Abstract
While histones are best known for DNA binding and transcription-regulating properties, they also have antimicrobial activity against a broad range of potentially pathogenic organisms. Histones are abundant in neutrophil extracellular traps, where they play an important role in NET-mediated antimicrobial killing. Here, we show anti-influenza activity of histones against both seasonal H3N2 and H1N1, but not pandemic H1N1. The arginine rich histones, H3 and H4, had greater neutralizing and viral aggregating activity than the lysine rich histones, H2A and H2B. Of all core histones, histone H4 is most potent in neutralizing IAV, and incubation with IAV with histone H4 results in a decrease in uptake and viral replication by epithelial cells when measured by qRT-PCR. The antiviral activity of histone H4 is mediated principally by direct effects on viral particles. Histone H4 binds to IAV as assessed by ELISA and co-sedimentation of H4 with IAV. H4 also induces aggregation, as assessed by confocal microscopy and light transmission assays. Despite strong antiviral activity against the seasonal IAV strains, H4 was inactive against pandemic H1N1. These findings indicate a possible role for histones in the innate immune response against IAV.
Keywords: Histones, innate immunity, influenza, pandemic, antimicrobial peptide
Introduction
Influenza virus is a human respiratory pathogen, responsible for major morbidity and mortality in yearly epidemics and pandemics. Influenza A viruses (IAVs) are particularly subject to frequent antigenic variations. Eighteen different hemagglutinin (HA) and 11 different neuraminidase (NA) subtypes of IAV have been identified so far 1 and these are capable of reassorting to generate new viral strains with pandemic potential. At present only the H3N2 and H1N1 subtypes are currently endemic in the human population. These IAV subtypes give rise to yearly epidemics as a result of minor variations in their envelope proteins. The presence of extensive animal reservoirs of IAV pose a constant threat of more radically novel viral strains entering the human population. In 2009 an H1N1 strain derived from re-assortment of human, avian and porcine gene segments was sufficiently different from seasonally circulating H1N1 strains to give rise to a pandemic.2 Other avian strains including H5N1, have infected humans through animal to human transmission generally with high mortality.1,3,4
Humans have evolved very complex and sophisticated innate immune barriers to IAV infection which serve to contain replication of novel viral strains during the first several days of infection, before adaptive immune responses develop.5 Our laboratory has focused specifically on the roles of soluble innate inhibitors of IAV in the respiratory tract, including lectin inhibitors (e.g. collectins like surfactant protein D and ficolins)6–8 and antimicrobial peptides (e.g. defensins and the cathelicidin LL-37),9–12 and on the role of neutrophils.13,14 Neutrophils are the predominant inflammatory cell initially recruited to the airway after IAV infection and they play an important role in containment of the virus.15
Neutrophils are directly activated by IAV and we recently showed that this results in neutrophils extracellular trap (NET) formation.13 NETs are a complex containing a mixture of DNA and several granular and cytoplasmic proteins (e.g. myeloperoxidase, elastase, histones and antimicrobial peptides), which are capable of trapping and killing a broad variety of pathogens. 6,17 All core histones, known as H2A, H2B, H3, and H4, are present in NETs and account for 70% of all NET-associated proteins.16 Histones are important in the anti-microbial function of NETs, as Abs against H2A and H2B prevent NET-mediated bacterial killing.17
The anti-bacterial activity of histones has been well investigated since the first report of their antimicrobial activity in 1942.18–25 Both full-length and fragmented histones display potent antimicrobial activity against a variety of both Gram-positive and Gram-negative bacteria. In addition, histones have been shown to be antimicrobial in the presence of fungi 26,27 and parasites.28 In contrast, the antiviral activity of histones is not as well studied. Calf and fowl histones reduce infectivity of Semliki virus,29 and histones can inhibit attachment of Norwalk virus by binding to both the viral particles and the cell membrane.30 In addition, histones can function as cytosolic sensors for viral dsDNA in human cells, resulting in activation of innate viral pathways and inhibition of viral multiplication upon recognition of viral nucleic acids.31
As IAV has been shown to stimulate in vitro and in vivo formation of NETs, it is likely that IAV can encounter histones during infection.13,32,33 We therefore investigated the anti-influenza infectivity of histones. Here, we report that arginine-rich histones possess potent anti-influenza activity against both seasonal H3N2 and H1N1 strains. In contrast, pandemic H1N1 of 2009 was not inhibited.
Materials and methods
Viral preparations
Philippines 82/H3N2 (Phil82) strain was kindly provided by Dr E Margot Anders (University of Melbourne, Melbourne, Australia) and grown in the chorioallantoic fluid of 10 d old chicken eggs and purified on a discontinuous sucrose gradient.14 For confocal microscopy and binding of Phil82 to H4, Phil82 was labeled with Alexa Fluor-594 using Alexa Fluor® 594 Protein Labeling Kit (Invitrogen, Carlsbad, CA). Aichi68/H3N2 was obtained from ATCC and prepared in eggs using similar techniques. For removal of sucrose, purified viruses were dialyzed against PBS and stored at −80°C. A/California/2009 H1N1 pandemic strain (Cal09), A/New York/2001 H1N1, Mex 1:7, Mex 2:6 were prepared by reverse genetics as previously described.6,9,34 The Mex 1:7 strain contains 7 gene segments from the seasonal NY01 strain and only the hemagglutinin (HA) from the Mexico 09/H1N1 pandemic strain. The Mex 2:6 strain is similar but has both the HA and neuraminidase (NA) from Mexico 09/H1N1. These proteins of Mexico 09 are nearly identical with those of Cal09.
Cell culture
MDCK (Madin-Darby canine kidney) cells were cultured in MEM medium (Corning, Corning, NY), supplemented with 10% FCS (Fisher Scientific, Pittsburgh, PA) and penicillin-streptomycin (PenStrep, Sigma Aldrich, St. Louis, MO). A549 (human lung epithelial carcinoma) cells were cultured in F12K medium (Corning, Corning, NY), supplemented with 10% FCS and PenStrep. All cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2 and passaged every 3–4d.
Infectious focus assay
A fluorescent focus assay was used to measure viral neutralization as previously described.35,36 In brief, MDCK cells and A549 cells were grown to monolayers of 100% confluency in 96-well plates (Corning). Cells were infected with IAV (MOI = 0.1) diluted in PBS in the presence of histones (Sigma Aldrich, St. Louis, MO and New England Biolabs, Ipswich, MA) in different concentrations at 37°C/5% CO2. After 45 min, plates were washed with MEM (MDCK) or F12K (A549) and incubated overnight at 37°C/5% CO2. IAV-infected cells were detected using nucleoprotein directed mAB (Millipore, Billerica, MA). IAV-infected cells were then counted using a fluorescent microscope. For some experiments, histone H4 was added for 45 min before or after infection with IAV.
