Abstract
MicroRNAs (miRNAs/miRs) are short non-coding RNAs (between 20 and 22 nucleotides) that regulate gene expression by binding to the 3′-untranslated region of target mRNA, and preventing protein translation or inducing mRNA destabilization. miRNAs are predicted to target ~60% of all mRNAs, therefore providing a marked degree of regulation of a number of cellular processes. In the present study, the expression of miR-548c-3p was determined by reverse transcription-quantitative polymerase chain reaction analysis and demonstrated to be markedly downregulated in clinical malignant glioma tissues and the glioma T98G cell line compared with normal human brain tissue. Transfection of miR-548c-3p inhibited cell proliferation by inducing G1 cell cycle arrest and also inhibited the migration of the T98G cells in vitro. Furthermore, a bioinformatic algorithm and a luciferase reporter assay identified proto-oncogene c-Myb (c-Myb) as a potential direct target of miR-548c-3p. Further experiments demonstrated that the inhibition of c-Myb by miR-548c-3p partially mediated the antitumor effect of miR-548c-3p. The results of the present study provide the novel insight that miR-548c-3p inhibits glioma tumorigenesis by targeting c-Myb. Therefore, miR-548c-3p may contribute to the development of improved glioma treatment.
Keywords: microRNA-548c-3p, proto-oncogene c-Myb, glioma, proliferation, migration
Introduction
Gliomas are the most common type of primary brain tumor (1,2). The World Health Organization classification system groups gliomas into four histological grades as follows: Astrocytomas, oligodendrogliomas, ependymomas and oligo-astrocytomas (mixed gliomas) (3). Astrocytomas are subdivided as follows: Pilocytic, grade I; diffuse, grade II; anaplastic, grade III; and glioblastoma multiforme, grade IV (3). Malignant gliomas account for 80% of all gliomas and are subcategorized into grade III/IV tumors (4). The incidence of gliomas has increased from 5.9/100,000 people in 1973 to 6.61/100,000 people in 2016, primarily due to improved radiological diagnosis (5,6). Despite combined treatment regimens, however, it remains an incurable disease and the prognosis is poor (7). Thorough investigation is required to improve our understanding of its biological characteristics and identify a novel molecular target for clinical therapy.
In 1993, the identification of a small endogenous regulatory RNA molecule in Caenorhabditis elegans led to the description of a family of numerous short single-stranded ribonucleic acids (between 20 and 22 nucleotides) termed microRNAs (miRNAs/miRs) (8). These molecules are critical post-transcriptional regulators of gene expression in complex organisms. It is not unexpected, therefore, that miRNAs are themselves tightly regulated to allow for gene expression to be shaped in a temporally restrained and tissue-specific manner, which is required for properly structured organismal development and growth (9–11). miR-548 is a poorly-conserved primate-specific miRNA gene family, which is involved in the regulation of the actin cytoskeleton, the mitogen-activated protein kinase signaling pathway, ubiquitin-mediated proteolysis, glioma, colorectal cancer and non-small cell lung cancer (12).
Upregulation of miR-548c-3p was also identified in human embryonic stem cells and in unfractionated castration-resistant prostate cancer (13). Overexpression of miR-548c-3p was also reported in the blood of patients with gastric cancer (14). The higher expression of miR-548c-3p may be more definite in the case of Helicobacter pylori-negative gastric cancer (15). These results suggested that miR-548c-3p is involved in tumor progression. In the present study, miR-548c-3p was identified to be downregulated in glioma tissues and cell lines; however, the underlying molecular mechanism remains unclear.
The proto-oncogene c-Myb (c-Myb) codes for a transcription factor that is a member of the Myb family (16,17). The transcription factor Myb has a key role in stem and progenitor cell regulation within the colonic crypts, bone marrow and a neurogenic region of the adult brain (16). In various types of human cancer, mutations have been identified in the Myb gene: Overexpression of c-Myb contributes to transformation in pediatric T-cell acute lymphocyte leukemia, pancreatic tumors and colon tumors (18,19). Rearrangements and amplifications of Myb have been identified in ~25% of diffuse cerebral gliomas (20). These results suggest that c-Myb is involved in glioma tumorigenesis.
