RCC tumors with mutant PBRM1 associates with longer PFS on Everolimus at 11.1 months than those with wild-type PBRM1 at 5.3 months (12). It implicated that mTOR pathway activation through various means not limited to mutations in MTOR/TSC1/TSC2 could be the preferred route for ccRCC pathogenesis after mutations of VHL and PBRM1.
Here, we employ three different ccRCC evolution scenarios that all start with VHL mutations and incorporate mTOR activation to illustrate the importance of knowing the spatiotemporal sequence of mTOR activation during pathogenesis, and surmise how sequences could impact treatment outcome (Fig. 1). Scenario (1) starts with VHL mutation, followed immediately by mTOR pathway mutation, which renders nearly all ccRCC subclones of this tumor addicted to the mTORC1 hyperactivity. Scenario (2) carries PBRM1 mutation as the early (second) driver event, followed by mTOR pathway activation as the intermediate (third) driver event, which renders major clones of this tumor sensitive to mTOR inhibitors. Scenario (3) starts with VHL mutation, followed by a few early, intermediate pathway convergences that do not involve PBRM1 mutations or direct activation of mTOR pathway, and only acquired mTOR pathway mutations late as late or speedy drivers, which renders limited or no benefit to mTOR inhibitors.
Figure 1.
Diagram exemplifies clear cell kidney cancer evolution initiated with the VHL mutation, which was followed by mTOR pathway activation that could take places at unique temporospatial sequence within individual tumors. The early acquisitions of mTOR pathway activation through either genetic or epigenetic means as illustrated scenarios (1) and (2) could render such tumors dependent on continuous mTOR pathway hyperactivation and thereby more susceptible to mTORC1 inhibitors. River branch image adapted from Wei and Hsieh (2).
Altogether, current report leaves us with the following conclusions. The advent of diagnostic molecular pathology in our clinics provides opportunity to study the RCC biology of individual cancers and can provide background to unusual outcomes. The armamentarium for advanced RCC continues to expand, targeted immunotherapy recently having been added as the third class of agents. With eight targeted agents approved for treating mRCC, it is self-evident that most patients will not receive all 8 drugs, and many patients may not even experience all three classes. Thus, the key question lies in front of us is “Can we sequence mTOR inhibitor therapy with VEGF or PD1 inhibitors in the near future based on cancer genomics?” The answer will be a positive one if we begin to incorporate NGS Matters into both trial-based and case-based studies.
Acknowledgments
Grant Support
J.J. Hsieh is supported by the NIH under award number R01CA138505, The J. Randall & Kathleen L. MacDonald Kidney Cancer Research Fund, and The Jill and Jeffrey Weiss Kidney Cancer Research Fund.
Footnotes
Disclosure of Potential Conflicts of Interest
M.H. Voss reports receiving commercial research grants from Bristol-Myers Squibb and Pfizer; speakers bureau honoraria from Novartis; and is a consultant/advisory board member for Bayer, Calithera BioSciences, Exelixis, GlaxoSmithKline, Natera, and Novartis. J.J. Hsieh reports receiving commercial research grants from Cancer Genetics, Novartis, and Pfizer and is as a consultant/advisory board member for Chugai Pharma, Eisai, and Novartis. No other potential conflicts of interest were disclosed.
References
- 1.Kwiatkowski DJ, Choueiri TK, Fay AP, Rini BI, Thorner AR, de Velasco G, et al. Mutations in TSC1, TSC2, and MTOR are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2016 Feb 1; doi: 10.1158/1078-0432.CCR-15-2631. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei EY, Hsieh JJ. A river model to map convergent cancer evolution and guide therapy in RCC. Nat Rev Urol. 2015;12:706–12. doi: 10.1038/nrurol.2015.260. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sankin A, Hakimi AA, Mikkilineni N, Ostrovnaya I, Silk MT, Liang Y, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 2014;3:1485–92. doi: 10.1002/cam4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakimi AA, Pham CG, Hsieh JJ. A clear picture of renal cell carcinoma. Nat Genet. 2013;45:849–50. doi: 10.1038/ng.2708. [DOI] [PubMed] [Google Scholar]
- 6.Voss MH, Hakimi AA, Pham CG, Brannon AR, Chen YB, Cunha LF, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res. 2014;20:1955–64. doi: 10.1158/1078-0432.CCR-13-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–63. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Jonasch E, Agarwal N, Beard C, Bhayani S, Bolger GB, et al. Kidney cancer, version 3.2015. J Natl Compr Cancer Netw. 2015;13:151–9. doi: 10.6004/jnccn.2015.0022. [DOI] [PubMed] [Google Scholar]
- 9.Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–16. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voss MH, Molina AM, Motzer RJ. mTOR inhibitors in advanced renal cell carcinoma. Hematol Oncol Clin North Am. 2011;25:835–52. doi: 10.1016/j.hoc.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, Barrios CH, Kim TM, Falcon S, Cosgriff T, Harker WG, et al. Phase II randomized trial comparing sequential first-line everolimus and second-line sunitinib versus first-line sunitinib and second-line everolimus in patients with metastatic renal cell carcinoma. J Clin Oncol. 2014;32:2765–72. doi: 10.1200/JCO.2013.54.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh JJ, Chen D, PW, Chen YB, Redzematovic A, Marker M. Identification of efficacy biomarkers in a large metastatic renal cell carcinoma (mRCC) cohort through next generation sequencing (NGS): results from RECORD-3. J Clin Oncol. 2015;33(suppl):abstr 4509. [Google Scholar]