LDH assay
Release of lactate dehydrogenase (LDH) was measured using LDH assay kit (Clontech, Mountain View, CA). Samples were added to cells in the same manner as with the fluorescent focus assays. LDH release was measured in cell supernatants after overnight incubation at 37°C/5% CO2. One hundred µl of cell supernatant was incubated with 100 µl reaction mixture prepared according to manufacturer’s instructions for 30min. Supernatant from cells incubated with lysis buffer (Roche, Branford, CT) was used as a positive control. Cell viability was obtained using the following formula: 100-(OD490sample-OD490negative control)/(OD490positive control - OD490negative control) × 100.
Hemagglutination (HA) inhibition assay
HA inhibition was measured by serially diluting histones in round bottom 96 well plates (Serocluster U-Vinyl plates; Costar, Cambridge, MA) using PBS as a diluent, followed by addition of a fixed concentration of IAV and human type O red cells as described.37
Neuraminidase (NA) inhibition assay
The neuraminidase (NA) assay was performed using a 2’-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) based influenza neuraminidase kit (Life technologies, USA) as per manufacturer’s instructions. The assay is based on quantitation of fluorogenic end product 4-methylumbelliferone released from non-fluorogenic MUNANA by neuraminidase. Various doses of peptides were incubated with IAV for 30 min, 37°C followed by addition of MUNANA substrate (provided with the kit). Samples were further incubated for another 1 h at 37°C. The reaction was then stopped and read using a POLARstar OPTIMA fluorescent plate reader (BMG Labtech, Durham NC).
Binding of IAV to H4
We first tested ability of IAV to bind to H4 coated on 96-well black clear-bottom plates (Scientific, Pittsburgh, PA). Plates were coated overnight with 10 µg/ml (890 nM) H4 and 15 mg/ml (890 nM) BSA (fraction V, fatty-acid-free and low endotoxin; Sigma Aldrich, St. Louis, MO) in PBS. For blocking, 100 µl of superblock (1:2 in H2O) (Pierce, Rockford, IL) was added to wells and immediately dumped, after which wells were incubated with 100 µl superblock (1:2 in H2O) for 5 min. Fifty µl of sample containing diluted Alexa Fluor 594-labeled Phil82 was added to each well for 45 min. Fluorescence was measured using a POLARstar OPTIMA fluorescent plate reader (BMG Labtek, Durham, NC).
We also tested binding of H4 to IAV by co-precipitation and SDS-PAGE. Two µg H4 was incubated with increasing amounts of Phil82 in PBS for 30 min at 37° C. After incubation, sample was centrifuged at 13,000 g for 15 min. Supernatant was transferred to another tube and all samples were reduced and denatured by incubating in β-mercaptoethanol and boiling of samples for 5 min at 95°C. Samples were separated by SDS-PAGE on Mini-Protean Any kD TGX gel (BIO-RAD, Hercules, CA). For staining, the gel was incubated with GelCode Blue Stain Reagent (Pierce, Rockford, IL) for 1 h.
Measurement of viral RNA
RNA was isolated from cell supernatant and cell lysate at 45 min or 24 h after infection (MOI = 0.1) using the Magmax viral RNA isolation kit (Applied Biosystems, Carlsbad, California). The yield and quality of RNA was assessed by NanoDrop spectrophotometer. cDNA was reverse transcribed from 200 ng of RNA using TaqMan reverse transcription reagents (Applied Biosystems, Carlsbad, California). Reaction mixture and cycle conditions were as indicated by manufacturers’ instructions. Expression of viral RNA was assessed using quantitative PCR as previously described using primers for the viral M protein.10 For conversion of the Ct values to RNA copy numbers/ml, a standard curve was plotted using the Ct values of the standard dilutions against the Log10 values of the known RNA copy numbers/ml. The Ct values of the samples were converted to Log10 values using the standard curve (y = ax + b with y as Ct value, a as slope and b as intercept). Log10 values of the samples were converted to RNA copy numbers/ml by calculating the antilog (10 log10 value).
Confocal microscopy
Alexa Fluor 594-labeled Phil82 was pre-incubated with histone H4 for 30min and added to MDCK cells. After 45 min, cells were washed and fixed with 1% paraformaldehyde. Wheat-germ agglutinin (WGA)-Oregon green 488 and DAPI 350 were used to stain the cell membrane and nucleus, respectively. Confocal microscopy was carried out with a Zeiss LSM510 microscope (LSEB) at [100×] resolution.
Measurement of viral aggregation by light transmission and electron microscopy
Viral aggregation caused by histones was measured by assessing light absorbance at 350 nM by suspensions of IAV. This was done using a Perkin Elmer Lambda 35 UV/Vis spectrophotometer as described.6 In addition, viral aggregation was assessed using electron microscopy (EM) as described.6 In brief, histones were incubated with IAV at 37°C for 30min, and a 4 µl sample was placed on each copper grid. After the unbound virus was blotted off, the grid was fixed with 4 µl of 2.5% glutaraldehyde for 5 min. Samples were stained with 1% sodium phosphotungstate (pH 7.3, Sigma-Aldrich, St. Louis, MO) for 10 s, and excess stain was blotted off. The grids were then air dried and stored in a grid box until examined with a Phillips 300 electron microscope (Rahwah, NJ).
Statistics
Statistical analysis was performed using the GraphPad Prism 6 software for Mac. Comparisons were made with the Kruskal-Wallis one-way analysis of variance and p values of <0.05 were taken as statistically significant.