In the present study, the potential effect of miR-548c-3p on glioma tumorigenesis was investigated. miR-548c-3p was identified to be downregulated in human malignant glioma tissues. Furthermore, c-Myb was identified to be the direct target of miR-548c-3p and mediated the biological effect of miR-548c-3p. These results suggest that miR-548c-3p may be important in the regulation of glioma development and may lead to clinical applications in the treatment of glioma.
Materials and methods
Cell culture
The human glioma T98G, U87 and U251 cell lines, and the human embryonic kidney (HEK)-293 cells (CRL-1573), were obtained from the American Type Culture Collection (Manassas, VA, USA) and grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare, Life Sciences Logan, UT, USA) at 37°C in a humidified atmosphere with 5% CO2. Human brain tissues and glioma specimens were obtained from 11 patients (5 males and 6 females; age, 35–55 years) treated at the Eye Hospital, Wenzhou Medical University (Wenzhou, China) between September 2012 and July 2013. All patients provided written informed consent. All studies and procedures involving human tissue were approved by the Wenzhou Medical University Institutional Review Board. Patient samples were used in accordance with The Declaration of Helsinki.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The T98G cells (1×105) were seeded in 6-well plates and total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A 10-ng sample of total RNA was transcribed into cDNA using a TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), and the miR-548c-3p expression level was quantified using a TaqMan® MicroRNA Assay kit (cat. no. 479537_MIR; Ambion; Thermo Fisher Scientific, Inc.), which included primers for has-miR-548c-3p. All procedures were performed according to the manufacturer's protocols. The thermocycling conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 5 sec, 58°C for 10 sec and 72°C for 5 sec. Each sample was analyzed in triplicate. qPCR was performed using a 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), the expression of miR-548c-3p was normalized to the expression of U6 small nuclear RNA, and relative expression levels were calculated using the 2−ΔΔCq method (21). The expression level of miR-548c-3p in the human brain was set as the wild-type control. To determine c-Myb expression in the T98G cells (1×105), the cells were transfected with miR-548c-3p using 1 µl Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cultured at 37°C for 24 h. c-Myb expression levels were then quantified by measuring cyanine dye incorporation (SYBR Green PCR Master mix; Applied Biosystems; Thermo Fisher Scientific, Inc.) using the 7500 Real-Time PCR system. The primer sequences for c-Myb were as follows: Forward, 5′-ACGAGGATGATGAGGACTTTGAG-3′; and reverse, 5′-TTTTCCCCAAGTGACGCTTT-3′.
Cell proliferation and colony formation assays. The T98G cells were plated at 3×103 cells/well in 96-well plates
All transfections were performed in triplicate. Cells in each well were transfected with 50 nM miR-548c-3p precursor molecule (cat. no. MC11455; Ambion; Thermo Fisher Scientific, Inc.) or a negative control (NC) precursor miRNA (cat. no. 17110; Ambion; Thermo Fisher Scientific, Inc.). Following 1–4 days of culture in a humidified atmosphere with 5% CO2 at 37°C, cell proliferation was assessed using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay, a colorimetric method for determining the number of viable cells, with a CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega Corp., Madison, WI, USA), according to the manufacturer's protocol. MTS solution was added to each well prior to incubation at 37°C for 3 h. Cell proliferation was assessed by measuring the absorbance at 490 nm using a microtiter plate reader (Molecular Devices, LLC, Sunnyvale, CA, USA). The T98G cells were transfected with 50 nM c-Myb-specific small interfering RNA (siRNA; 50 nM; Ambion; Thermo Fisher Scientific, Inc.) or NC siRNA (cat. no. 4392420; Ambion; Thermo Fisher Scientific, Inc.) using Lipofectamine® 2000 reagent. The mock control group was untreated T98G cells cultured under normal conditions. The MTS assay was performed 3 days after transfection. To evaluate the colony formation ability, T98G cells transfected with miR-548c-3p or NC siRNA were seeded in 3.5-cm plates (1,000 cells/dish). The colonies were fixed with 10% formalin at room temperature for 30 min, then stained with 0.1% crystal violet (Sigma; Merck Millipore, Darmstadt, Germany) at room temperature for 30 min. Colonies with >50 cells were counted using a light microscope (Carl Zeiss, Axio Observer D1, Germany) after 6 days.
