Abstract
Glycosylation is critical for the regulation of several cellular processes. One glycosylation pathway, the unusual O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation) has been shown to be required for proper mitosis, likely through a subset of proteins that are O-GlcNAcylated during metaphase. As lectins bind glycosylated proteins, we asked if specific lectins interact with mitotic O-GlcNAcylated proteins during metaphase to ensure correct cell division. Galectin-3, a small soluble lectin of the Galectin family, is an excellent candidate, as it has been previously described as a transient centrosomal component in interphase and mitotic epithelial cells. In addition, it has recently been shown to associate with basal bodies in motile cilia, where it stabilizes the microtubule-organizing center (MTOC). Using an experimental mouse model of chronic kidney disease and human epithelial cell lines, we investigate the role of Galectin-3 in dividing epithelial cells. Here we find that Galectin-3 is essential for metaphase where it associates with NuMA in an O-GlcNAcylation-dependent manner. We provide evidence that the NuMA-Galectin-3 interaction is important for mitotic spindle cohesion and for stable NuMA localization to the spindle pole, thus revealing that Galectin-3 is a novel contributor to epithelial mitotic progress.
Introduction
Glycosylation encompasses a broad and varied range of post-translational modifications (PTMs)1. While other PTMs such as phosphorylation are simple, glycosylation PTMS have more complex structures with a highly variable composition and arrangement, and it is thus difficult to infer a single function for a specific glycosylation. While most glycosylations occur in the Golgi apparatus, an unusual type of glycosylation, called O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation), occurs in the cytosol in a ubiquitous and reversible manner, similar to phosphorylation2. Two proteins control O-GlcNacylation: the O-GlcNAmine transferase, to add the O-GlcNAc moiety, and the O-GlcNAminase, to remove it. It is not yet completely understood how these proteins are regulated, nor how they target or ignore glycosylation sites in cytosol substrates3, 4. A large panel of proteins are O-GlcNAcylated, and the PTM is dynamic, changing in response to glucose intake or different signalling pathways4, 5. One current hypothesis is that this PTM would act as a tuning mechanism to regulate signalling pathway activity, in part by competing with phosphorylation, for their shared threonine/serine modification sites5, 6. However, recent studies have suggested that O-GlcNAcylation serves an active role beyond blocking phosphorylation. In addition, O-GlcNAcylation has been reported to be required for proper mitosis7–9, and only a specific subset of substrate proteins are O-GlcNAcylated during metaphase8. This finding indicates clearly that glycosylation contributes to the regulation or progression of cell division process, and suggest that the link between cancer establishment/progression and glycosylation may occur through direct action on mitosis via modification of M-phase proteins.
Lectins are glycosylation “readers”, but, so far, no lectin has been described to recognize O-GlcNAcylated motifs. A small number of lectins are found soluble in the cytosol, and the Galectin family is among them. Over the years, Galectins have been implicated in a wide range of implications in both extra- and intracellular biologic processes10. The Galectin family is highly conserved and is comprises 15 members, initially described based on their affinity for ß-galactosides. They are small (from 14 to 36 kDa), non-glycosylated, and soluble, and they are found inside and outside of cells. Three different structural configurations can be distinguished: the prototypic Galectins (Galectin-1, -2, -5, -7, -10, -11, -13, -14, -15), which are exclusively composed of a pair of unique Galectin carbohydrate-recognition domain (CRD); the tandem-repeat Galectins (Galectins-4, -6, -8, -9, -12), which have two CRDs linked through a short linker peptide, and the chimeric Galectin (Galectin-3), with one CRD domain and a unique N-terminal tail. Galectin-3 is highly expressed in epithelial cells and is involved in many processes, including intracellular trafficking and cell cycle signaling11, 12. It has been notably described as a transient centrosomal component in interphase and mitotic epithelial cells13, 14. In addition, it has recently been shown to associate with basal bodies in motile cilia, where it tightly associates with and stabilizes the microtubule-organizing center (MTOC)15. Importantly, perturbation of Galectin-3 expression has been associated with epithelial cancer progression16, 17. The correlation between Galectin-3 and epithelial cancer, the localization of Galectin-3 at the MTOC and the apparent importance of glycosylation in cell division process, prompted us to hypothesize that Galectin-3 is required for proper mitosis in a glycosylation-dependent manner.
Here we show that the absence of Galectin-3 leads to severe defects in epithelial tissue architecture, at least in part due to cell division failure. Further investigation highlighted the association of Galectin-3 with the mitotic spindle pole, a robust structure that ensures proper sister chromatide alignment18–20. We also find that Galectin-3 interacts with a key mitotic regulator, NuMA (Nuclear Mitotic Apparatus protein), which is required for the establishment and the cohesion of the spindle pole21. We report that the Galectin-3/NuMA interaction is functionally important for the spindle pole organization. Altogether our results provide new mechanistic insight into the role of glycosylation in controlling epithelial cell division.
Results
Galectin-3 is required for normal tissue organization in mouse kidney after nephron reduction
To first test the importance of Galectin-3 in mitosis in vivo, we analysed mitotic defects in the mouse kidney, where Galectin-3 has been reported to associate with centrosomes and centrosome-derived organelles13, 14. Since the renal epithelium is only renewed at a slow rate, we challenged gal3−/− mutant mice with a pathological renal stress to stimulate cell proliferation, using the established experimental model of chronic kidney disease designated “nephron reduction” or “partial nephrectomy” (Nx)22, 23. This model, which consists in the excision of 75% of the total renal mass (Fig. 1a), is characterized first by a phase of compensatory growth, then the progressive development of renal lesions, leading to deterioration of the remaining kidney tissue. An increase in epithelial cell proliferation accompanies both the compensatory and the deterioration phase22, 23. We previously showed that these events are genetically determined and identified mouse strains that are resistant to renal lesion development, such as the 129/sv background mice used in this study24, 25. As expected, remnant kidneys of 129/sv wild-type (Nx wt) mice underwent compensatory growth, but did not displayed renal lesions the 3 months following Nx24 (Fig. 1b,c). However, we observed that following Nx, the lack of Galectin-3 (Nx gal3−/−) leads to increased kidney weight increase (Fig. 1d) and severe tubular alterations, specifically cyst development, unlike their cyst-free wt littermates (Fig. 1b,c and e). When we assessed renal function by measuring plasma creatinine, we observed a stronger elevation of creatinine concentration occurs in Nx gal3−/− mice compared to Nx wt mice, demonstrating renal failure (Fig. 1f). These kind of defects in the morphogenesis and function of renal tubules have also been observed in the context of cystic kidney phenotypes, in both acquired cystic kidney disease (ACKD) and genetic polycystic kidney disease (PKD), where they correlate with defects in cell proliferation, growth of primary cilium and perturbation of mitosis26–30. As in these other cystic kidney tissues, we found that Nx gal3−/− kidneys exhibit increased cell proliferation (Fig. 1g). The primary cilium growth also overelongates in Nx gal3−/− renal interphase cells (Fig. 1h-i), as previously observed in MDCK cyst cultures14. In addition, Nx gal3−/− cells display numerous pericentrin accumulations in contrast with pericentrin localization in wt cells, suggesting that the absence of Galectin-3 may lead to a centrosomal perturbation with PCM fragmentation or centrosome duplication (Fig. 1j). Interestingly, Nx gal3−/− cells show a dramatic increase dysmorphic metaphases plates (i.e. fragmented metaphase plate, triskelion shaped metaphase plate, …). Whereas control metaphase cells display regular metaphase plates, gal3−/− metaphase cells harbour ill-shaped metaphase plate in a triskelion-like shape or individual chromosomes escaping the metaphase plate, suggesting the presence of multipolar cells and defective organisation of spindles in Nx gal3−/− cells (Fig. 1k-l). These data demonstrate that loss of Galectin-3 leads to severe tissue disorganization during renal recovery and strongly suggest that Galectin-3 promotes robust cell division in sensitized tissue in vivo.
Figure 1.