Results
Histones neutralize H3N2 viral strains
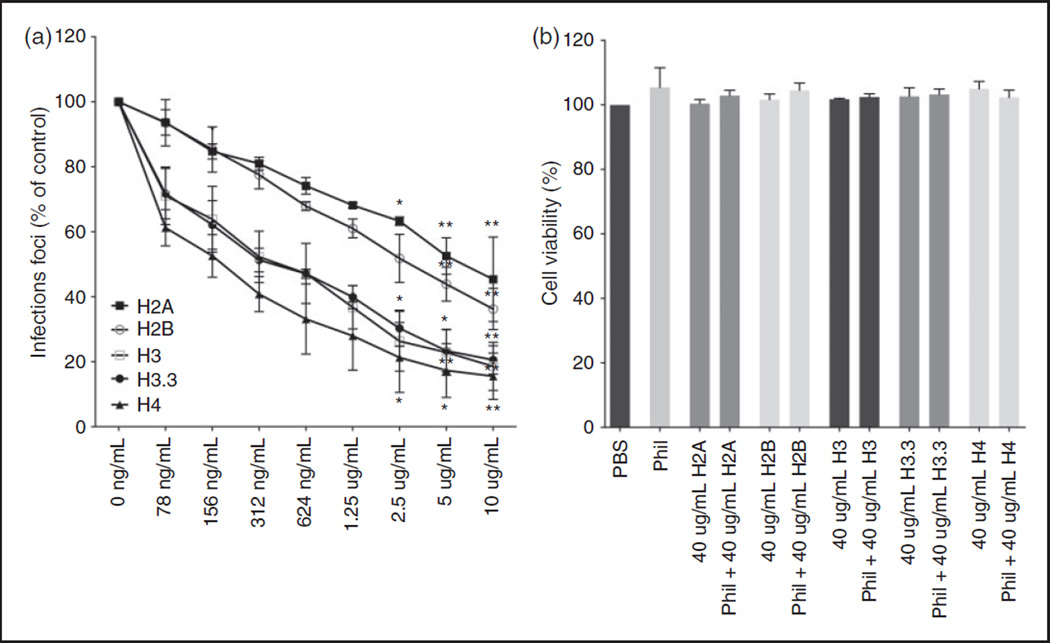
To investigate if histones could inhibit the infectivity of seasonal H3N2 strains, we pre-incubated the influenza A H3N2 strain Phil82 with increasing concentrations of all core histones, followed by infection of A549 cells with the viral samples. Cells were incubated with virus and histones for 45 min, and after 18 h, the amount of infected cells was quantified by immunofluorescent labeling of influenza nuclear protein (Figure 1A). While all histones inhibit infectivity of Phil82 without causing significant cell cytotoxicity (Figure 1B), the lysine rich histones H2A and H2B are less potent compared to the arginine rich histones H3, H3.3, and H4. Of all tested histones, histone H4 seemed to be the most efficient in inhibiting infection of A549 cells by Phil82. For comparison, we also tested a preparation of calf thymus histones. This preparation also had some neutralizing activity but it was relatively modest as compared to H3 or H4 histones (e.g. 10 µg/ml of the calf thymus histones reduced infectious foci to 64 ±17% of control; n = 4; P <0.01 vs control) (data not shown).
Figure 1.
Histones neutralize IAV H3N2 strain Phil82. A. Viral neutralization was measured by immunofluorescent detection of IAV nuclear protein after infection of A549 monolayers with IAV pre-incubated with histones. Unless otherwise indicated, all data points are P ≤ 0.001. (ns: P > 0.05. *: P ≤ 0.05. **: P ≤ 0.01). B. Release of LDH was assessed as a measure of cell cytotoxicity. Results are mean ± SD in panel A and mean + SD in panel B. Three separate experiments were performed (each variable run in duplicate in each experiment). (*P ≤ 0.05. **P ≤ 0.01. ***P ≤ 0.001).
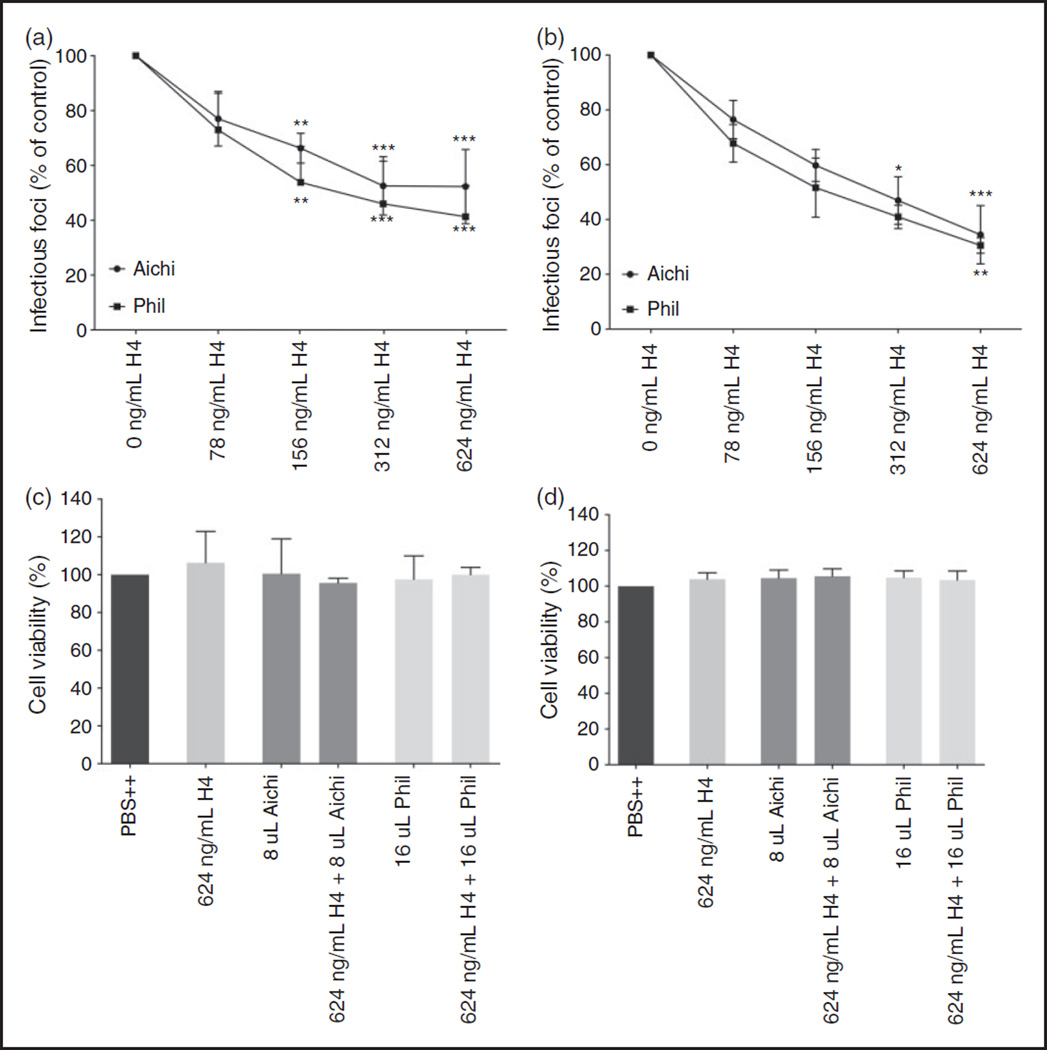
The same assay was also performed with histone H4 and the pandemic H3N2 strain (Aichi68) on both A549 and MDCK cells (Figure 2A/2B). The lowest dose (78 ng/mL) shows a significant decrease in influenza A nuclear protein production for both the Aichi68 and Phil82 strain on MDCK cells, as well as A549 cells, indicating that the anti-influenza effect of H4 is neither IAV strain nor cell line specific. LDH assays were performed and did not show any evident cellular toxicity from the histones (Figure 2C/2D).
Figure 2.