Flow cytometric analysis of the cell cycle
The T98G cells were plated into 60-mm dishes and cultured at 37°C until between 50 and 70% confluence for each transfection. Each cell line was transfected with 50 nM miR-548c-3p precursor molecule or NC siRNA. A total of 48 h after transfection, the cells were collected, washed with phosphate-buffered saline (PBS) and stained with propidium iodide using the Cycletest™ Plus DNA kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer's protocol. Stained cells (1×105) were analyzed for DNA content using a FACSCalibur flow cytometer with CellQuest Pro software (version 6.0) (both BD Biosciences).
Bioinformatics prediction and luciferase reporter assays
TargetScan (www.targetscan.org) was used to predict the direct targets of miR-548c-3p. The 3′-untranslated region (UTR) of human c-Myb was amplified from human genomic DNA and individually cloned into a pMIR-REPORT vector (Ambion; Thermo Fisher Scientific, Inc.) using directional cloning. Seed regions were mutated to remove all complementarity to nucleotides 1 to 7 of miR-548c-3p using a QuikChange XL Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA). The HEK-293 cells were co-transfected with 0.4 µg firefly luciferase reporter vector and 0.02 µg control vector containing Renilla luciferase (pRL-SV40 vector) (both Promega Corp.), together with 50 nM miR-548c-3p precursor molecule or NC precursor miRNA, using Lipofectamine® 2000 in 24-well plates. Each transfection was performed with four replicate wells. Luciferase assays were performed 24 h after transfection using the Dual-Luciferase Reporter Assay system (Promega Corp.) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Transwell migration assays
The T98G cells were transfected with 50 nM miR-548c-3p precursor molecule or NC. After 24 h, the cells were harvested by trypsinization and washed once with Hanks' balanced salt solution (Invitrogen; Thermo Fisher Scientific, Inc.). Transwell culture inserts (pore size, 8-µm; Costar; BD Biosciences) were placed into the wells of 24-well culture plates, separating the upper and the lower chambers. DMEM (400 µl) was added in the lower chamber and 1×105 cells were added to the upper chamber. A total of 24 h after incubation at 37°C and 5% CO2, the cells that adhered to the inserts were fixed with 70% methanol at room temperature for 30 min and then stained with 0.1% crystal violet for 30 min. The number of cells that had migrated through the pores was quantified by counting 10 independent visual fields using a light microscope and a ×20 objective.
Western blot analysis
The T98G cells (1×105) were transfected with the miR-548c-3p precursor molecule or NC. A total of 24 h after transfection, the cells were washed with ice-cold PBS and subjected to lysis in a lysis buffer (50 mM Tris-HCl, 1 mM EDTA, 20 g/l SDS, 5 mM dithiothreitol and 10 mM phenylmethylsulfonyl fluoride). Protein concentration of whole cell lysates was assessed using the Pierce™ BCA Protein Assay Kit (Pierce; Thermo Fisher Scientific, Inc.). Protein lysates (50 µg each) were separated by 10% SDS-PAGE, then electrotransferred onto nitrocellulose membranes. The membranes were blocked with a buffer containing 5% skimmed milk powder in PBS with 0.05% Tween-20 for 2 h and incubated overnight with primary antibodies (described below) at 4°C. Antibodies directed against c-Myb (1:1,000; cat. no. 12319), protein lin-28 homolog B (Lin28b; 1:1,000; cat. no. 4196), Myc proto-oncogene protein (Myc; 1:1,000; cat. no. 9402) and GAPDH (1:2,000; cat. no. 2118) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Following a second wash with PBS containing 0.05% Tween-20, the membranes were incubated at room temperature for 1 h with horseradish peroxidase-conjugated anti-rabbit Immunoglobulin G secondary antibody (dilution, 1:4,000; cat. no. 7074; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and developed with an enhanced chemiluminescence detection kit (Pierce; Thermo Fisher Scientific, Inc.). GAPDH was used as a loading control.
Caspase activity assay
Apoptosis in the T98G cells was determined using the Caspase-Glo® 3/7 assay kit (Promega Corp.) according to the manufacturer's protocol. The T98G cells were plated in triplicate in 96-well plates and transfected with miR-548c-3p as aforementioned. Samples were then incubated at room temperature with the caspase substrate (provided in the kit) for 2 h followed by measurement of the optical density at 560 nm using a microtiter plate reader (Molecular Devices, LLC, Sunnyvale, CA, USA).