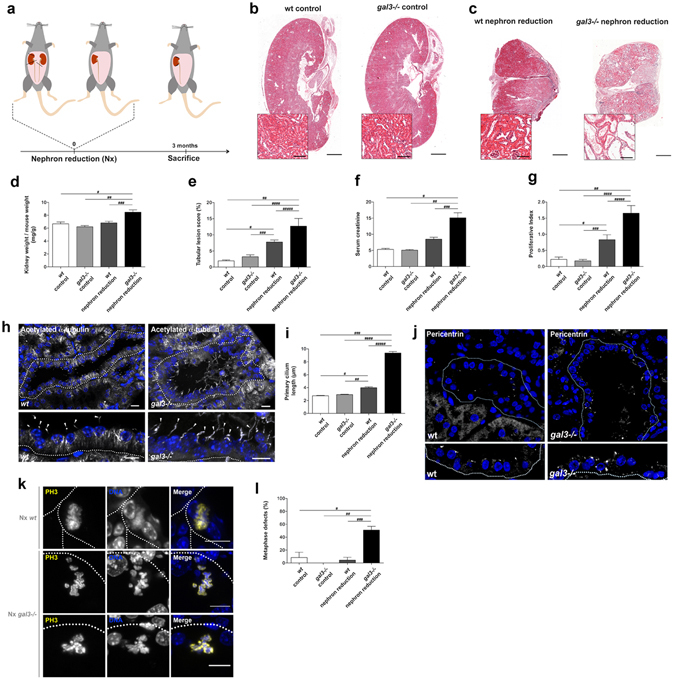
Galectin-3 loss leads to severe tissue disorganization during renal recovery. (a) Scheme showing the nephronic reduction procedure. (b–c) Histological analyses of sham-operated (Control) (b) and Nx (c) wt and gal3−/− kidneys 3 months after surgery. Scale bars, 1 mm; inserts, 100 µm. (d) Statistical analysis of the ratio kidney weight over mouse weight in control and Nx, wt and gal3−/− kidneys. (e) Statistical analysis of tubular dilatations. (f) Statistical analysis of the amount of serum creatinine in control and Nx wt and gal3−/− kidneys. (g) Statistical analysis of the proliferative index in control and Nx wt and gal3−/− kidneys. (h) Confocal microscopy analysis and 3D rendering of primary cilia (white arrow arrowhead) immunostained with monoclonal anti-acetylated α-tubulin antibody (white) in Nx wt and Nx gal3−/− kidneys. Scale bars, 10 µm. (i) Statistical analysis of the primary cilium length in control and Nx kidneys in wt and gal3−/− mice. (j) Confocal microscopy analysis of the distribution of pericentrin (white) in Nx wt and Nx gal3−/− kidneys. N(Nx wt) = 6 mice, N(Nx gal3−/−) = 6. (k) Confocal microscopy analysis of mitoses with PH3 immunostaining (yellow) in Nx kidneys. Arrowheads, irregular metaphasic plate; arrows, lagging chromosomes. (l) Statistical analysis of the percentage of cells with unconventional metaphasic plate morphology. In the Fig. 1, data are means ± SEM. Renal epithelium was demarcated with a dotted white line. Nuclei were detected by Hoechst 33342 staining (blue). Detailed information about quantifications is available in Supplementary Information.
Galectin-3 is required for normal cell division in epithelial cell cultures
We next sought to determine if Galectin-3 promotes normal mitosis. Since the in vivo system was limiting for this study, we switched to in vitro with the use of HeLa cells, a non-ciliated epithelial cell line. We silenced Galectin-3 by transient RNA interference (siRNA; Supplementary Fig. S1a,b). We found that a significant increase in mitosis-related defects occurred in the absence of Galectin-3 as described thereafter18, 31. Galectin-3-depleted cells displayed an increased frequency of multinucleated cells (Supplementary Fig. S2a,b) and of supernumerary centrioles at interphase (Supplementary Fig. S3a,b), when compared with controls. Looking more precisely at metaphase cells, multipolar mitoses were more frequent in Galectin-3-depleted cells than in control cells (22.59 ± 2.79% and 22.99 ± 1.97% in both Galectin-3-siRNA cells versus 7.67 ± 1.84% in control cells; Fig. 2a,b). Surprisingly, Galectin-3-silenced cells showed multipolar mitoses with acentrosomal poles more often than control cells (77.59 ± 10.74% and 79.76 ± 4.02% in both Galectin-3-siRNA cells versus 33.92 ± 2.52% in control cells; Fig. 2c,d). These data indicate that the increase in multipolar mitoses in Galectin-3-silenced cells is probably be due to defects in the level of the pericentrosomal matrix (PCM), an improper centrosome clustering or in spindle pole stability rather than due to centrosomal overduplication18. To test this, we next analysed the distribution of PCM proteins. We examined ninein and γ-tubulin (Fig. 2e–h), which bind and nucleates microtubules respectively. Ninein appeared less focused at spindle poles and more scattered in the cytoplasm (Fig. 2e,f). γ-tubulin spread along MTs at the level of the spindle pole and spindle in Galectin-3-depleted cells (Fig. 2i,j). We conclude that Galectin-3 is instrumental for normal mitotic spindle formation, and we speculate that it promotes PCM stability at the spindle pole.
Figure 2.
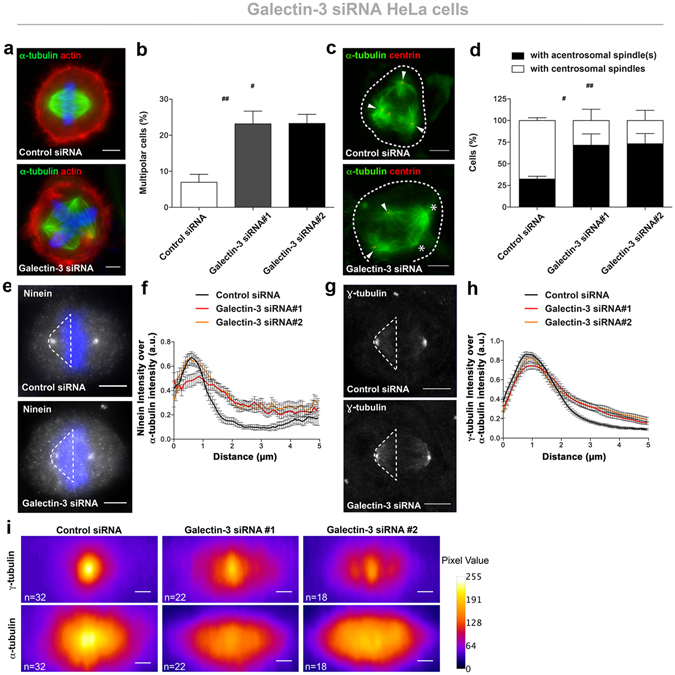
Galectin-3 is required for correct mitoses in epithelial cell cultures. (a) Confocal microscopy analysis of mitotic poles after α-tubulin immunostaining and actin detection in Hela cells. Scale bars, 5 µm. (b) Statistical analysis of the number of multipolar mitosis. n(Control siRNA) = 1406 cells, n(Galectin-3 siRNA #1) = 1291, n(Galectin-3 siRNA #2) = 1158. Data are means ± SEM. (c) Confocal microscopy analysis of centrosomal (arrowheads) and acentrosomal (stars) mitotic spindles. Cells were immunostained for α-tubulin (green) and centrin-3 (red). Scale bars, 5 µm. (d) Statistical analysis of the number of centrosomal or acentrosomal mitotic spindles. n (Control siRNA) = 48 cells; n (Galectin-3 siRNA #1) = 55; n (Galectin-3 siRNA #2) = 53. Data are means ± SEM. (e) Fluorescence microscopy analysis of ninein in control and Galectin-3 siRNA cells. Nuclei were detected by Hoechst 33342 staining (blue). A white dotted line specifies the area which has been used for ninein intensity quantification in (f). Scale bars, 5 µm. (f) Statistical analysis of the intensity of ninein fluorescence over α-tubulin fluorescence intensity along the mitotic spindle in control (Control siRNA, black line) and Galectin-3-depleted (Galectin-3 siRNA#1, red line and Galectin-3 siRNA#2, orange line) metaphase Hela cells. n (Control siRNA) = 19 cells; n (Galectin-3 siRNA#1) = 19 cells; n (Galectin-3 siRNA#2) = 13 cells. (g) Fluorescence microscopy analysis of γ-tubulin in control and Galectin-3 siRNA cells. A white dotted line specifies the area which has been used for γ-tubulin intensity quantification in (h). Scale bars, 5 µm. (h) Statistical analysis of the intensity of γ-tubulin fluorescence over α-tubulin fluorescence intensity along the mitotic spindle in control (black line) and Galectin-3-depleted (red and orange lines) metaphase Hela cells. n(Control siRNA) = 26 cells; n(Galectin-3 siRNA#1) = 19; n(Galectin-3 siRNA#2) = 21. (i) Fluorescence microscopy analysis of α-tubulin and γ-tubulin (in pseudocolor spectrum) at spindle pole in bipolar mitosis. Images are mean projection of xz spindle centered through DNA staining. n(Control siRNA) = 32 cells; n(Galectin-3 siRNA#1) = 22; n(Galectin-3 siRNA#2) = 18. Detailed information about quantifications is available in Supplementary Information.
