Abstract
Two-component systems are crucial for signal perception and modulation of bacterial behavior. Nevertheless, to date, very few ligands have been identified that directly interact with histidine kinases. The histidine kinase/response regulator system YehU/YehT of Escherichia coli is part of a nutrient-sensing network. Here we demonstrate that this system senses the onset of nutrient limitation in amino acid rich media and responds to extracellular pyruvate. Binding of radiolabeled pyruvate was found for full-length YehU in right-side-out membrane vesicles as well as for a truncated, membrane-integrated variant, confirming that YehU is a high-affinity receptor for extracellular pyruvate. Therefore we propose to rename YehU/YehT as BtsS/BtsR, after “Brenztraubensäure”, the name given to pyruvic acid when it was first synthesized. The function of BtsS/BtsR was also assessed in a clinically relevant uropathogenic E. coli strain. Quantitative transcriptional analysis revealed BtsS/BtsR importance during acute and chronic urinary-tract infections.
Introduction
Exponential growth of bacteria in complex, nutrient-rich media usually ends when at least one nutrient has been used up. We recently reported that the histidine kinase/response regulator system YehU/YehT of E. coli, belongs to the LytS/LytTR family and presumably plays a role in tuning bacterial exploitation of available carbon sources1. Strikingly, the YehU/YehT system is the most widespread representative of its family found in γ-proteobacteria – and many LytS/LytTR-type systems regulate crucial host-specific mechanisms during infection of human or plant hosts by members of this bacterial clade2. This system is conserved in non-pathogenic as well as pathogenic E. coli.
Our previous studies on YehU/YehT in E. coli identified yjiY as its sole target gene3 (Fig. 1). This gene codes for the putative carbon starvation transporter YjiY, which is homologous (61.1% identity) to CstA4 and was found to be expressed in cells that were grown in complex media containing a high content of amino acids, such as LB or CAA (casamino acids), as well as in minimal medium supplemented with certain carbon sources, such as gluconic or glucuronic acid. Studies in E. coli revealed that YehT-mediated yjiY transcription is also regulated by the cAMP/CRP complex3 (Fig. 1), and down-regulated in the presence of energetically favorable carbon sources like glucose. Furthermore, YjiY is subject to translational control via the Csr regulatory circuit5 (Fig. 1), which synchronizes the output of E. coli central carbohydrate metabolism (glycolysis versus gluconeogenesis) with YjiY production1, 6. Finally, yjiY transcription is under positive feedback regulation by a second two-component system, YpdA/YpdB, and its gene product YhjX1 (Fig. 1).
Figure 1.
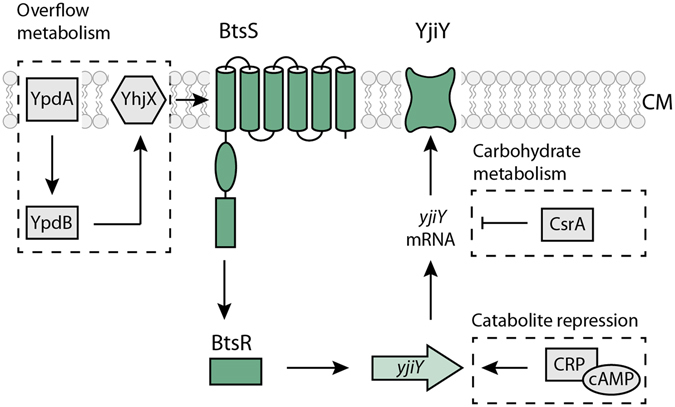
Schematic depiction of the BtsS/BtsR (YehU/YehT) system in Escherichia coli. The scheme summarizes the regulatory network associated with signal transduction by the BtsS/BtsR two-component system, the influence of two-component system YpdA/YpdB and the global regulators CsrA and CRP. Membrane proteins are integrated in the cytoplasmic membrane (CM). Activating (↑) and inhibitory (⊥) effects are indicated. See text for details.
Here, we performed a comprehensive in vivo characterization of yjiY expression in order to identify the primary stimulus sensed by the histidine kinase YehU. We found that the YehU/YehT system responds to depletion of nutrients specifically serine and the concomitant presence of extracellular pyruvate. Biochemical studies revealed that pyruvate binds specifically to the extracellular side of the membrane-spanning domain of YehU. We therefore renamed the system BtsS/BtsR, for “Brenztraubensäure”, the original name given by Jöns Jakob Berzelius to the compound when he first synthesized pyruvic acid in 18357. Finally, we found that the BtsS/BtsR system of uropathogenic E. coli may contribute to acute urinary tract infection.