Wound healing assay
A total of 1.5×105 T98G cells/well were cultured in a 12-well plate for 24 h prior to transfection with miR-548c-3p or NC. After 2 days, the cells were scratched with a 100-µl pipette tip, washed with Hanks' balanced salt solution three times, and cultured in serum-free DMEM at 37°C and 5% CO2. Images were captured 0, 1 and 3 days subsequent to the wound being made.
Statistical analysis
SPSS software (version 20.0; IBM SPSS, Inc., Armonk, NY, USA) was used for statistical analysis. Student's t-test was used to determine the statistical significance of differences between groups. All results are presented as the mean ± standard error of the mean from experiments performed at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-548c-3p is downregulated in glioma tissues and glioma cell lines
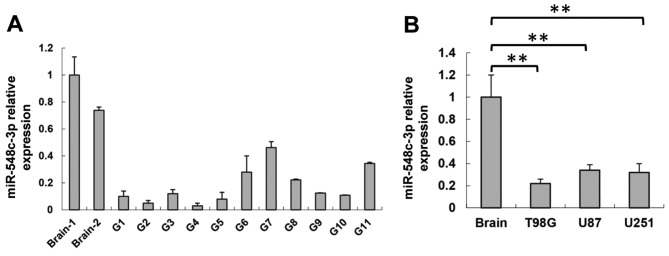
To determine whether miRNA was involved in the regulation of glioma tumorigenesis, the expression of miR-548c-3p was determined in glioma and wild-type brain tissues. Total RNA was extracted and RT-qPCR analysis was performed. miR-548c-3p expression was decreased significantly in all 11 glioma tissue samples compared with two wild-type brain samples (Fig. 1A). Consistent with the data from glioma tissues, miR-548c-3p expression was demonstrated to be significantly downregulated in the U87, U251 and T98G glioma cell lines compared with the brain tissue (P<0.01; Fig. 1B). These results indicated that miR-548c-3p serves certain roles in glioma development.
Figure 1.
miR-548c-3p expression is downregulated in human glioma tissues and human glioma cell lines. (A) qPCR analysis of miR-548c-3p in human glioma tissues (G1-G11) compared with two wild-type human brain tissues. (B) qPCR analysis of miR-548c-3p in human glioma U87, U251 and T98G cell lines compared with human brain tissue. **P<0.01. qPCR, quantitative polymerase chain reaction; miR, microRNA.
miR-548c-3p inhibits T98G cell proliferation and migration
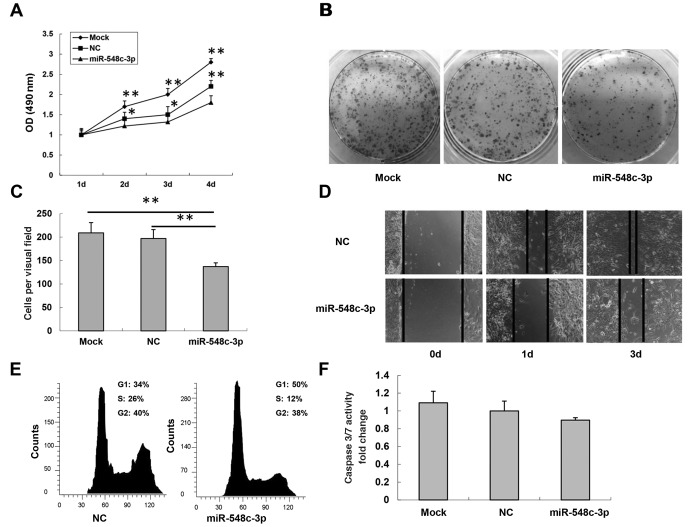
As miR-548c-3p expression was downregulated in glioma, its biological effects on a glioma cell line were investigated. The T98G cells were transfected with the miR-548c-3p precursor molecule or NC. An MTS assay was performed to assess growth inhibition 1, 2, 3 and 4 days after transfection. Transient transfection of miR-548c-3p into T98G cells caused a significant inhibition of proliferation at day 4 compared with that of the NC and mock cells (25±6.2% inhibition; P<0.01; Fig. 2A). To further verify miR-548c-3p-mediated inhibition of cell proliferation, a colony formation assay was used to determine visually the effect of miR-548c-3p transfection on T98G cells (Fig. 2B). Cell migration, a prerequisite for malignant transformation and metastasis, was assessed using Transwell migration and wound healing assays. In the Transwell assay, T98G cells were transfected with either the miR-548c-3p precursor or an NC precursor. The cells were seeded on culture inserts, and the ability of cells to migrate to the underside of the inserts was determined. As presented in Fig. 2C, migration of miR-548c-3p-transfected cells was significantly decreased compared with mock- and NC-transfected cells (mock, 205±45; NC, 197±49; miR-548c-3p, 137±38; both P<0.01). The results of the wound healing assay were consistent with the Transwell migration assay. Migration of miR-548c-3p-transfected T98G cells was slower than that of NC-transfected T98G cells 1 and 3 days after transfection (Fig. 2D). Overall, the introduction of miR-548c-3p resulted in reduced cell motility.