Galectin-3 associates with spindle poles
To understand how Galectin-3 promotes normal spindle pole formation, we examined its subcellular localization during mitosis (Fig. 3). Although Galectin-3 is primarily cytosolic during mitosis, it is transiently enriched at the spindle pole during prophase and metaphase, and at the cleavage furrow during cytokinesis (Fig. 3a,b, arrows), showing that Galectin-3 exhibits a dynamic distribution, similar to other MTOC-associated proteins during mitosis. For a more detailed understanding of Galectin-3 localization at spindle poles, we used high-resolution microscopy (3D-Structured Illumination Microscopy, 3D-SIM) (Fig. 3c–f) and observed Galectin-3 colocalized with the spindle pole (Fig. 3d, arrow), close to γ-tubulin MT-nucleation complexes (Fig. 3f, arrow). To confirm Galectin-3 association with the PCM, its localization was followed after nocodazole treatment and wash out. Early during metaphase MT regrowth, Galectin-3 localizes close to MT nucleation complexes at the base of MT asters (Supplementary Fig. 4a, arrowheads) and to γ-tubulin foci (Supplementary Fig. 4b, arrowheads). These results show that Galectin-3 closely associates with the spindle MTOC, and we propose that Galectin-3 likely functions at the spindle pole to promote normal spindle formation.
Figure 3.
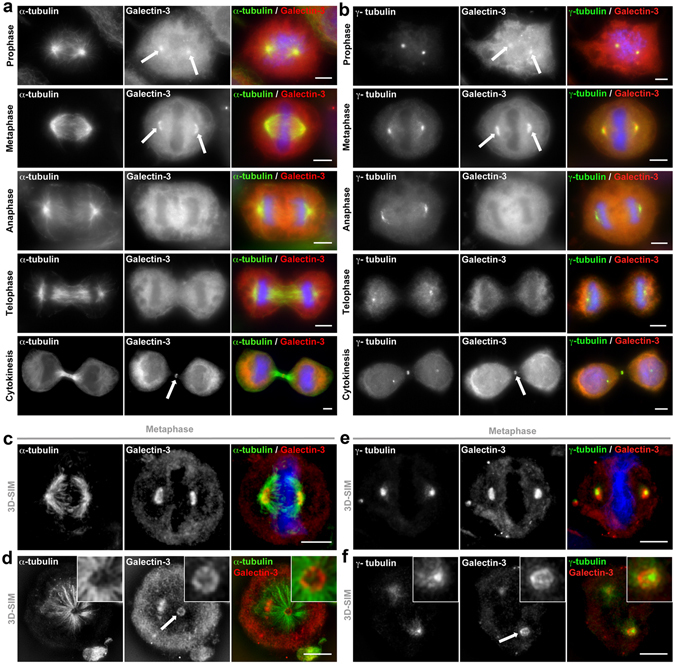
Galectin-3 associates with spindle poles. (a,b) Epifluorescence microscopy analysis of Galectin-3 distribution during mitosis. Hela cells were immunostained for Galectin-3 (red) and either α-tubulin (a) or γ-tubulin (b) (green). Galectin-3 is located at the spindle poles in prophase and metaphase (arrows), and with cleavage furrow in cytokinesis (arrows). Nuclei were detected by Hoechst 33342 staining (blue). Scale bars, 5 µm. (c–f) 3D-SIM microscopy analysis of Galectin-3 localization at spindle poles. Hela cells were immunostained for Galectin-3 (red) and either α-tubulin (c,d) or γ-tubulin (e,f) (green). Longitudinal (c and e) and top (d and f) spindle views are shown and close-ups on spindle pole top views are presented in inserts. Nuclei were detected by Hoechst 33342 staining (blue). Galectin-3 is associated with (−) end MTs neighbouring γ-TURC complexes at spindle poles (arrows). Scale bars, 5 µm.
Galectin-3 and NuMA interact at spindle poles through lectin activity of Galectin-3
To further investigate the molecular mechanism of Galectin-3 function, we looked for its protein interactors. We identified a small number of candidates via mass spectrometry that co-immunoprecipitated with Galectin-3 in metaphase enriched cell lysates (Supplementary Fig. S5). Since Galectin-3 is best characterized as a lectin, we focused our analysis on glycoproteins. NuMA (Nuclear Mitotic Apparatus protein) interacts with MT (−)-ends after nuclear breakdown and forms a lattice that stabilizes and strengthens radial spindle MT arrays. NuMA was recently reported to be glycosylated in mitotic cells and it co-immunoprecipitated with Galectin-3 in our assays, making NuMA an excellent candidate interactor of Galectin-38, 21. First, we observed that transfected form of NuMA-GFP and Galectin-3 colocalize at spindle poles in metaphase HeLa cells, and vice versa (Fig. 4a). Second, their association was analysed more thoroughly by immunoprecipitation followed by Western blot. We found that Galectin-3 co-immunoprecipitated with NuMA, and vice versa (Fig. 4b). We next tested whether the Galectin-3/NuMA association depends on Galectin-3 glycan recognition by performing Galectin-3 immunoprecipitation in the presence of sugar agonists (lactose and galactose) or sugar for which it has no affinity (glucose)32 (Fig. 4c). Co-immunoprecipitations in the presence of glucose had no effect, but NuMA’s association with Galectin-3 was abolished when lactose or galactose were added, suggesting that their association is a glycan-dependent interaction. To test the importance of Galectin-3 lectin activity, we analysed the behaviour of sugar binding incompetent variant form and consequences on mitosis. Arg186 is particularly required for sugar binding of the Galectin-3’s CRD domain. Substitution of Arg186 by serine (R186S) has been previously reported to impair Galectin-3 binding to common glycoproteins, whereas the mutation of a close amino acid, i.e. substitution of Gly182 by alanine (G182A), has little effect on CRD activity32, 33. Endogenous Galectin-3 was depleted using CRISPR-Cas9 strategy (Supplementary Fig. S6), and wt Galectin-3-GFP, Galectin-3(R186S)-GFP and Galectin-3(G182A)-GFP constructs were introduced by transient transfection in Galectin-3-depleted Hela cells. Removal of Galectin-3 using CRISPR-Cas9 strategy caused an increase of multipolar mitoses in comparison with control CRISPR cells (Fig. 4d,e), as in Galectin-3 siRNA transfection experiments (Fig. 2a,b). While the expression of Galectin-GFP or Galectin-3(G182A)-GFP constructs rescued a bipolar state, Galectin-3(R186S)-GFP transfection did not (Fig. 4d,e), showing that Galectin-3 CRD activity is required for Galectin-3 function in metaphase. Moreover, Galectin-3(R186S)-GFP levels at spindle pole were decreased (Fig. 4d and f), suggesting that glycan binding helps recruit Galectin-3 to the spindle pole. This observation was corroborated by biochemical analysis. NuMA failed to co-immunoprecipitate with Galectin-3(R186S)-GFP form (Fig. 4g). These data indicate that Galectin-3 lectin activity is required to bind NuMA and to promote proper metaphase.
Figure 4.