Histone H4 neutralizes H3N2 IAV strains Phil82 and Aichi. A-B. Viral neutralization of Aichi68 and Phil82 pre- incubated with histone H4 on both MDCK (a) and A549 (b) cells. Neutralization was assessed by immunofluorescent detection of IAV nuclear protein. C-D. Release of LDH was assessed as a measure of cell cytotoxicity for both MDCK (c) and A549 (d) cells. Results are mean±SD in panels A and B and mean + SD in panels B and C. Three separate experiments were performed (each variable run in duplicate in each experiment). (*: P ≤ 0.05. **: P ≤ 0.01. ***: P ≤ 0.001).
Histone H4 or the other histones did not inhibit hemagglutination (HA) activity of Phil82 IAV as assessed by HA inhibition assay (data not shown). In addition, H4 did not inhibit, but actually significantly increased, viral neuraminidase activity (Table 1).
Table 1.
Effects of histone H4 on neuraminidase activity of IAV.
| Inhibitor | % of Control neuraminidase activity |
|---|---|
| Histone H4 0.5 µg/ml | 117± 13 |
| Histone H4 5 µg/ml | 135 ± 12* |
| Histone H4 10 µg/ml | 131 ± 2* |
| Oseltamivir 6.25 µg/ml | 4±1.5# |
Indicates significant increase in neuraminidase activity (P < 0.05; n = 3)
Indicates significant reduction in neuraminidase activity (P < 0.01; n = 3)
Results shown are mean ± SD
Histone H4 inhibits viral uptake at 45 min after infection and release of virus from infected cells after 24 h on infection
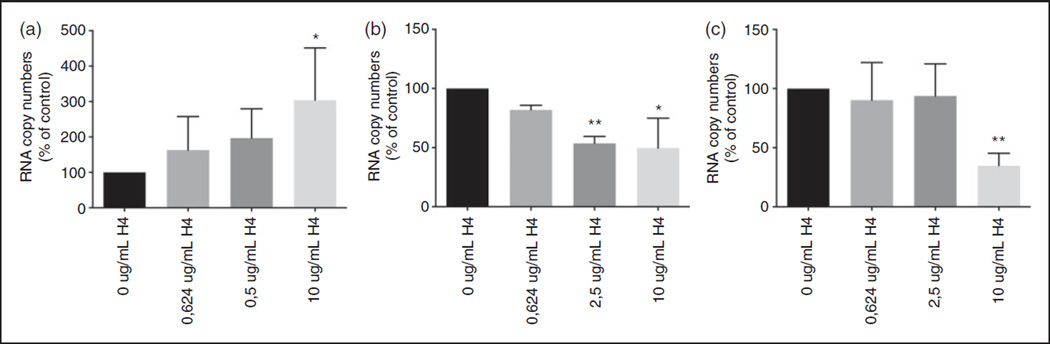
We next used RT-qPCR to confirm that less virus particles were taken up in the presence of histone H4 (Figure 3A/3B). RNA was isolated from cells infected with IAV pre-incubated with increasing concentrations of histone H4 for 45 min. With increasing amounts of histone H4, less viral RNA could be detected in the cell lysates, while the amount of viral RNA increased in the supernatants, indicating that uptake of viral particles by MDCK cells is reduced in the presence of H4. To see if viral replication was inhibited by histone H4, we expressed the amount of viral RNA present in both the supernatant and cell lysate after 24 h as a percentage of the amount isolated from the cell lysate at 45 min, thereby correcting for the difference in uptake after 45 min. Figure 3C shows the marked decrease in viral RNA after 24 h in the presence of histone H4.
Figure 3.
Histone H4 inhibits viral uptake and replication. Aichi68 was pre-incubated with increasing concentrations of histone H4 for 30min, and incubated with MDCK monolayers for 45 min. Cell supernatants were then collected for RNA isolation, and after washing, cells were lysed for collection of RNA. For RNA isolation after 24 h, cell free virus was washed off after the initial 45 min incubation and then cultured in MEM for an additional 24 h and then the same procedure was followed for RNA isolation. A. Mean RNA copy number isolated from supernatant at 45 min. B. Mean RNA copy number isolated from cell lysates at 45 min. C. Combined mean RNA copy number isolated from both the supernatant and cell lysates at 24 h. Results are mean + SD of 5 separate experiments (each variable run in duplicate in each experiment) and are expressed as % of control (e.g. untreated virus). (*: P ≤ 0.05. **: P ≤ 0.01. ***: P ≤ 0.001).
Histone H4 binds to IAV and inhibits replication by direct effects on the virus
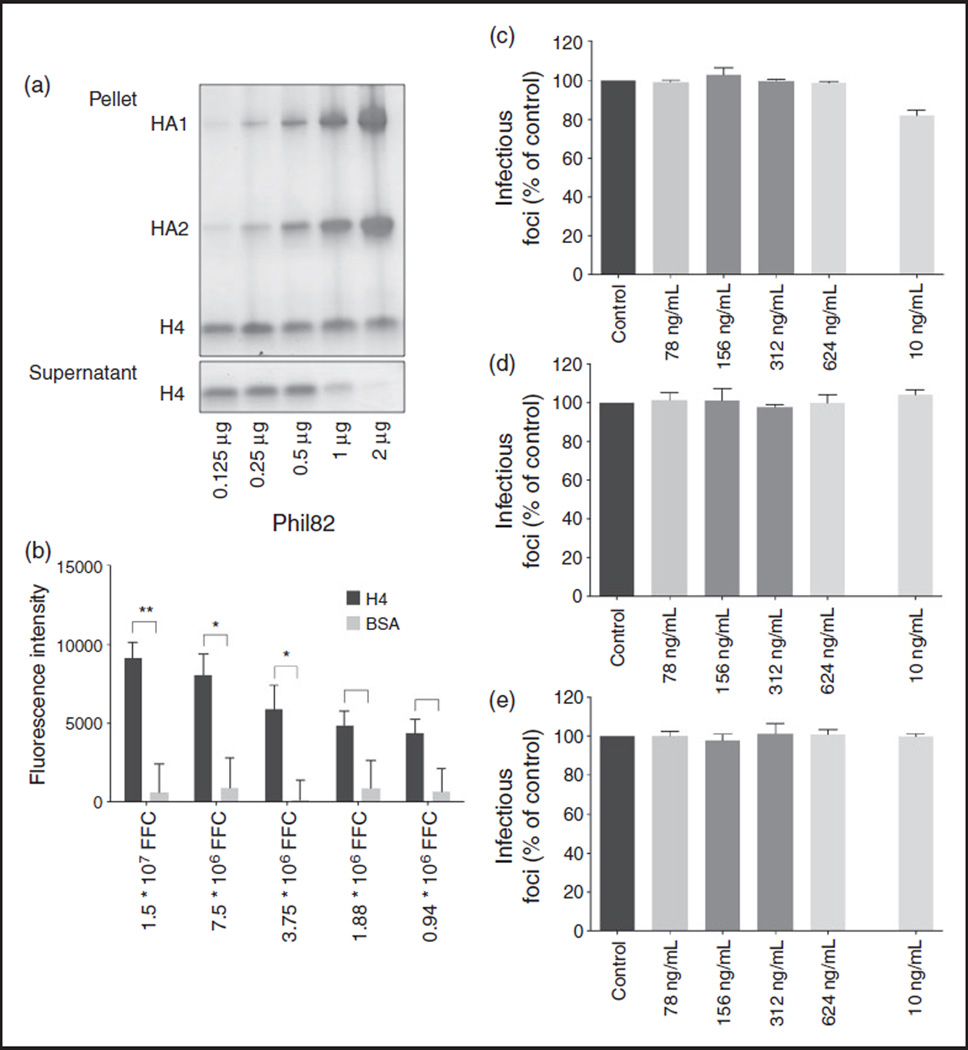
To further study the mechanism by which H4 inhibits IAV infectivity, we investigated if IAV binds to H4. IAV spins down when centrifuged at 13,000 g. We incubated H4 with increasing amounts of IAV, and after centrifugation, separated the pellet from the supernatant to see which fraction contained more H4. While H4, to some extent, also spins down in the absence of IAV, increasing amounts of virus result in a smaller fraction of H4 in the supernatant, suggesting that H4 binds to IAV. For unclear reasons there was not a clear increase in H4 in the pellet in presence of virus. It is possible that the H4 is partly included in viral protein bands or components of the virus that did not enter the gel. In any case, to confirm that H4 binds to IAV, we also coated 96-well plates with H4 and BSA (as a negative control), and incubated wells with Alexa-594 labeled Phil82. As can be seen in Figure 4b, Phil82 bound to H4 in a dose-dependent manner, while there was no binding of Phil82 to BSA.