Figure 2.
miR-548c-3p inhibits T98G cell proliferation and migration. (A) An MTS cell proliferation assay was performed 1, 2, 3 and 4 days after transfection of T98G cells with either miR-548c-3p or NC. (B) Colony formation assay of T98G cells transfected with miR-548c-3p or NC. (C) Transwell migration assay of miR-548c-3p- or NC-transfected T98G cells. (D) Wound healing assay of T98G cells at 0, 1 and 3 days after transfection with miR-548c-3p or NC. (E) Flow cytometry assay of T98G cells with miR-548c-3p or NC. (F) Caspase 3/7 activity assay of T98G cells transfected with either NC or miR-548c-3p. Caspase 3/7 activity is relative to that of NC, set as 1. *P<0.05; **P<0.01. miR, microRNA; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; NC, negative control scrambled oligonucleotide; OD, optical density.
The cell cycle was investigated using flow cytometry. In T98G cells transfected with miR-548c-3p, 50% accumulated in the G1 phase compared with 34% of cells for NC-transfected cells (Fig. 2E). To further evaluate miR-548c-3p-mediated inhibition of cell proliferation, caspase activity was investigated to determine the involvement of apoptosis. No significant difference was observed in caspase 3/7 activity between miR-548c-3p- and NC-transfected cells (Fig. 2F). Therefore, these results indicated that T98G cell growth was inhibited by miR-548c-3p expression via cell cycle G1 arrest rather than the induction of apoptosis.
c-Myb is a target of miR-548c-3p
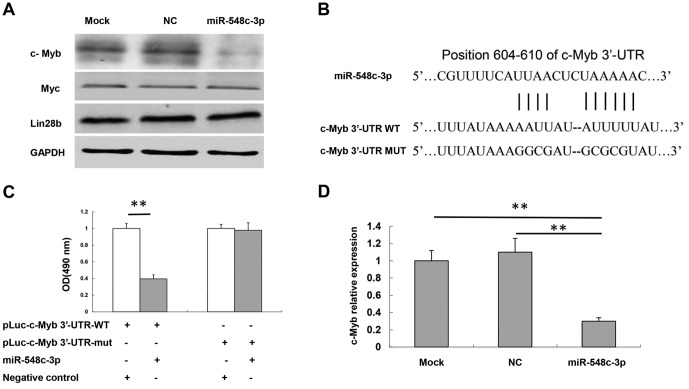
Analysis using TargetScan (www.targetscan.org) was conducted for miR-548c-3p target prediction to investigate the underlying molecular mechanisms of miR-548c-3p-mediated cell proliferation and migration. The potential targets associated with tumorigenesis were identified as c-Myb, Myc and Lin28b. The effect of miR-548c-3p on the expression of these genes was investigated, with alteration of c-Myb being the most marked (Fig. 3A). The potential binding site of miR-548c-3p was predicted in the 3′-UTR of c-Myb mRNA. Alignment between the predicted miR-548c-3p target sites and miR-548c-3p, the conserved 7-bp seed sequence for miR-548c-3p-mRNA pairing, is presented in Fig. 3B. To evaluate the specific regulation of c-Myb through the predicted binding site, the c-Myb 3′-UTR sequence was amplified and inserted downstream of the firefly luciferase coding region of a pMIR-Luc vector. Mutants of the putative binding site were also prepared.