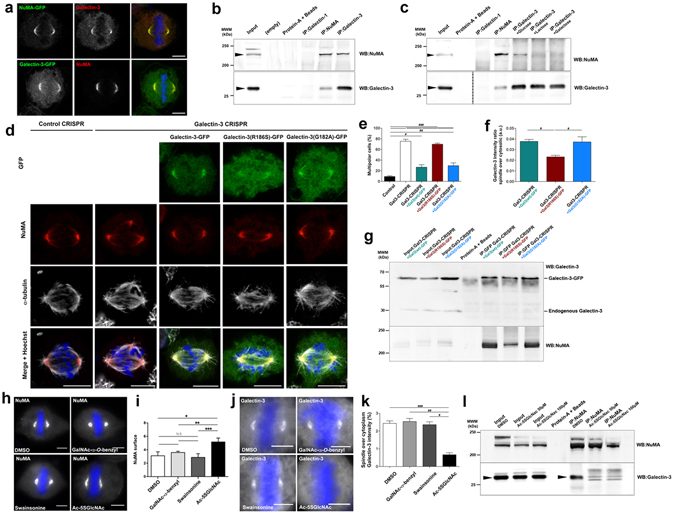
Galectin-3 and NuMA interaction relies on Galectin-3 lectinic activity. (a) Confocal microscopy analysis of endogenous Galectin-3 and transfected NuMA-GFP, and vice versa. Scale bars, 5 µm. (b) Western blot detection of NuMA (Top blot, black arrowhead) or Galectin-3 (Bottom blot, black arrowhead) after immunoprecipitation from mitotic extracts. Input, 10% of total cell lysate. (c) Western blot detection of NuMA (Top blot, black arrowhead) and Galectin-3 (Bottom blot, black arrowhead) after immunoprecipitation of NuMA or Galectin-3 in presence of Galectin-3-competitive sugars. Input, 10% of total cell lysate. (d) Confocal microscopy analysis of NuMA and α-tubulin in control and Galectin-3-depleted cells using CRISPR-Cas9 strategy, after transfection of Galectin-3-GFP, Galectin-3-R186S-GFP or Galectin-3-G182A-GFP. Scale bars, 5 µm. (e) Statistical analysis of the number of multipolar cells after transfection of Galectin-3-GFP constructs in control or Galectin-3–depleted cells using CRISPR-Cas9 strategy. (f) Statistical analysis of the intensity of Galectin-3-GFP constructs over cytosol. (g) Western blot detection of Galectin-3 (Top blot) and NuMA (Bottom blot) after the immunoprecipitation of GFP from mitotic extracts of Galectin-3-depleted cells using CRISPR-Cas9 strategy, and transfected with Galectin-3-GFP constructs. Input, 10% of total cell lysate. (h) Fluorescence microscopy analysis of NuMA distribution in metaphase cells after DMSO or glycosylation inhibitor treatments. Scale bars, 5 µm. (i) Statistical analysis of the volume occupied by NuMA at spindle poles. (j) Fluorescence microscopy analysis of Galectin-3 distribution in metaphase cells after DMSO or glycosylation inhibitor treatments. Scale bars, 5 µm. (k) Statistical analysis of the intensity of Galectin-3 fluorescence at spindle poles over cytoplasmic Galectin-3 fluorescence intensity. (l) Western blot detection of NuMA (Top blot) and Galectin-3 (Bottom blot, black arrowhead) after the immunoprecipitation of NuMA from mitotic extracts, treated with DMSO or Ac-5SGlcNAc. Input, 10% of total cell lysate. Detailed information about quantifications is available in Supplementary Information.
We next asked which kind of glycan chains mediates Galectin-3 association with NuMA. HeLa cells were treated with different glycosylation inhibitors: N-glycosylation (swainsonine), GalNAc-type of O-glycosylation (GalNAcα-O-benzyl) or GlcNAc-type of O-glycosylation (Ac-5SGlcNAc)34. NuMA or Galectin-3 localization was only disturbed after GlcNAc-type O-glycosylation inhibition, with significant spreading of NuMA along spindle MTs (Fig. 4h,i) and decreased Galectin-3 enrichment at spindle poles (Fig. 4j,k). In addition, NuMA and Galectin-3 no longer co-immunoprecipitated in Ac-5SGlcNAc-treated cells (Fig. 4l). Together, these results suggest that the NuMA/Galectin-3 association occurs through an O-GlcNAc modification and that it is important for localizing both proteins at the spindle pole.
Galectin-3 and NuMA interact at spindle poles through an O-GlcNAc modification of NuMA
A single O-GlcNAc modification site on Ser1844 has been previously reported for NuMA8 (Fig. 5a). To test whether this specific glycosylation site is implicated in NuMA interaction with Galectin-3, we tested two GFP-tagged mutants forms of NuMA: (i) a glycosylation-blocked mutant form, where the serine is substituted with an alanine and which cannot be O-GlcNAc-glycosylated (NuMA-S1844A-GFP), and as a control (ii) a glycosylation-permissive mutant form, which can still be O-GlcNAc-glycosylated, by substituting the serine with a threonine (NuMA-S1844T-GFP) (Fig. 5b,c). Endogenous NuMA was depleted using CRISPR-Cas9 strategy (Supplementary Fig. S7), and wt NuMA-GFP, NuMA(S1844A)-GFP and NuMA(S1844T)-GFP constructs were introduced by transient transfection in NuMA-depleted Hela cells. Removal of NuMA using CRISPR-Cas9 strategy caused an increase of multipolar mitoses in comparison with control CRISPR cells (Fig. 5d,e). While the expression of NuMA-GFP or NuMA(S1844T)-GFP constructs rescued a bipolar state, NuMA(S1844A)-GFP transfected cells displayed an increase in multipolar mitoses similar to NuMA-depleted cells (Fig. 5d,e), indicating that indicating that NuMA glycosylation is important for spindle pole formation and stability. Moreover, the glycosylation mutant form was sufficient to induce the formation of cytosolic NuMA foci (Fig. 5f; Supplementary Fig. S8a–c). The NuMA glycosylation mutant was also defective in Galectin-3 recruitment to spindle poles (Fig. 5f,g). We next tested whether the Galectin-3/NuMA interaction depends on NuMA glycosylation state by performing immunoprecipitation. We found that Galectin-3 co-immunoprecipitated with NuMA-GFP and NuMA(S1844T)-GFP, but not with NuMA(S1844A)-GFP (Fig. 5h), showing that Galectin-3 associates with NuMA through its glycan chains. Our results reveal that Galectin-3 interacts with NuMA at the spindle pole through O-GlcNAc modifications and that O-GlcNAc modification of NuMA is required for its own focusing, as well as Galectin-3 positioning, at the spindle pole during metaphase.
Figure 5.
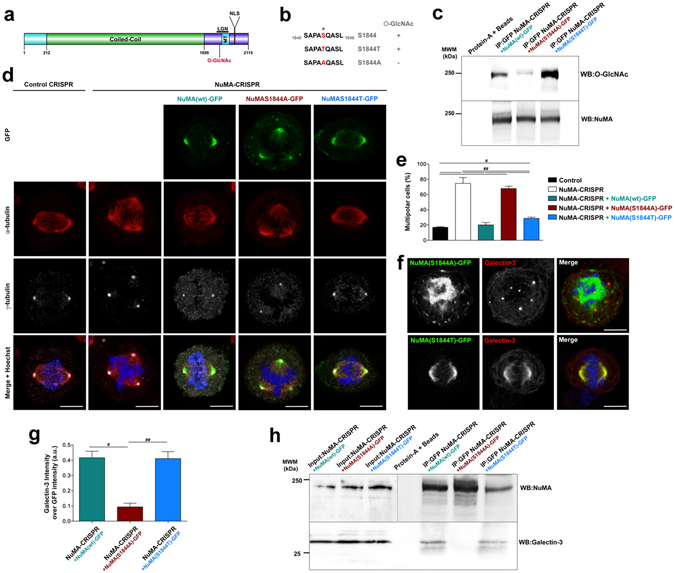
Galectin-3 and NuMA interaction relies on O-GlcNAc modification of NuMA. (a) Schemes showing NuMA functional domains along its amino acid sequence: in light blue the N-terminal globular domain, in green the central coiled-coil domain and in dark blue the C-terminal globular domain (based on UniprotKB_Q14980). (b) Schemes showing the point mutations generated at NuMA O-GlcNAc glycosylation site. (c) Western blot analysis of NuMA (Bottom blot) and O-GlcNAc motifs (Top blot) after the immunoprecipitation of GFP from mitotic extracts of NuMA-depleted cells using CRISPR-Cas9 strategy, and transfected with NuMA-GFP constructs. (d) Confocal microscopy analysis of α-tubulin and γ-tubulin in control and NuMA-depleted cells using CRISPR-Cas9 strategy, after transfection of NuMA-3-GFP, NuMA-S1844A-GFP or NuMA-S1844T-GFP plasmids. Scale bars, 5 µm. (e) Statistical analyses of the number of multipolar cells after transfection of NuMA-GFP constructs in control or NuMA-depleted cells using CRISPR-Cas9 strategy. (f) Confocal microscopy analysis of mutated forms of NuMA and Galectin-3 in metaphase cells. Scale bars, 5 µm. Nuclei were detected by Hoechst 33342 staining (blue). (g) Statistical analysis of the Galectin-3 intensity over NuMA intensity after transfection of NuMA-GFP constructs in NuMA-depleted cells using CRISPR-Cas9 strategy. (h) Western blot analysis of NuMA (Top blot) and Galectin-3 (Bottom blot) after the immunoprecipitation of GFP from mitotic extracts of NuMA CRISPR KO cells transfected with NuMA-GFP constructs. Input, 10% of total cell lysate.