Figure 4.
Effect of H4 on IAV is mediated through direct effects on virus (a). H4 was incubated with increasing amounts of Phil82. After centrifugation, pellet and supernatant were run on gel separately and stained with Gelcode blue. Results are representative of three similar experiments. (b). 96 well plates were coated with H4 or BSA and incubated with Alexa-594 labeled Phil82. Results are mean ±SEM of three separate experiments (each variable run in duplicate in each experiment). C-E. H4 was added to MDCK cells for 45 min before (c), 45 min after (d), or overnight after (e) incubation with virus. Results are mean + SD of three separate experiments (each variable run in duplicate in each experiment) (* P < 0.05 ** P < 0.01 *** P < 0.001).
In addition, we used the infectious focus assay to determine the extent to which neutralization depended on pre-incubation of IAV with H4. We found that adding H4 before or after cells have been infected with IAV did not seem to affect viral infection, as shown in Figure 4C–E, strengthening the conclusion that antiviral effects of H4 are mediated via direct effects on the virus.
Histone H4 induces viral aggregation as assessed by confocal microscopy, light absorbance, and EM
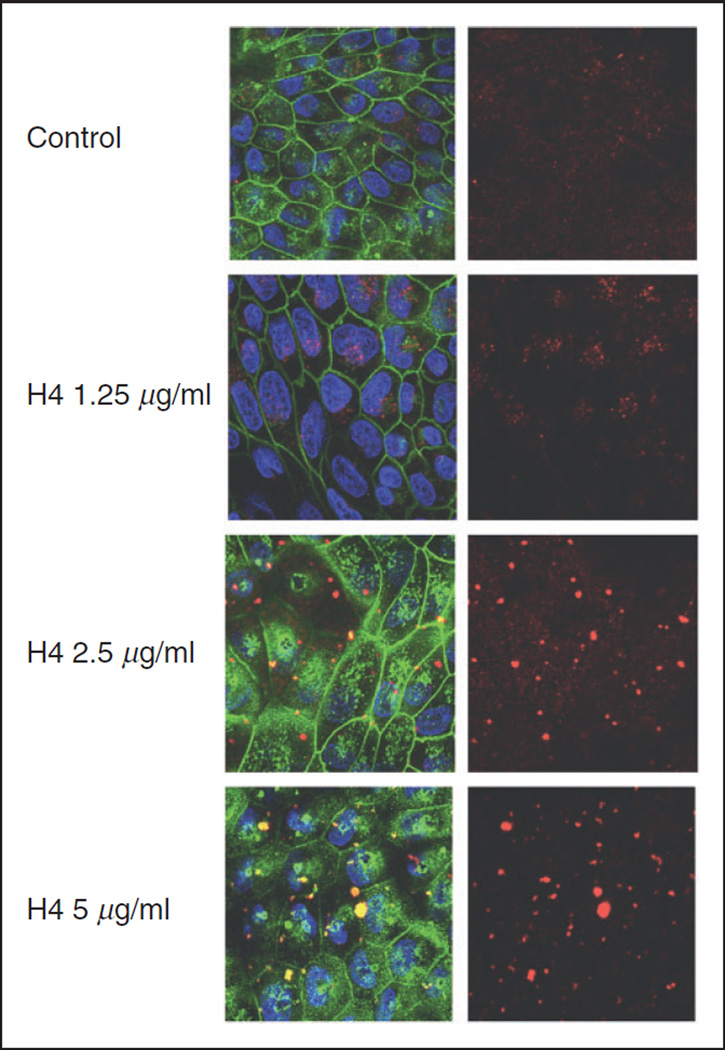
We next used confocal microscopy to investigate the interaction of H4 and IAV with MDCK cells in more detail (Figure 5). Cells were infected with Alexa Fluor 594-labeled Phil82 in the presence of histone H4. With increasing amounts of histone, aggregates of IAV are evident.
Figure 5.
Effects of H4 on viral interactions with MDCK cells under confocal microscopy. Alexa Fluor 594-labeled Phil82. IAV was pre-incubated with increasing concentrations of histone H4, and incubated with MDCK monolayers for 45 min. IAV is shown in red, cell nuclei were stained with DAPI 350 and appear blue, cell membranes were labeled with WGA-Oregon green 488 and are visible in green. The pictures on the left side were taken by combining wavelengths of virus, cell membranes and nuclei. Pictures on the right were visualized at wavelength of IAV alone. All pictures were taken at 100× magnification.