Figure 3.
c-Myb is a target of miR-548c-3p. (A) Protein expression levels of Myc, Lin28b and c-Myb in T98G cells following transfection with miR-548c-3p were determined using western blotting. GAPDH was used as an internal control. (B) Specific locations of the binding sites. Alignment between the predicted miR-548c-3p target sites, miR-548c-3p and c-Myb 3′-UTR-WT or 3′-UTR-mut, the conserved ‘seed’ sequence of between 7 and 8 bp, for miR-548c-3p-mRNA pairing, is indicated. (C) Human embryonic kidney-293 cells were co-transfected with 50 nM miR-548c-3p, pLuc-c-Myb 3′-UTR-WT or pLuc-c-Myb 3′-UTR-mut along with a pRL-SV40 reporter plasmid. After 24 h, luciferase activity was measured. (D) c-Myb mRNA expression levels in T98G cells following transfection with miR-548c-3p were determined using the quantitative polymerase chain reaction. U6 small nuclear RNA was used as an internal control. **P<0.01. c-Myb, proto-oncogene c-Myb; miR, microRNA; Myc, Myc proto-oncogene protein; Lin28b, protein lin-28 homolog B; UTR, untranslated region; WT, wild-type; mut, mutant; OD, optical density.
As indicated, introduction of miR-548c-3p in HEK-293 cells with the wild-type 3′-UTR (pLuc-c-Myb 3′-UTR) construct significantly inhibited the luciferase activity compared with the NC group (P<0.01; Fig. 3C). Mutation of the binding site, using a mutant vector (pLuc-MET 3-UTR-Mut), completely eliminated the ability of miR-548c-3p to regulate luciferase expression. These results demonstrated that c-Myb was a potential target of miR-548c-3p. RT-qPCR assay results also demonstrated that miR-548c-3p was able to inhibit c-Myb mRNA expression compared with NC and mock cells (P<0.01; Fig. 3D).
c-Myb rescues the effect of miR-548c-3p overexpression
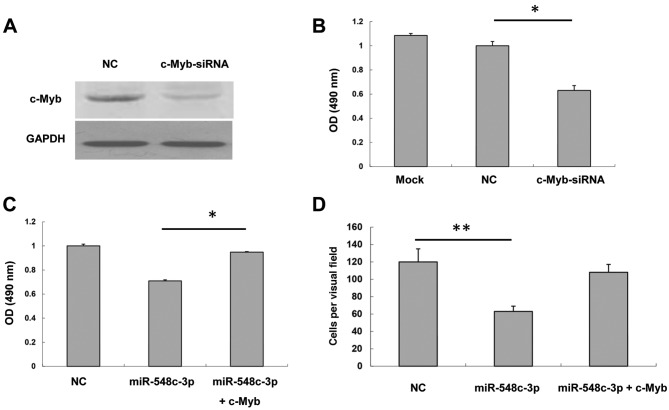
To confirm that c-Myb was responsible for miR-548c-3p inhibition of T98G proliferation and migration, c-Myb was knocked down and the knockdown efficiency of c-Myb siRNA was analyzed. A western blot assay demonstrated that c-Myb siRNA was able to inhibit c-Myb expression to a marked extent (Fig. 4A). An MTS assay was performed to test the effect of c-Myb siRNA on T98G proliferation. The proliferation of T98G cells transfected with c-Myb siRNA was inhibited compared with the NC (Fig. 4B). Furthermore, cells were co-transfected with miR-548c-3p and c-Myb to evaluate the effects of c-Myb on miR-548c-3p-overexpressed cells. As presented in Fig. 4C, miR-548c-3p inhibited T98G cell proliferation; however, cells co-transfected with miR-548c-3p and c-Myb rescued proliferation markedly compared with NC. This suggested that miR-548c-3p may exert its effect through the inhibition of c-Myb. The Transwell assay demonstrated that cells co-transfected with miR-548c-3p and c-Myb rescued the inhibitory effect of miR-548c-3p on T98G migration (Fig. 4D). Therefore, these data indicated that c-Myb is responsible for the inhibition by miR-548c-3p of the proliferation and migration of T98G cells.
Figure 4.