Galectin-3 acts to organize NuMA at the spindle pole
We next evaluated a functional interaction between Galectin-3 and NuMA. We found that siRNA silencing of NuMA displaced Galectin-3 from the spindle pole in HeLa cells (Supplementary Fig. S9; Fig. 6a). Conversely, siRNA reduction of Galectin-3 led to NuMA mislocalization (Fig. 6b). NuMA normally localizes in a concise ring at mitotic poles and dimly at the plasma membrane in control cells (Fig. 6b; Supplementary Fig. S10a,b). However, Galectin-3 silencing leads to NuMA spreading along spindle MTs (Fig. 6b,c). In addition, the NuMA lattice at the spindle was deformed (Fig. 6d). Whereas the NuMA ring occupied a mean volume of 19.6 ± 2.1 µm3 in control cells, it increased to 35.2 ± 3.6 and 32.5 ± 2.3 µm3 in Galectin-3-depleted cells (Fig. 6e). Defective NuMA localization was further analysed with live cell imaging. We observed that NuMA-GFP shuttled between the spindle poles and the cell cortex in control cells (Fig. 6f, Supplementary movie 1), but larger foci of NuMA (as observed previously with GFP-mutant version of NuMA in NuMA KO cells, Supplementary Fig. S8) and unshaped spindle poles appeared in Galectin-3-depleted cells (Fig. 6f, Supplementary Fig. S11 and Supplementary movie 2). These results show that NuMA and Galectin-3 are functionally interdependent for their spindle pole localization in metaphase cells.
Figure 6.
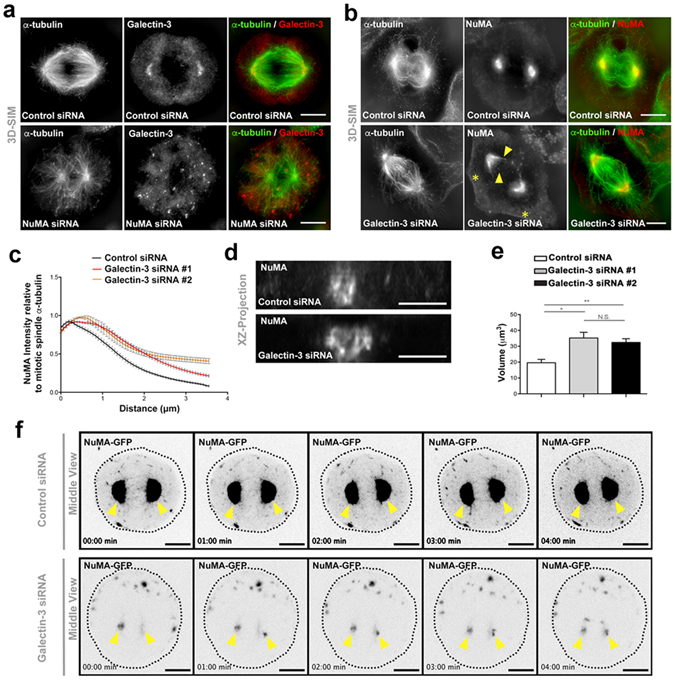
Galectin-3 acts in the organisation of NuMA at the spindle pole. (a) 3D-SIM microscopy analysis of α-tubulin (green) and Galectin-3 (red) distribution in control (Control siRNA, upper panel) and NuMA-silenced (NuMA siRNA, lower panel) Hela cells. Deprivation of NuMA leads to the loss of Galectin-3 recruitment at spindle poles and perturbation of mitotic spindles. Scale bars, 5 µm. (b) 3D-SIM microscopy analysis of α-tubulin (green) and NuMA (red) distribution in control (Control siRNA, upper panel) and Galectin-3 silenced (Galectin-3 siRNA, lower panel) Hela cells. NuMA is no longer focused at spindle poles and instead spreads along aberrant mitotic spindles (arrowheads) in the absence of Galectin-3. In addition, a cortical NuMA signal appears in Galectin-3 siRNA cells (stars). Scale bars, 5 µm. (c) Statistical analysis of the intensity of NuMA fluorescence over α-tubulin fluorescence intensity along the mitotic spindle in control (Control siRNA, black line) and Galectin-3-depleted (Galectin-3 siRNA#1, red line and Galectin-3 siRNA#2, orange line) metaphase Hela cells. n (Control siRNA) = 24 cells; n (Galectin-3 siRNA#1) = 24 cells; n (Galectin-3 siRNA#2) = 24 cells. (d) Z-projection of spindle poles immunostained for NuMA in control (Control siRNA, upper panel) and Galectin-3-depleted (Galectin-3 siRNA, lower panel) metaphase Hela cells. (e) Statistical analysis of the volume occupied by NuMA at the spindle pole in control (Control siRNA, white bar) and Galectin-3-depleted (Galectin-3 siRNA#1, grey bar and Galectin-3 siRNA#2, black bar) metaphase Hela cells. (f) Spinning-disc videomicroscopy analysis of NuMA-GFP localization in control (Control siRNA, upper panels) and Galectin-3-silenced (Galectin-3 siRNA, lower panels) metaphase Hela cells. Middle views going through the spindle poles (yellow arrowheads) are shown. Time frames from 0 to 4 min are presented.
Galectin-3 participates in spindle pole cohesion and pericentrosomal matrix stabilization
To study the consequence of the loss of Galectin-3 to spindle and pole formation, we looked at spindle pole reformation after MT depolymerisation by nocodazole treatment and wash out (Fig. 7a). In control cells, small MT arrays form all over at NuMA foci one minute after nocodazole wash out (Fig. 7a, left panels). By contrast, Galectin-3-depleted cells show disorganized and fewer MTs and disorganized arrangements (Fig. 7a,b, right panels). Five minutes after nocodazole wash out, an unshaped spindle appeared with characteristic astral and interpolar MTs in control cells (Fig. 7a, left panels). However, multiple small spindles remained, as well as collection of MTs and NuMA foci in Galectin-3 depleted cells compared to control cells (Fig. 7a,b, right panels). Finally, after twenty minutes, control cells displayed normal spindle poles and MT arrays (Fig. 7a,b, left panels), but Galectin-3 depleted cells displayed incomplete reformed spindle poles (Fig. 7a,b, right panels). We also noticed that Galectin-3-depleted spindles were defective, either improperly shaped or with scattered spindle poles (Fig. 7a, right panels). These data suggest that Galectin-3 acts as an organizer of the NuMA network at spindle poles. In addition, the absence of Galectin-3 induces a delay in the gathering of individual MTs in foci, impairing the proper formation of spindle poles and the metaphase plate (Fig. 7c).
Figure 7.
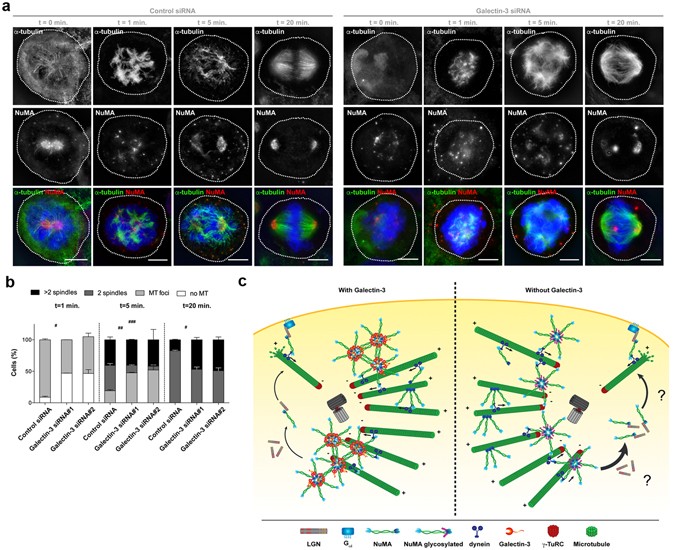
Galectin-3 participates in spindle pole cohesion and pericentrosomal matrix distribution. (a) 3D-SIM microscopy analysis of α-tubulin (green) and NuMA (red) after nocodazole treatment, wash out and MT regrowth for 0, 1, 5 or 20 min in control (Control siRNA, left panels) or Galectin-3-deprived (Galectin-3 siRNA, right panels). Nuclei were detected by Hoechst 33342 staining (blue). Scale bars, 5 µm. (b) Statistical analysis of metaphase morphology during MT regrowth at different time point (from left to right, separated by a doted-line, 1, 5 and 20 min after nocodazole washout/MT regrowth) in control and Galectin-3-depleted HeLa cells. Metaphase morphology has been set in four different categories; no MT (white): no MT appeared to be grown; MT foci (light grey): MT are organized in bouquet originating from a single γ-tubulin/NuMA foci; 2 spindles (dark grey): metaphase presented a classical bipolar organization; > 2 spindles (black): metaphase presented a multipolar profile. Data are means ± SEM. (c) Scheme showing the predicted mechanism of action of Galectin-3 in the stabilization of NuMA complexes and spindle pole cohesion in metaphase epithelial cells. As previously proposed47, NuMA oligomerizes through its C-terminal globular domain (dark blue) to form radial structures that would interact altogether to form higher structure. NuMA interacts directly with MTs and dynein through its C-terminal globular domain (dark blue) and N-terminal region (light blue), respectively. The glycosylation (purple) is present on the C-terminal globular domain (dark blue). As Galectin-3 would recognize the glycosylation on the C-terminal globular domain, we hypothesize that it would facilitate the oligomerization of NuMA. This will contribute to a correct organization of spindle pole (left panel). We hypothesize that the absence of Galectin-3 might disturb NuMA oligomerized structures, affecting the structure of the spindle pole (right panel). Moreover, perturbation of the oligomerization of NuMA might lead to an increase of NuMA shuttling to the cortex through dynein-based transport and LGN interaction at plasma membrane (question mark, right panel). Detailed information about quantifications is available in Supplementary Information.