Results
Elucidation of the stimulus for BtsS/BtsR (YehU/YehT) by in vivo yjiY expression analyses
It was previously shown that cultivation of E. coli in amino acid-rich media leads to activation of the BtsS/BtsR system and transient expression of the target gene yjiY in the late-exponential growth phase3. To identify a potential quorum sensing-like molecule, we used a combination of chemical fractionation of the medium and measurements of reporter strain activity. For this purpose, we cultivated E. coli MG1655 ΔyjiY/pBBR yjiY-lux in M9-minimal medium with gluconic acid as sole carbon source3. Shortly before the induction of yjiY we removed the cells and fractionated the supernatant by high pressure liquid chromatography. All fractions were analyzed for their potential to induce yjiY using reporter strain E. coli KX1468 pBBR yjiY-lux grown in minimal medium and succinate as C-source. After several rounds of fractionation/freeze-drying we found that the fraction with the highest induction potential contained a high concentration of gluconic acid, the initial carbon source (data not shown). This result ruled out the possibility that E. coli produces and senses a quorum sensing-like molecule. Then we quantified yjiY expression as a function of nutrient levels. For this purpose, we cultivated the reporter strain E. coli MG1655/pBBR yjiY-lux in LB medium with decreasing amounts of nutrients (1.0x, 0.5x, 0.4x, 0.3x LB, 0.2x LB and 0.1x LB), keeping the osmolarity of the medium constant. The growth rates (µ) of E. coli cells decreased with the dilution of LB medium, and exponential growth ceased at different time points (Table S1). Strikingly, expression of yjiY always began shortly before the onset of stationary phase (Fig. S1), and E. coli cells grown in 0.1x LB did not express yjiY. These results suggested that BtsS/BtsR somehow responds to nutrient limitation. We reasoned that supplying the relevant nutrient(s) in excess should suppress or postpone yjiY induction. Therefore, the reporter strain was grown in LB media supplemented with an excess of each individual L-amino acid (Fig. 2A). Particularly, the addition of L-serine delayed the expression of yjiY by almost two doubling periods (Fig. 2A). Subsequently, we tested different concentrations of L-serine in the reporter assay and found a concentration-dependent delay in yjiY expression, accompanied by a decrease in peak expression levels (Fig. 2B). The addition of serine does not influence the growth of E. coli and does not delay the onset of stationary phase (Fig. 2C). Although L-serine is not a preferred carbon source for E. coli and high external concentrations are actually toxic to the organism, it is the first amino acid to be consumed when mixtures of amino acids are available8. These data suggest that BtsS/BtsR responds to depletion of nutrients, specifically serine.
Figure 2.
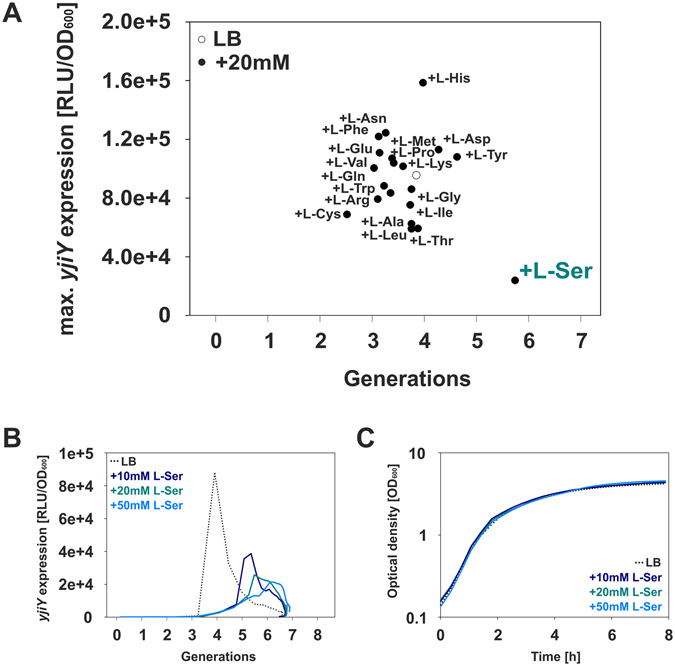
Effects of an excess of individual amino acids on the expression of yjiY. (A) E. coli MG1655/pBBR yjiY-lux was grown in LB medium supplemented with one of the indicated amino acids (at 20 mM), and growth and luminescence were monitored over time. Maximal yjiY expression values are depicted. The open circle provides a benchmark and indicates yjiY expression in E. coli grown in LB medium. (B) Expression of yjiY in LB (dotted line) supplemented with increasing L-serine concentrations. (C) Corresponding E. coli growth curves in LB media supplemented with increasing L-serine concentrations. Experiments were performed at least three times (standard deviation <10%), and results of a representative experiment are shown.