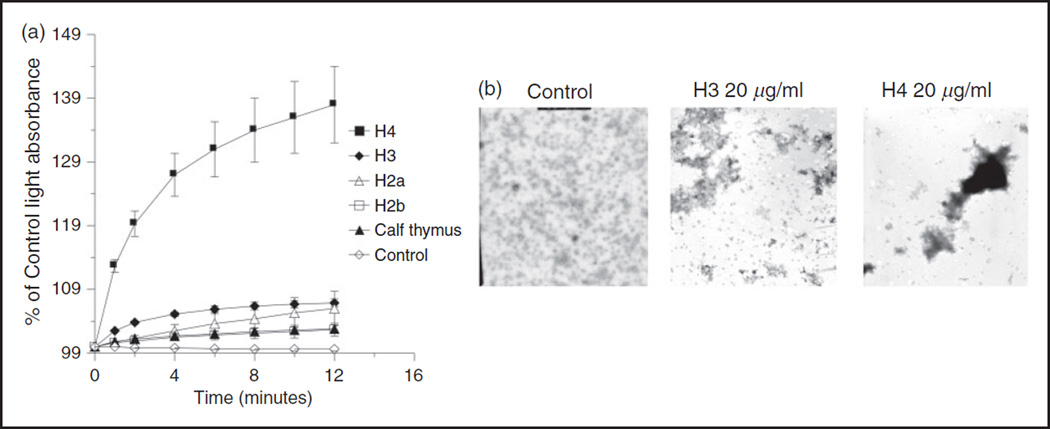
To quantitatively assess the relative ability of the different histones to cause viral aggregation we performed light absorption assays. In these assays H4 caused the greatest degree of aggregation (Figure 6a), followed by H3. Figure 6a compares the histones at the same concentration (10 µg/ml). We also tried increasing the concentration of H3 and calf thymus histones to 20 and 40 µg/ml and found dose related increases in aggregation. For instance, 40 µg/ml of H3 and calf thymus histones resulted in 124 ± 0.5 and 113 ± 1% increases in light absorption after 12 min incubation with virus (n = 3; P < 0.02 for either one) (data not shown). Viral aggregation induced by H3 and H4 histones was readily apparent on EM as well (Figure 6b).
Figure 6.
Histones induce viral aggregation. Viral aggregation was measured by increased light absorption through stirred suspensions of IAV (Phil82 strain) (panel a) and by EM (panel b). After 12 min of incubation of the histones with IAV (Phil 82 H3N2 strain) light absorption was significantly greater than control (virus alone incubated in control buffer) for all the histone preparations (P < 0.01 for all; n = 3 or more experiments). Results shown are mean ± SD. Panel B is representative of three experiments.
Histone H4 inhibits seasonal H1N1 IAV but not the 2009 pandemic H1N1
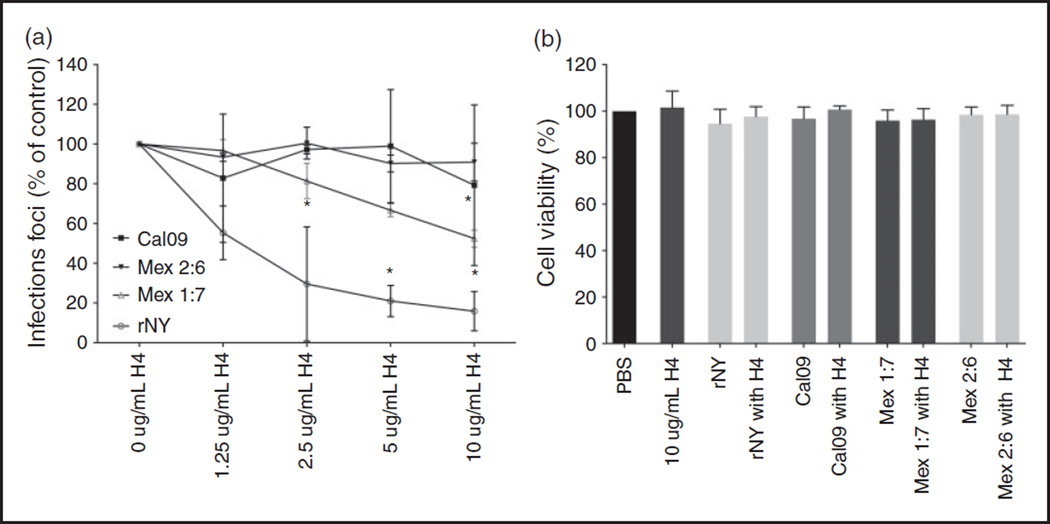
To investigate whether the anti-influenza activity of H4 can also be extended to neutralize influenza A H1N1 strains, we infected MDCK cells with rNY (seasonal H1N1) and Cal09 (pandemic H1N1). Histone H4 could inhibit infectivity of rNY to the same extent as with Phil82 and Aichi, but Cal09 could not be neutralized with concentrations of up to 10 µg/ml (Figure 7).
Figure 7.
Histone H4 neutralizes seasonal H1N1, but not pandemic H1N1. A. IAV H1N1 strains were pre-incubated with H4 for 30 min and added to MDCK cells for 45 min. Viral neutralization was measured by immunofluorescent detection of influenza nuclear protein. B. Release of LDH was assessed as a measure of cell cytotoxicity. Results are mean ± SD in panel A and mean + SD in panel B. Four separate experiments were performed for infectious focus assays and three experiments for LDH assays. Each variable run in duplicate in each experiment. (*P < 0.05. **P < 0.01. ***P < 0.001).
To investigate which properties of Cal09 are responsible for the absence of inhibitory activity of H4, we used seasonal H1N1 strains that have been modified to express either HA (Mex 1:7) or both HA and NA (Mex 2:6) from Cal09 while retaining the internal genes of rNY (Figure 7). H4 is still mildly effective against Mex 1:7, but Mex 2:6 is as resistant to H4 as Cal09, suggesting that the HA and NA are involved in resistance of pandemic H1N1 to H4. We have recently obtained similar results using the cathelicidin, LL-37.38
Discussion
To the best of our knowledge, our report is the first showing anti-influenza activity of histones. We showed activity against both influenza A H3N2 and H1N1 strains, and found that the anti-influenza activity is mainly mediated via direct effects on the virus. This is similar to our findings with defensins,11,12,39 LL-3710 and Alzheimer’s associated β-amyloid protein.33 The relative potency of H3 or H4 histones was comparable or greater than that of the other antimicrobial peptides. Like with the defensins and β-amyloid (but unlike with LL-37), histones induced viral aggregation. Based on the RT-PCR and confocal microscopy assays higher concentrations of H4 acted by reducing viral uptake by target cells. Since we found no significant effect of adding histones to cells after virus infection, and no inhibition of neuraminidase activity effects on later stages of viral replication are likely minimal. The histones do not act by inhibiting either HA binding or NA enzymatic activity. We have obtained similar results with the other antimicrobial peptides (references above and unpublished data). Of interest, H4 actually significantly increased NA activity. We cannot explain this at present, although we speculate that it may result from alteration of viral membranes allowing greater access of the substrate to the active site of the neuraminidase. At this point we cannot determine which viral components are bound by histones.