Discussion
Cells have evolved many molecular mechanisms to precisely control the sequence of mitotic events and avoid aneuploidy. Spindle pole integrity is a critical feature that ensures correct DNA segregation. Here we confirm and extend the importance of an unconventional O-GlcNAc-glycosylation during metaphase, and identify and characterize the role of the lectin Galectin-3. While most glycosylation occurs in the Golgi apparatus, O-GlcNAc-glycosylation takes place in the cytosol under control of O-linked N-acetylglucosamine transferase (OGT)35. Hart and colleagues demonstrated that OGT enzyme and O-GlcNAc-modified proteins are enriched at mitotic spindles in HeLa cells, and modulation of OGT expression leads to metaphase and cytokinesis defects7, 8. Notably, O-GlcNAc modification is critical to the activity of NuMA, a key player in the establishment and the maintenance of spindle poles, i.e. NuMA21, 36, 37. In this study, we found that Galectin-3 is a functional partner of NuMA, binding through O-GlcNAc moieties during metaphase. This interaction is important for mitosis as it promotes NuMA focusing at spindle poles. Moreover, Galectin-3 silencing mimics the metaphase perturbation observed when OGT expression is reduced8. These results suggest a model in which Galectin-3 interacts with glycosylated NuMA to organize the spindle pole (Fig. 7c). At the molecular level, it is well-established that Galectin-3 acts as an oligomerization factor when bound to its ligands, and it mediates the formation of lattices that stabilize or gather many different proteins38. For instance, Galectin-3 ensures the polymerization of Hensin, an extracellular matrix protein implicated in epithelial cells differentiation39. The existence of an oligomerization factor has been hypothesized to promote the formation of the NuMA matrix, but this factor had not been identified21. This work suggests that Galectin-3 could physically bind NuMA and thereby trigger the formation and/or the stabilization of a NuMA lattice at spindle poles (Fig. 7c). NuMA-Galectin-3 association is critical to stabilize the NuMA matrix at spindle poles as well as for MT dynamics at spindles, validating the longstanding hypothesis that post-translational modification of NuMA and an additional NuMA-binding factor promote NuMA lattice ordering at the spindle pole17. In addition, the Ser1844 O-GlcNAc-glycosylation site is located close to the LGN binding domain in the NuMA sequence40 (Fig. 3i). Disrupting the Galectin-3/NuMA association induces a weakening of the NuMA lattice at the spindle, but it also causes an increased pool of NuMA to localize to the cell cortex (Fig. 4b; Supplementary Fig. S10). Could O-GlcNAc-glycosylation influence NuMA interaction with LGN? We speculate that stabilization of NuMA at the spindle pole by Galectin-3 may also prevent the accumulation NuMA dimers at the cortex by LGN for correct progression through metaphase (Fig. 5b).
Galectin-3 modulates the organization of MT (−)-ends via NuMA at spindle poles, and this may be similar to the recently proposed function of Galectin-3 recently proposed at basal bodies of multiciliated cells7. The lectin ensures the recruitment of MTs and consequently the maintenance of the basal body MTOC, through the stabilization of γ-tubulin at basal foot cap7. It will be interesting to investigate whether Galectin-3 functions generally in the organization of the MTOC and MT arrays in different types of epithelial cells. Galectin-3 is mostly expressed in epithelia and is considered as a multifunctional protein, due to the diverse cellular functions that it promotes41.
Defects in Galectin-3 expression (either loss or over) are correlated with cancer progression and are actually correlated with poor diagnosis16, 42, 43. Two roles of Galectin-3 in immune response and neoangiogenesis have been largely described17, 44, but prior to this work, a direct role for Galectin-3 in cell division had not been investigated. Although Raz and colleagues showed that the lectin helps modulate cell cycle progression, through indirect repression of cyclin-E and A and inducing cyclin D1 expression in breast cancer cells in vitro 45. Here we show that Galectin-3 directly participates in proper completion of epithelial mitosis. We were surprised to find that gal3−/− mice do not display any obvious phenotypic and morphological abnormalities under physiological conditions and do not spontaneously develop tumors (Fig. 1b)46. However, they are sensitive to additional stresses such as tissue damage/loss or genetic mutations. Thus loss of Galectin-3 could sensitize cells to the loss of additional cell division regulators, which could trigger morphogenetic alterations and/or epithelial tumor development.
Material and Methods
Materials
Rabbit antibodies directed against Galectin-3 (1:200) and Galectin-1 (1:200) were kindly provided by Dr. H. P. Elsaesser (Philipps University, Marburg, Germany) and Dr. D. N. W. Cooper (University of California, San Francisco, California, USA), respectively. Mouse anti-α-tubulin (T9026; 1:100), anti-acetylated-α-tubulin (T6793; 1:100), anti-γ-tubulin (T6557; 1:200), and rabbit anti-γ-tubulin (T3559; 1:200) antibodies were from Sigma-Aldrich (St Louis, Missouri, USA). Rabbit anti-ninein (ab4447; 1:100), rabbit anti-NuMA (ab109262; 1:200) and mouse anti-O-GlcNAc (ab2739; 1/500) antibodies were from Abcam (Cambridge, UK). Mouse anti-dynein intermediate chains, cytoplasmic (MAB1618; 1:100) and rabbit anti-phosphorylated (Ser10) histone H3 (06-570; 1:200) antibodies were from EMD Millipore (Billerica, Massachusetts, USA). Rabbit anti-NuMA (PAB12087; 1:200) antibody was from Abnova (Taipei, Taiwan). Mouse anti-actin (8691001) antibody was from MP Biomedicals (Santa Ana, California, USA). Human anti-CREST (15-235; 1:200) antibody was from Antibodies Incorporated (Sacramento, California, USA). Rabbit anti-pericentrin (PRB432C; 1:100) antibody was from Covance (Princeton, New-Jersey, USA). Mouse anti-GFP antibody (#11.814.460.001, clones 7.1 and 13.1; 1/500) was from Roche (Basel, Switzerland). Alexa-488, -568, or -633 secondary antibodies (1:250), Hoechst 33342 (H3570) and Alexa-647-Phalloidin (A22287; 1:50) were from Life Technologies (Paisley, UK). O-linked N-acetylglucosamine inhibitor, Ac-5S-GlcNAc34 was provided by Dr. D. Vocadlo (Simon Fraser University, Burnaby, Canada). Benzyl 2-acetamido-2-deoxy-α-D-galactopyranoside (GalNAc-α-O-Benzyl; B4894), Protein-A/Sepharose beads (P9424), nocodazole (M1404), swainsonine (S9263) and HRP-labelled secondary antibodies were from Sigma-Aldrich.
Mice and preparation of tissue samples
Mice were housed in EOPS (Environment without Specific Pathogenic Organisms) environment, and handled in accordance with French regulation for animal care. Experimental procedures were performed in accordance with Departmental Director of “Services Vétérinaires de la Préfecture de Police de Paris”. Nephron reduction experimental protocol was approved by the ethical committee of the Paris Descartes University (approval number: A75-15-34).
2-month-old Galectin-3−/− 129/Sv mice46 and 129/Sv wild-type littermates were subjected to 75% nephrectomy (Nx) or sham operation (Control), as previously described22. After surgery, mice were fed a defined diet containing 30% casein and 0.5% sodium. Mice were sacrificed by cervical dislocation 90 days after surgery and weighed. Plasma samples were collected at the time of sacrifice and plasma creatinine was measured using Konelab Analyzer (Thermo Fisher Scientific, Waltham, MA, USA). Kidneys were harvested and weighed prior to analyses.
For immunohistochemistry, half-kidneys from 7 sham-operated wt, 5 sham-operated gal3−/−, 23 Nx wt and 20 Nx gal3−/− mice were fixed by successive 1 hour incubations in cold 70, 90, and 100% methanol solutions and then paraffin-embedded. Half kidneys were fixed overnight in 4% paraformaldehyde. The tissues were then processed for paraffin embedding as previously described6. Paraffin sections were prepared on poly-L-lysine hand-coated microscope slides for analyses. For cell proliferation analyses, 200 μg EdU (Thermo Fischer Scientific) in 100 μl sterile PBS was delivered 4 h prior mouse sacrifice by intraperitoneal injection.