Expression levels of c-Myb are able to rescue miR-548c-3p function on proliferation and migration. (A) c-Myb protein expression levels in T98G cells following transfection with c-Myb siRNA were determined by western blotting. GAPDH was used as an internal control. (B) An MTS cell proliferation assay was performed 2 days after transfection of T98G cells with either c-Myb siRNA or NC. (C) An MTS cell proliferation assay was performed 2 days after transfection of T98G cells with miR-548c-3p or co-transfection of T98G cells with miR-548c-3p and c-Myb. (D) A Transwell migration assay of T98G was performed. T98G glioma cells were transfected with miR-548c-3p or co-transfected with miR-548c-3p and c-Myb. *P<0.05; **P<0.01. c-Myb, proto-oncogene c-Myb; miR, microRNA; siRNA, small interfering RNA; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; NC, negative control; OD, optical density.
Discussion
Following the identification of the miRNA let-7, a number of miRNAs have been linked to oncogenes and tumor suppressor genes, including the Ras proto-oncogene, the anti-apoptotic gene BCL2 and the potent p53 tumor suppressor gene (22,23). As downregulation of miRNA has been observed in various types of tumor, miRNAs function as tumor suppressors or oncogenes by interacting with their corresponding targets. However, it was previously demonstrated that upregulation of miRNA occurs in certain types of tumor (24). It was hypothesized that miRNAs are able to act as oncogenes or as tumor suppressors, depending on the type of tissue and the context in which they are expressed (25,26). Knowledge of the underlying molecular mechanism of miRNAs in cancer remains limited. miR-548c-3p has been investigated in certain types of cancer, including breast, prostate and H. pylori-negative gastric cancer (13,15,27). Mutation of miR-548c-3p may be an important driving force in tumorigenesis. Furthermore, it was reported that downregulation of miR-34b and miR-548c-3p, which target high-mobility group protein A1 (HMGA1), may account for the overexpression of HMGA1 protein detected in the majority of human pituitary adenomas (28). In the present study, the expression level of miR-548c-3p was investigated, and downregulation of miR-548c-3p in glioma tissues and the T98G cell line was demonstrated. Furthermore, miR-548c-3p inhibited T98G cell proliferation and migration. These results suggested that miR-548c-3p serves an important role in glioma.
To identify the potential targets of miR-548c-3p, candidate genes identified using TargetScan were screened. c-Myb was selected due to its involvement in numerous cancer types (29,30). c-Myb has the potential to induce cell proliferation and is likely to serve a role in stimulating progression through the cell cycle in wild-type cells. Furthermore, interactions with cell cycle regulators, including cyclin D1, may be important for its oncogenic activity (31). In the present study, the c-Myb 3′-UTR was inserted downstream of the firefly luciferase coding region of a pMIR-Luc vector. Using western blot analysis, it was demonstrated that c-Myb expression was markedly reduced when transfected with miR-548c-3p. These results identified that c-Myb was the target of miR-548c-3p. miR-548c-3p induced T98G arrest at G1 phase, which was possibly associated with the activation of genes required for the G1/S phase transition by c-Myb. However, elucidation of the signaling pathways involved requires further study.
It was previously demonstrated that the modulation of c-Myb levels in a number of tumor lines was frequently associated with increased migration and invasion capabilities (32). Tanno et al (33) demonstrated that the expression of neuronal cadherin was increased in c-Myb-expressing HEK-293 and LAN-5 cells, and may be responsible for the biological function of c-Myb. In the present study, bioinformatic prediction and experimental data indicated that miR-548c-3p is able to target c-Myb. Overexpression of c-Myb rescued the effect of miR-548c-3p. Therefore, this revealed that miR-548c-3p exerts its effect on cell proliferation and migration though downregulation of c-Myb; however, the underlying molecular mechanism remains unclear.
In summary, miR-548c-3p was identified as one of the key regulators of c-Myb involved in the tumorigenesis of glioma. Reconstruction of miR-548c-3p or inhibition of c-Myb function may be a promising therapeutic strategy for the treatment of glioma.
Acknowledgements
The authors would like to thank Dr Yihong Wang (Sir Run Run Shaw Hospital, Zhejiang University College of Medicine, Hangzhou, Zhejiang, China) for providing the clinical diagnosis of the glioma tissues. The present study was partially supported by the Natural Science Foundation of China (grant nos. NSFC81201588) the Key Projects in the National Science & Technology Pillar Program (No. 2015BAI09B01) and the Natural Science Foundation of Zhejiang Province (grant no. LQ12H16003).