Changes in extracellular serine and pyruvate concentrations during growth of E. coli
When E. coli is grown in amino acid-rich media, 50.7% of L-serine is converted directly to pyruvate, whereas 36.3% is used for glycine synthesis, 6.5% for cell biomass, and the remainder for other metabolites9. Its central role in pyruvate supply provides one explanation for the importance of L-serine in growing E. coli 10. We therefore monitored the changes in extracellular serine and pyruvate concentrations during growth in LB medium, and found that extracellular levels of serine decreased at a constant rate (Fig. 3). The starting concentration of serine in the medium (approximately 200 µM) was completely exhausted after 120 min of growth at the late-exponential growth phase. At the same time, the abundance of extracellular pyruvate peaked (approx. 500 µM) and shortly after yjiY expression reached its maximum level (Fig. 3). It was previously shown that the external pyruvate derives from overflow metabolism in E. coli during growth in amino acid-rich media11, which was confirmed by monitoring the intracellular concentrations of serine and pyruvate (Fig. S2).
Figure 3.
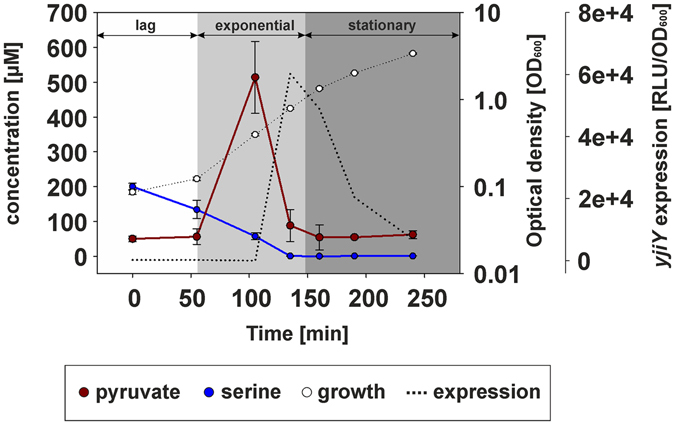
Determination of changes in extracellular concentrations of serine and pyruvate during growth of E. coli. E. coli MG1655/pBBR yjiY-lux was cultivated in LB medium, and growth (OD600) and luminescence were monitored. At the times indicated, cells were harvested, and serine and pyruvate levels were quantified by hydrophilic interaction liquid chromatography. All experiments were performed in triplicate, and the error bars indicate the standard deviation of the means. The growth phases of E. coli are marked as following: lag phase (white), exponential growth (light grey) and stationary phase (dark grey).
These data reveal that induction of BtsS-dependent yjiY expression coincides with the decline of serine in the medium and an extracellular accumulation of pyruvate (Fig. 3).
Extracellular pyruvate triggers yjiY expression under nutrient limitation
In the next experiment we tested the influence of serine, pyruvate and related metabolites on yjiY expression in E. coli cells growing in low-nutrient environment. Since E. coli harbors a second two-component system, YpdA/YpdB, which responds to high concentrations of extracellular pyruvate (the threshold concentration that leads to induction was determined to be 600 µM) and positively regulates the BtsS/BtsR system11, we modified our reporter strain by deleting yhjX, which is sufficient to interrupt the feedback loop1. The resulting strain was then cultivated in 10-fold diluted (0.1x) LB medium for 1 h. At this time point, cells do not induce expression of yjiY, but experience soon carbon limitation (Fig. 4A, Fig. S1). However, expression of yjiY was rapidly triggered upon addition of pyruvate, and the induction level increased linearly with increasing pyruvate concentration (Fig. 4A). Addition of L-serine also induced yjiY expression, but only after a 20-min delay (Fig. 4B). Under these conditions the growth of E. coli did not differ significantly by addition of pyruvate or serine (Fig. S3). Only supplementation of 10 mM serine prolonged the exponential growth phase. Moreover, higher serine concentrations delayed yjiY expression for even longer, and decreased the level of induction attained (Fig. 4B). The threshold concentration of pyruvate required for detectable yjiY expression was 10 µM, and that for L-serine 50 µM. None of the other tested compounds (each of the other 19 amino acids, phosphoenolpyruvate, lactate, oxaloacetate, α-ketoglutarate, valeriate, propionate, acetate, malate) were able to induce yjiY in this context. These results suggest that extracellular pyruvate acts as a direct stimulus for BtsS/BtsR-mediated yjiY expression, whereas delayed yjiY induction in response to L-serine may depend on uptake of the amino acid, its conversion to pyruvate and excretion of the pyruvate into the culture medium.