Histological and immunohistological analyses
For histology, 5 µm paraffin sections were stained with Hematoxylin and Eosin (H&E) as previously described6. Mosaics of H&E-stained kidney sections were created under a 10x lens using a motorised Zeiss Axiovert200 microscope equiped with a color camera AxioCam HRc and Zeiss AxioVision software (Zeiss, Oberkochen, Germany). Zooms were taken under a 40x lens. The degree of tubular dilation was automatically quantified using a Nikon digital camera Dx/m/1200 and NIS software (Laboratory Imaging Ltd). Ten randomly selected microscopic fields (X200) were scored. Surgical scars were excluded from analysis.
For immunostaining, 8μm (cilium and centrosome analyses) or 30 µm (mitosis analyses) paraffin sections were dewaxed in a xylene bath and rehydrated once in isopropanol and in increasing ethanol solutions. Sections were saturated in 10% goat serum (Dako, Carpinteria, California, USA) for 30 min. Primary antibodies incubations were performed at 4 °C for 12 h in 10% goat serum solution. Secondary antibody incubations were done at room temperature for 2 hours in 10% goat serum solution. Tissue sections were mounted in Mowiol 4-88 (EMD Millipore) solution. To label proliferative cells, paraffin sections were stained with the Click-iT Edu Alexa Fluor 488 Imaging kit (Thermo Fisher Scientific).
Cell culture
HeLa cells were maintained in culture in DMEM High-Glucose (11965-092; Life Technologies) supplemented with 10% FBS and 1% Penicillin-Streptomycin (Life Technologies). To investigate metaphase plate position, the cells were fixed for 1 min in 0.025% glutaraldehyde, 0.05% Triton X-100 followed by 10 min in 1% glutaraldehyde and 45 min in 0.5 mg/mL NaBH4 all in cytoskeletal buffer48. For other immunostainings, the cells were fixed with pre-cooled 100% methanol (−20 °C) for 5 min. The cells were then processed for immunofluorescence microscopy as previously described14 and mounted in Mowiol 488 (EMD Millipore).
Transfections were carried out using Lipofectamine 2000 reagent (Life Technologies) following the manufacturer’s recommendations. Galectin-3 or NuMA reduction were performed by transfecting a mix of siRNA duplexes directed against human Galectin-3 (gal3 siRNA #1: 5′-AUAAGGCACAAUCAGUGGC-3′/5′-GCCACUGAUUGUGCCUUAU-3′, gal3 siRNA #2: 5′-ACCCAAACCCUCAAGGAUGTT-3′/5′-CCAUCUUCUGGACCAGCCAA-3′), or human NuMA1 (NuMA siRNA #1: 5′-CCUCACCGAGAAGGAUGCACAGAUA-3′/5′-UAUCAGUGCAUCCUUCUCGGUGAGG-3′, NuMA siRNA #2: 5′-CAAAGAGCUGCGAGCUGAAGCUGAA-3′/5′-UUCAGCUUCAGCUCGCAGCUCUUUG-3′) (LifeTechnologies). Control transfections were performed with luciferase siRNA (Dharmacon, GE-healthcare, Chalfont St. Giles, UK). Phenotypes of Galectin-3 or NuMA reduction was analyzed 48 to 72 h later.
For rescue experiments, Galectin-3 or NuMA depletion were performed by transfecting Galectin-3 CRISPR/Cas9 Knockout or NuMA CRISPR/Cas9 Knockout plasmids purchased from Santa Cruz (Dallas, Texas, USA). CRISPR/Cas9 plasmids were co-transfected with puromycin resistance gene coding plasmid. Co-transfected cells were selected in DMEM supplemented with 1 μg/ml puromycin (Invivogen, San Diego, CA, USA).
Plasmids
Galectin-3-GFP, Galectin-3-R186S-GFP and Galectin-3-G182A-GFP were generated as previously described33. NuMA-GFP construct was purchased from Addgene (Dr. M. Mancini (Baylor College of Medicine, Texas, USA). Mutated forms of NuMA-GFP were generated by site-directed mutagenesis using a Phusion-site directed mutagenesis kit (Thermo Scientific) with forward primers for the S1844T form 5′-GCTCCTGCTACTCAGGCTAGCCTG-3′ and for the S1844A form 5′-GCTCCTGCTGCTCAGGCTAGCCTG-3′ and as reverse primer 5′-AGCAGGAGCAGACCGCGTGCTGTAG-3′.
Drug Treatment
For microtubule regrowth experiments, HeLa cells were incubated in 10 ng/ml nocodazole for 4 h at 37 °C, followed by a 30 min at 4 °C. Microtubule regrowth was initiated by with pre-warmed complete media and incubation at 37 °C for several time points. Cells were then analyzedafter fixation in −20 °C pre-cooled methanol for 5 min. To inhibit O-linked N-acetylgalactosamine addition, cells were treated with 4 mM GalNAc-α-O-benzyl for 48 h. To inhibit N-linked glycosylation, cells were treated with 10 µg/ml swainsonine for 24 h. To inhibit O-linked N-actylglucosamine addition, cells were treated with 50 or 100 µM Ac-5S-GlcNAc for 48 h.
Biochemistry
Cell lysates were prepared as previously described14. For immunoprecipitation, cell extracts were prepared from enriched metaphase cells by mitotic shake-off. Mitotic cell lysates were precleared by incubation with protein-A/Sepharose beads, and then antibodies were applied. Immunoprecipitation of Galectin-1 and incubation of protein-A coated beads alone served as controls. Immunocomplexes were recovered by addition of Protein-A/Sepharose beads overnight at 4 °C, followed by three washes in PBS. Finally, the beads were resuspended in Laemmli buffer for Western blot. For the identification of Galectin-3 binding proteins, beads were submitted to proteolytic digestion and the peptides were analyzed by mass spectrometry (ESI-LTQ-Orbitrap spectrometer) coupled with nano LC chromatographic separation system (Proxeon) in the IJM Proteomic facility. For immunoprecipitation in presence of sugar agonists, glucose, lactose or galactose was added to the lysis buffer to a final concentration of 300, 100 and 300 mM, respectively. Samples were loaded in NuPAGE precast polyacrylamide gels (Thermo Scientific). Primary antibodies were detected using HRP-conjugated secondary antibodies and SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Scientific), visualized on a ImageQuant LAS4000 (GE-Healthcare). Signal quantification was performed using ImageJ.
Image acquisition
Immunofluorescent images were acquired on a Zeiss Axio Imager.Z1 microscope (Zeiss) equipped with an AxioCam digital camera using a 100x NA 1.4 objective. When applied, deconvolution was performed as previously described49. Confocal image sections were acquired on Leica TCS SP5 microscope (Leica Microsystems, Deerfield, Illinois, USA) using a 63x NA 1.4 and 100x NA 1.4 lenses from the Imagoseine plateform (IJM). 3D-SIM images were acquired on the Nikon N-SIM system using a 100x APO TIRF NA 1.49 objective in the NIKON Imaging Center at the Curie Institute (Paris, France). The images reconstruction based on the Gustafsson methods50 were performed on the NIS-Elements Ar software. Videomicrosocopy were performed with a spinning-disk confocal set-up on a Nikon TiE inverted microscope equiped with a CoolSNAP HQ2 camera operating under Metamorph software. Images were then adjusted for brightness and contrast using ImagJ software.
For ciliary length measurement, primary cilia were detected using acetylated-α-tubulin immunostaining. 3D reconstruction of confocal Z-stacks acquired was performed and ciliary length was measured using the filament option of Imaris Software.
For the enrichment of Galectin-3 at the spindle, ImagJ was used to manually measure the total pixel intensity of Galectin-3 at the spindle pole, which was reported to the total pixel intensity of Galectin-3 in the cytoplasm. The measurement of NuMA volume along the spindle was performed on ImageJ as previously described51. Spindle pole was manually isolated using α-tubulin staining. Line scan was performed for each isolated spindle for α-tubulin and the corresponding marker analyzed. Volume analysis was performed using 3D Object Counter plugin52 on ImageJ.
Statistical analyses
Statistical analyses have been performed using Prism (GraphPad software, La Jolla, CA, USA).