Out of all core histones, histone H4 was the most potent at inhibiting viral infectivity, closely followed by histone H3 and the histone variant H3.3 which differ in only 5 amino acids.40 Both H2A and H2B (and a mixture of calf thymus histones) had less inhibitory activity. The only other known study about histone antiviral activity did not show a difference in activity when comparing all core histones.30 Interestingly, our findings do not correspond with the antibacterial activity of histones, where H2A and H2B in general are more active than H3 and H4.18,20 Both H2A and H2B are classified as lysine-rich, whereas H3 and H4 are more arginine-rich, suggesting that this structural difference could account for the variation in antiviral activity.
We cannot as yet explain why pandemic H1N1 of 2009 was not inhibited by histone H4. Seasonal and pandemic H3N2 strains were inhibited as was the seasonal H1N1 strain NY01. Recombinant NY01 strains expressing HA or both HA and NA from a pandemic strain (Mex09 which is nearly identical to Cal09) showed a decrease in sensitivity for histone-mediated inhibition. With only the HA from the pandemic strain, the virus was inhibited, although less so than intact NY01. However, when both HA and NA are replaced with those of pandemic H1N1, the virus was no longer inhibited. This suggests that H4 interactions with the pandemic HA and NA are mediating the resistance, even though H4 did not inhibit HA or NA functional activities. Further studies will be needed to clarify the mechanisms through which histones bind to viral particles and the mechanism of resistance of pandemic H1N1 to histones. In any case, pandemic H1N1 has also been found to be resistant to some other innate inhibitors including collectins, pentraxins and LL-734,38,41 and this property could account for rapid spread of pandemic IAV (i.e. because of ability to bypass initial innate barriers to infection).
In conclusion, we show here for the first time that histones can neutralize H3N2 and H1N1 seasonal strains, but not pandemic H1N1 strains. Out of all core histones, histone H4 was most potent and inhibited viral uptake and replication by directly interacting with viral particles. Based on our findings, we propose that histones could play a role in the innate immune response against IAV. Although NETs and histones contribute in a positive way to innate defense, they are also damaging in a variety of settings through adverse pro-inflammatory effects.42
Histones can act as alarm signals and trigger cell activation through toll like receptors.43 In particular extracellular histones are implicated as a cause of mortality in sepsis and acute lung injury in sepsis, trauma and post-transfusion settings.42,44–48 Of interest, histones H3 and H4 have been found to be particularly contributory to injury in some of these studies. It should be noted that our cell viability assays were done at the same time point as the viral neutralization assays to confirm that antiviral effects were not the result of reduced cell viability. However, it is possible that longer times of incubation of histones with cells (in presence or absence of virus) could cause cell injury. We will evaluate this aspect of histones in the context of IAV in future studies.
Acknowledgments
This work was supported by NHLBI grant number R01 HL 069031.
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Footnotes
Conflict of interest
The authors do not have any potential conflicts of interest to declare.
References
- 1.Belser JA, Lu X, Maines TR, et al. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J Virol. 2007;81:11139–11147. doi: 10.1128/JVI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Riel D, Munster VJ, de Wit E, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 4.Ling MT, Tu W, Han Y, et al. Mannose-binding lectin contributes to deleterious inflammatory response in pandemic H1N1 and avian H9N2 infection. J Infect Dis. 2012;205:44–53. doi: 10.1093/infdis/jir691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tripathi S, White MR, Hartshorn KL. The amazing innate immune response to influenza A virus infection. Innate Immun. 2015;21:73–98. doi: 10.1177/1753425913508992. [DOI] [PubMed] [Google Scholar]
- 6.Verma A, White M, Vathipadiekal V, et al. Human H-ficolin inhibits replication of seasonal and pandemic influenza A viruses. J Immunol. 2012;189:2478–2487. doi: 10.4049/jimmunol.1103786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartshorn KL. Role of surfactant protein A and D (SP-A and SP-D) in human antiviral host defense. Front Biosci (Schol Ed) 2010;2:527–546. doi: 10.2741/s83. [DOI] [PubMed] [Google Scholar]
- 8.Hartshorn KL, Crouch EC, White MR, et al. Evidence for a protective role of pulmonary surfactant protein D (SP-D) against influenza A viruses. J Clin Invest. 1994;94:311–319. doi: 10.1172/JCI117323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathi S, White MR, Wang G, Hartshorn K. LL-37 modulates human phagocyte responses to influenza A virus. J Leukoc Biol. 2014;96:531–938. doi: 10.1189/jlb.4A1113-604RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi S, Tecle T, Verma A, Crouch E, White M, Hartshorn KL. The human cathelicidin LL-37 inhibits influenza A viruses through a mechanism distinct from that of surfactant protein D or defensins. J Gen Virol. 2013;94:40–49. doi: 10.1099/vir.0.045013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doss M, Ruchala P, Tecle T, et al. Hapivirins and diprovirins: novel theta-defensin analogs with potent activity against influenza A virus. J Immunol. 2012;188:2759–2768. doi: 10.4049/jimmunol.1101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartshorn KL, White MR, Tecle T, Holmskov U, Crouch EC. Innate defense against influenza A virus: activity of human neutrophil defensins and interactions of defensins with surfactant protein D. J Immunol. 2006;176:6962–6972. doi: 10.4049/jimmunol.176.11.6962. [DOI] [PubMed] [Google Scholar]
- 13.Tripathi S, Verma A, Kim EJ, White MR, Hartshorn KL. LL-37 modulates human neutrophil responses to influenza A virus. J Leukoc Biol. 2014 doi: 10.1189/jlb.4A1113-604RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartshorn KL, Collamer M, Auerbach M, Myers JB, Pavlotsky N, Tauber AI. Effects of influenza A virus on human neutrophil calcium metabolism. J Immunol. 1988;141:1295–1301. [PubMed] [Google Scholar]