References
- 1.Stupp R, Tonn JC, Brada M, Pentheroudakis G. High-grade malignant glioma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21:v190–v193. doi: 10.1093/annonc/mdq187. (Suppl 5) [DOI] [PubMed] [Google Scholar]
- 2.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleihues P, Ohgaki H. Primary and secondary glioblastomas: From concept to clinical diagnosis. Neuro Oncol. 1999;1:44–51. doi: 10.1215/15228517-1-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher JL, Schwartzbaum JA, Wrensch M, Wiemels JL. Epidemiology of brain tumors. Neurol Clin. 2007;25:867–890. doi: 10.1016/j.ncl.2007.07.002. vii. [DOI] [PubMed] [Google Scholar]
- 6.McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34:981–998. doi: 10.1016/j.ncl.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 9.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 10.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanno B, Cesi V, Vitali R, Sesti F, Giuffrida ML, Mancini C, Calabretta B, Raschellà G. Silencing of endogenous IGFBP-5 by micro RNA interference affects proliferation, apoptosis and differentiation of neuroblastoma cells. Cell Death Differ. 2005;12:213–223. doi: 10.1038/sj.cdd.4401546. [DOI] [PubMed] [Google Scholar]
- 12.Liang T, Guo L, Liu C. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J Biomed Biotechnol. 2012;2012:679563. doi: 10.1155/2012/679563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rane JK, Scaravilli M, Ylipää A, Pellacani D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T, Maitland NJ. MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur Urol. 2015;67:7–10. doi: 10.1016/j.eururo.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 15.Chang H, Kim N, Park JH, Nam RH, Choi YJ, Lee HS, Yoon H, Shin CM, Park YS, Kim JM, Lee DH. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver. 2015;9:188–196. doi: 10.5009/gnl13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rushton JJ, Davis LM, Lei W, Mo X, Leutz A, Ness SA. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene. 2003;22:308–313. doi: 10.1038/sj.onc.1206131. [DOI] [PubMed] [Google Scholar]
- 17.Rushton JJ, Ness SA. The conserved DNA binding domain mediates similar regulatory interactions for A-Myb, B-Myb and c-Myb transcription factors. Blood Cells Mol Dis. 2001;27:459–463. doi: 10.1006/bcmd.2001.0405. [DOI] [PubMed] [Google Scholar]
- 18.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Ness SA. Myb proteins: Angels and demons in normal and transformed cells. Front Biosci (Landmark Ed) 2011;16:1109–1131. doi: 10.2741/3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Schumacher SE, McKenna A, Lawrence MS, Bergthold G, Brastianos PK, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci USA. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 23.Cho WC. OncomiRs: The discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobson JR, Taipaleenmäki H, Hu YJ, Hong D, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Pratap J. hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3. Cancer Cell Int. 2014;14:73. doi: 10.1186/s12935-014-0073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107:505–510. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 26.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 29.Zhou YE, O'Rourke JP, Edwards JS, Ness SA. Single molecule analysis of c-myb alternative splicing reveals novel classifiers for precursor B-ALL. PLoS One. 2011;6:e22880. doi: 10.1371/journal.pone.0022880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brill LB, II, Kanner WA, Fehr A, Andrén Y, Moskaluk CA, Löning T, Stenman G, Frierson HF., Jr Analysis of MYB expression and MYB-NFIB gene fusions in adenoid cystic carcinoma and other salivary neoplasms. Mod Pathol. 2011;24:1169–1176. doi: 10.1038/modpathol.2011.86. [DOI] [PubMed] [Google Scholar]
- 31.Ganter B, Fu S, Lipsick JS. D-type cyclins repress transcriptional activation by the v-Myb but not the c-Myb DNA-binding domain. EMBO J. 1998;17:255–268. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ameh EA, Mshelbwala PM, Nasir AA, Lukong CS, Jabo BA, Anumah MA, Nmadu PT. Surgical site infection in children: Prospective analysis of the burden and risk factors in a sub-Saharan African setting. Surg Infect (Larchmt) 2009;10:105–109. doi: 10.1089/sur.2007.082. [DOI] [PubMed] [Google Scholar]
- 33.Tanno B, Sesti F, Cesi V, Bossi G, Ferrari-Amorotti G, Bussolari R, Tirindelli D, Calabretta B, Raschellà G. Expression of Slug is regulated by c-Myb and is required for invasion and bone marrow homing of cancer cells of different origin. J Biol Chem. 2010;285:29434–29445. doi: 10.1074/jbc.M109.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]