Electronic supplementary material
Acknowledgements
We thank Valérie Doye (Institut Jacques Monod, IJM) for precious advices, René-Marc Mège (IJM) and Anne-Lise Haenni (IJM) for critical reading the manuscript, David Vocadlo for sharing of the O-GlcNAc inhibitor and Vincent Leguillier (IJM) for technical help. We acknowledge the ImagoSeine facility (IJM), also associated to IBiSA and France BioImaging infrastructures, where confocal microscopy analyses were performed. We are grateful to all the persons working in the Laboratoire d’Expérimentation Animale et Transgénèse (LEAT) facility for technical assistance. High-resolution 3D-SIM analyses were acquired at the Nikon center imaging facility (Curie Institute, Paris). Mass spectrometry analyses were done at proteomic facility (IJM). This work was supported by grants from the INSERM (to F.T.), Université Paris Descartes (to F.T.), the Agence Nationale pour la Recherche (to F.T.), the GEFLUC Paris-Il-de-France “Les entreprises contre le cancer” (to D.D.), from the “Association pour la Recherche contre le Cancer” (to D.D.) and from “La Ligue, Comité de Paris” (to D.D.).
Author Contributions
J.M., L.S., A.V., A.M., T.D., M.B., F.T. and D.D. performed the experiments. J.M., F.P., F.T. and D.D. designed the experiments. U.J.N. and H.L. provided Galectin-3 mutant constructs. J.M., L.S., A.V., M.B., F.T. and D.D. performed analyses. J.M., F.T. and D.D. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01614-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nature reviews. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cecioni S, Vocadlo DJ. Tools for probing and perturbing O-GlcNAc in cells and in vivo. Curr Opin Chem Biol. 2013;17:719–728. doi: 10.1016/j.cbpa.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. The Journal of biological chemistry. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagel AK, Ball LE. O-GlcNAc transferase and O-GlcNAcase: achieving target substrate specificity. Amino Acids. 2014;46:2305–2316. doi: 10.1007/s00726-014-1827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. The Journal of cell biology. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. Journal of cell science. 2010;123:13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slawson C, et al. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. The Journal of biological chemistry. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2–ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan EP, Caro S, Potnis A, Lanza C, Slawson C. O-linked N-acetylglucosamine cycling regulates mitotic spindle organization. The Journal of biological chemistry. 2013;288:27085–27099. doi: 10.1074/jbc.M113.470187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabinovich GA, Thijssen VL. Introduction to special issue: Galectins go with the flow. Glycobiology. 2014;24:885–885. doi: 10.1093/glycob/cwu081. [DOI] [PubMed] [Google Scholar]
- 11.Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochimica et biophysica acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Delacour D, Koch A, Jacob R. The role of galectins in protein trafficking. Traffic (Copenhagen, Denmark) 2009;10:1405–1413. doi: 10.1111/j.1600-0854.2009.00960.x. [DOI] [PubMed] [Google Scholar]
- 13.Chiu MG, et al. Galectin-3 associates with the primary cilium and modulates cyst growth in congenital polycystic kidney disease. The American journal of pathology. 2006;169:1925–1938. doi: 10.2353/ajpath.2006.060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch A, Poirier F, Jacob R, Delacour D. Galectin-3, a novel centrosome-associated protein, required for epithelial morphogenesis. Molecular biology of the cell. 2010;21:219–231. doi: 10.1091/mbc.E09-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clare DK, et al. Basal foot MTOC organizes pillar MTs required for coordination of beating cilia. Nature communications. 2014;5:4888. doi: 10.1038/ncomms5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Califice S, Castronovo V, Van Den Brule F. Galectin-3 and cancer (Review) International journal of oncology. 2004;25:983–992. [PubMed] [Google Scholar]
- 17.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 18.Maiato H, Logarinho E. Mitotic spindle multipolarity without centrosome amplification. Nature cell biology. 2014;16:386–394. doi: 10.1038/ncb2958. [DOI] [PubMed] [Google Scholar]
- 19.Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130459–20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavali PL, Peset I, Gergely F. Centrosomes and mitotic spindle poles: a recent liaison? Biochemical Society transactions. 2015;43:13–18. doi: 10.1042/BST20140269. [DOI] [PubMed] [Google Scholar]
- 21.Fant X, Merdes A, Haren L. Cell and molecular biology of spindle poles and NuMA. International review of cytology. 2004;238:1–57. doi: 10.1016/S0074-7696(04)38001-0. [DOI] [PubMed] [Google Scholar]
- 22.Pillebout E, et al. Proliferation and remodeling of the peritubular microcirculation after nephron reduction: association with the progression of renal lesions. The American journal of pathology. 2001;159:547–560. doi: 10.1016/S0002-9440(10)61726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillebout E, et al. JunD protects against chronic kidney disease by regulating paracrine mitogens. The Journal of clinical investigation. 2003;112:843–852. doi: 10.1172/JCI200317647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laouari D, et al. TGF-alpha mediates genetic susceptibility to chronic kidney disease. J Am Soc Nephrol. 2011;22:327–335. doi: 10.1681/ASN.2010040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laouari D, et al. A transcriptional network underlies susceptibility to kidney disease progression. EMBO Mol Med. 2012;4:825–839. doi: 10.1002/emmm.201101127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin F, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel V, et al. Acute kidney injury and aberrant planar cell polarity induce cyst formation in mice lacking renal cilia. Human molecular genetics. 2008;17:1578–1590. doi: 10.1093/hmg/ddn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdeguer F, et al. A mitotic transcriptional switch in polycystic kidney disease. Nature medicine. 2010;16:106–110. doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. The New England journal of medicine. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. International journal of cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 32.Delacour D, et al. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic (Copenhagen, Denmark) 2007;8:379–388. doi: 10.1111/j.1600-0854.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 33.Salomonsson E, et al. Mutational tuning of galectin-3 specificity and biological function. The Journal of biological chemistry. 2010;285:35079–35091. doi: 10.1074/jbc.M109.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gloster TM, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Molecular biology of the cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang CH, Lambie EJ, Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. The Journal of cell biology. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Compton DA, Cleveland DW. NuMA is required for the proper completion of mitosis. The Journal of cell biology. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boscher C, Dennis JW, Nabi IR. Glycosylation, galectins and cellular signaling. Current opinion in cell biology. 2011;23:383–392. doi: 10.1016/j.ceb.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Hikita C, et al. Induction of terminal differentiation in epithelial cells requires polymerization of hensin by galectin 3. The Journal of cell biology. 2000;151:1235–1246. doi: 10.1083/jcb.151.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends in cell biology. 2010;20:214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krzeslak A, Lipinska A. Galectin-3 as a multifunctional protein. Cellular & molecular biology letters. 2004;9:305–328. [PubMed] [Google Scholar]
- 42.Sakaki M, et al. Clinical significance of Galectin-3 in clear cell renal cell carcinoma. J Med Invest. 2010;57:152–157. doi: 10.2152/jmi.57.152. [DOI] [PubMed] [Google Scholar]
- 43.Straube T, et al. Changes in the expression and subcellular distribution of galectin-3 in clear cell renal cell carcinoma. J Exp Clin Cancer Res. 2011;30:89. doi: 10.1186/1756-9966-30-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nangia-Makker P, et al. Galectin-3 induces endothelial cell morphogenesis and angiogenesis. The American journal of pathology. 2000;156:899–909. doi: 10.1016/S0002-9440(10)64959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HR, Lin HM, Biliran H, Raz A. Cell cycle arrest and inhibition of anoikis by galectin-3 in human breast epithelial cells. Cancer research. 1999;59:4148–4154. [PubMed] [Google Scholar]
- 46.Colnot C, Ripoche MA, Scaerou F, Foulis D, Poirier F. Galectins in mouse embryogenesis. Biochemical Society transactions. 1996;24:141–146. doi: 10.1042/bst0240141. [DOI] [PubMed] [Google Scholar]
- 47.Harborth J, Wang J, Gueth-Hallonet C, Weber K, Osborn M. Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 1999;18:1689–1700. doi: 10.1093/emboj/18.6.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Small, J., Rottner, K., Hahne, P. & Anderson, K. I. Visualising the actin cytoskeleton. Microsc Res Tech47, 3–17, doi:10.1002/(SICI)1097-0029 (19991001)47:1<3::AID-JEMT2>3.0.CO;2-2 (1999). [DOI] [PubMed]
- 49.Sibarita JB. Deconvolution microscopy. Adv Biochem Eng Biotechnol. 2005;95:201–243. doi: 10.1007/b102215. [DOI] [PubMed] [Google Scholar]
- 50.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophysical journal. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lecland N, Luders J. The dynamics of microtubule minus ends in the human mitotic spindle. Nature cell biology. 2014;16:770–778. doi: 10.1038/ncb2996. [DOI] [PubMed] [Google Scholar]
- 52.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