- 15.Tate MD, Ioannidis LJ, Croker B, Brown LE, Brooks AG, Reading PC. The role of neutrophils during mild and severe influenza virus infections of mice. PLoS One. 2011;6:e17618. doi: 10.1371/journal.pone.0017618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urban CF, Ermert D, Schmid M, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans . PLoS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 18.Morita S, Tagai C, Shiraishi T, Miyaji K, Iwamuro S. Differential mode of antimicrobial actions of arginine-rich and lysine-rich histones against Gram-positive Staphylococcus aureus . Peptides. 2013;48:75–82. doi: 10.1016/j.peptides.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 19.Pavia KE, Spinella SA, Elmore DE. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Biochim Biophys Acta. 2012;1818:869–876. doi: 10.1016/j.bbamem.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagai C, Morita S, Shiraishi T, Miyaji K, Iwamuro S. Antimicrobial properties of arginine- and lysine-rich histones and involvement of bacterial outer membrane protease T in their differential mode of actions. Peptides. 2011;32:2003–2009. doi: 10.1016/j.peptides.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Tsao HS, Spinella SA, Lee AT, Elmore DE. Design of novel histone-derived antimicrobial peptides. Peptides. 2009;30:2168–2173. doi: 10.1016/j.peptides.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Koo YS, Kim JM, Park IY, et al. Structure-activity relations of parasin I, a histone H2A–derived antimicrobial peptide. Peptides. 2008;29:1102–1108. doi: 10.1016/j.peptides.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Kim HS, Park CB, Kim MS, Kim SC. cDNA cloning and characterization of buforin I, an antimicrobial peptide: a cleavage product of histone H2A. Biochem Biophys Res Commun. 1996;229:381–387. doi: 10.1006/bbrc.1996.1814. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch JG. Bactericidal action of histone. J Exp Med. 1958;108:925–944. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller BF, Abrams R, Dorfman A, Klein M. Antibacterial Properties of Protamine and Histone. Science. 1942;96:428–430. doi: 10.1126/science.96.2497.428. [DOI] [PubMed] [Google Scholar]
- 26.Urban CF, Reichard U, Brinkmann V, Zychlinsky A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 2006;8:668–676. doi: 10.1111/j.1462-5822.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 27.Gadebusch HH, Johnson AG. Natural host resistance to infection with Cryptococcus neoformans V. The influence of cationic tissue proteins upon phagocytosis and on circulating Ab synthesis. J Infect Dis. 1966;116:566–572. doi: 10.1093/infdis/116.5.566. [DOI] [PubMed] [Google Scholar]
- 28.Guimaraes-Costa AB, DeSouza-Vieira TS, Paletta-Silva R, Freitas-Mesquita AL, Meyer-Fernandes JR, Saraiva EM. 3’-nucleotidase/nuclease activity allows Leishmania parasites to escape killing by neutrophil extracellular traps. Infect Immun. 2014;82:1732–1740. doi: 10.1128/IAI.01232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connolly JH. Effect of histones and protamine on the infectivity of Semliki Forest virus and its ribonucleic acid. Nature. 1966;212:858. doi: 10.1038/212858a0. [DOI] [PubMed] [Google Scholar]
- 30.Tamura M, Natori K, Kobayashi M, Miyamura T, Takeda N. Inhibition of attachment of virions of Norwalk virus to mammalian cells by soluble histone molecules. Arch Virol. 2003;148:1659–1670. doi: 10.1007/s00705-003-0143-4. [DOI] [PubMed] [Google Scholar]
- 31.Kobiyama K, Takeshita F, Jounai N, et al. Extrachromosomal histone H2B mediates innate antiviral immune responses induced by intracellular double-stranded DNA. J Virol. 2010;84:822–832. doi: 10.1128/JVI.01339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narasaraju T, Yang E, Samy RP, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol. 2011;179:199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White M, Kandel R, Tripathi S, et al. Alzheimer’s associated beta amyloid protein inhibits influenza A virus and modulates viral interactions with phagocytes. PLoS One. 2014;9:e101364. doi: 10.1371/journal.pone.0101364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi L, Kash JC, Dugan VG, et al. The ability of pandemic influenza virus hemagglutinins to induce lower respiratory pathology is associated with decreased surfactant protein D binding. Virology. 2011;412:426–434. doi: 10.1016/j.virol.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reading PC, Morey LS, Crouch EC, Anders EM. Collectin-mediated antiviral host defense of the lung: evidence from influenza virus infection of mice. J Virol. 1997;71:8204–8212. doi: 10.1128/jvi.71.11.8204-8212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White MR, Crouch E, Chang D, et al. Enhanced antiviral and opsonic activity of a human mannose-binding lectin and surfactant protein D chimera. J Immunol. 2000;165:2108–2115. doi: 10.4049/jimmunol.165.4.2108. [DOI] [PubMed] [Google Scholar]
- 37.Hartshorn KL, Sastry KN, Chang D, White MR, Crouch EC. Enhanced anti-influenza activity of a surfactant protein D and serum conglutinin fusion protein. Am J Physiol Lung Cell Mol Physiol. 2000;278:L90–L98. doi: 10.1152/ajplung.2000.278.1.L90. [DOI] [PubMed] [Google Scholar]
- 38.Tripathi S, Wang G, White M, Qi L, Taubenberger J, Hartshorn K. Antiviral activity of the human cathelicidin, LL-37, and derived peptides on seasonal and pandemic influenza A virus. PLoS One. 2015;10(4):e0124706. doi: 10.1371/journal.pone.0124706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doss M, White MR, Tecle T, et al. Interactions of alpha-, beta-, and theta-defensins with influenza A virus and surfactant protein D. J Immunol. 2009;182:7878–7887. doi: 10.4049/jimmunol.0804049. [DOI] [PubMed] [Google Scholar]
- 40.Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends in biochemical sciences. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Job ER, Deng YM, Tate MD, et al. Pandemic H1N1 influenza A viruses are resistant to the antiviral activities of innate immune proteins of the collectin and pentraxin superfamilies. J Immunol. 2010;185:4284–4291. doi: 10.4049/jimmunol.1001613. [DOI] [PubMed] [Google Scholar]
- 42.Cheng OZ, Palaniyar N. NET balancing: a problem in inflammatory lung diseases. Front Immunol. 2013;4:1. doi: 10.3389/fimmu.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, Zhang X, Monestier M, Esmon NL, Esmon CT. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J Immunol. 2011;187:2626–2631. doi: 10.4049/jimmunol.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrams ST, Zhang N, Manson J, et al. Circulating histones are mediators of trauma-associated lung injury. Am J Respir Crit Care Med. 2013;187:160–169. doi: 10.1164/rccm.201206-1037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang H, Chen HW, Evankovich J, et al. Histones Activate the NLRP3 Inflammasome in Kupffer Cells during Sterile Inflammatory Liver Injury. J Immunol. 2013;191:2665–2679. doi: 10.4049/jimmunol.1202733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakahara M, Ito T, Kawahara K, et al. Recombinant Thrombomodulin Protects Mice against Histone-Induced Lethal Thromboembolism. PLoS One. 2013;8:e75961. doi: 10.1371/journal.pone.0075961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bosmann M, Grailer JJ, Ruemmler R, et al. Extracellular histones are essential effectors of C5aR- and C5L2-mediated tissue damage and inflammation in acute lung injury. FASEB J. 2013;27:5010–5021. doi: 10.1096/fj.13-236380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Zhang X, Pelayo R, et al. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15:1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]