Figure 4.
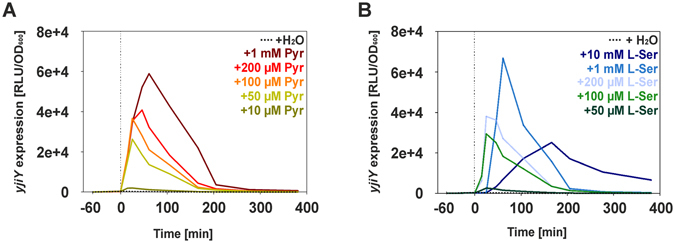
Stimulus-dependent yjiY expression under nutrient-limiting conditions. E. coli MG1655 mutant ΔyhjX harboring pBBR yjiY-lux was cultivated in 0.1x LB medium to establish low nutrient conditions. After 1 h (time point 0), the indicated concentration of pyruvate (A), or L-serine (B), or the equivalent volume of water was added. Experiments were performed at least three times (standard deviation <10%), and results of a representative experiment are shown.
The sensor histidine kinase BtsS binds pyruvate with high affinity
BtsS cannot be autophosphorylated, possibly owing to a defective ATP-binding site within the G1 box3 (data not shown). Therefore, we used the method of differential radial capillary action of ligand assays (DRaCALA)12 to determine whether BtsS physically interacts with pyruvate as ligand. The technique is based on the ability of proteins that have been immobilized on a nitrocellulose membrane to bind a radiolabeled ligand, whereas unbound ligands undergo radial diffusion. DRaCALA allows rapid detection of both the total ligand and the ligand sequestered by proteins. The fraction of ligand bound to the protein, defined as FB, is calculated from the signal intensity of the area with protein (inner circle) and the total signal intensity of the area (outer circle)12. For the DRaCALA we used unsealed membrane vesicles prepared from E. coli cells overproducing BtsS (MV BtsS) and calculated an FB value of >0.15 (Fig. 5A). Control membrane vesicles (MV, lacking overproduced BtsS) were used and the low value of FB (0.05) reflected only minor, non-specific binding. Therefore this assay was judged to be suitable for membrane vesicles, and it clearly indicated binding of radiolabeled pyruvate to BtsS. We also tested 3H-serine binding to BtsS in membrane vesicles using the DRaCALA technique. However, we only observed unspecific binding (data not shown).
Figure 5.
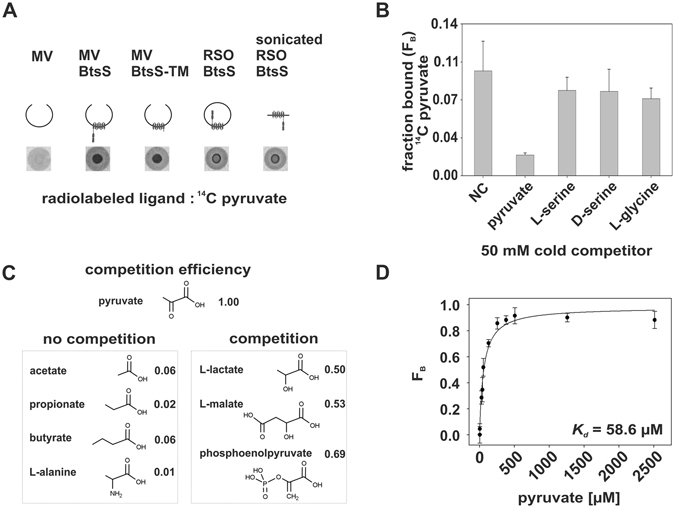
Analysis of the interaction of BtsS with selected ligands by DRaCALA. (A) A mixture of membrane vesicles (MV) or right-side-out vesicles (RSO) enriched with the corresponding proteins (indicated by graphical representations) and radiolabeled 14C pyruvate (5 µM) is dropped onto a nitrocellulose membrane, and ligand migration via capillary action is analyzed. (B) Competition assays. Binding of radiolabeled pyruvate (5 µM) to BtsS in MVs was analyzed in the presence of various unlabeled competitors (each 50 mM). NC, no competitor. (C) Relative efficiency of competition by various carboxylic acids. Binding of radiolabeled pyruvate (5 µM) to BtsS in MVs was analyzed in the presence of various carboxylic acids (each 50 mM). The efficiency of competition by cold pyruvate was set to 1.00, and the effect of the indicated compounds was calculated accordingly. (D) Determination of the dissociation constant (K d) for pyruvate to BtsS using DRaCALA. For each reaction radiolabeled pyruvate was used at 5 µM. Normalized FB values [FB = (FB(NC) − FB(pyr))/FB(NC), see Methods for details] were plotted as function of the pyruvate concentration. The best-fit line was determined by nonlinear regression using the equation y = Bmax * x/(K d + x).
In addition, we tested MVs harboring a membrane-integrated truncated variant of BtsS (MV BtsS-TM) lacking its soluble domains, and found that this truncated sensor also binds pyruvate (FB > 0.15). Furthermore, we prepared sealed right-side-out vesicles (RSO BtsS), in which membrane proteins retain their native orientation13. BtsS in these vesicles also showed pyruvate binding, and no significant change in binding was observed when these vesicles were fragmented by sonication and had a random orientation (sonicated RSO BtsS) (FB values of each 0.18) (Fig. 5A). These data reveal that pyruvate binds to the external side of the membrane-spanning domain of BtsS.
The specificity of pyruvate binding to BtsS was then addressed by using a competition assay (Fig. 5B), in which several competitors were added in excess to the reaction mixture. Only unlabeled (“cold”) pyruvate was able to prevent binding of radiolabeled pyruvate. L-serine, D-serine or glycine did not interfere with pyruvate binding to BtsS, suggesting that pyruvate binds specifically to BtsS.
Next we investigated the effects of varying the length and side-chain charge on ligand binding by BtsS. Using competition assays, we found that acetate, propionate and butyrate all failed to compete with pyruvate for binding, indicating the importance of the C-α keto group of pyruvate (Fig. 5C). The influence of polarity at the C-α position was then tested by addition of lactate and malate, which contain C-α hydroxyl groups. These molecules were able to decrease pyruvate binding by about 50% suggesting that a negative charge seems to be recognized. It should be noted that these compounds were tested in a 104-fold excess over pyruvate. Phosphoenolpyruvate, which harbors a phosphoryl group at the C-α position, also competed with pyruvate, reducing binding by approximately 50%. In contrast, a positive charge at the C-α position in the form of the amino group in alanine had no effect, reducing pyruvate binding by 1% (Fig. 5C). Notably, none of these compounds were able to induce yjiY expression in vivo, which emphasizes the specificity of BtsS for pyruvate. The dissociation constant (K d) for pyruvate binding to the histidine kinase was found to be 58.6 ± 8.8 µM (Fig. 5D).
To our knowledge this is the first application of DRaCALA to measure the interaction between a ligand and a membrane-embedded protein. Moreover, these assays unambiguously demonstrated that pyruvate binds specifically and with high affinity to BtsS.
BtsS/BtsR importance during urinary tract infection
To understand the potential implications of BtsS/BtsR for pathogenesis, we therefore turned our attention to uropathogenic E. coli (UPEC) that colonize a nutritionally demanding environment – the mammalian bladder. However, responses to nutritional stress are of the utmost importance for the survival of both commensal and pathogenic bacteria within a given host.
UPEC strains are the primary causative agent of urinary-tract infections (UTIs) worldwide, accounting for over 85% of all reported episodes14. In the bladder, bacteria adhere to the apical surface of the epithelium and are internalized, before replicating to form biofilm-like pods within host bladder cells15, 16. Subsequent to this transient intracellular cascade, adverse outcomes such as chronic colonization can ensue as a result of an aberrant host immune response17. Previous studies have demonstrated that successful UTI requires aerobic respiration and amino acid utilization18–20. To investigate the significance of BtsS/BtsR in bladder infection, we first created a btsS/btsR mutant in the UPEC cystitis isolate UTI89. Using the same reporter fusion, we demonstrated that this strain, unlike the wild-type parental UTI strain, failed to induce transient expression of yjiY (Fig. 6A). Both pyruvate and L-serine are present in human and murine urine, and levels are elevated in diabetic populations21–23. Given that BtsS/BtsR responds directly to extracellular pyruvate levels and indirectly to L-serine, we asked whether BtsS/BtsR is active in the bladder lumen during acute and chronic UTI.
Figure 6.
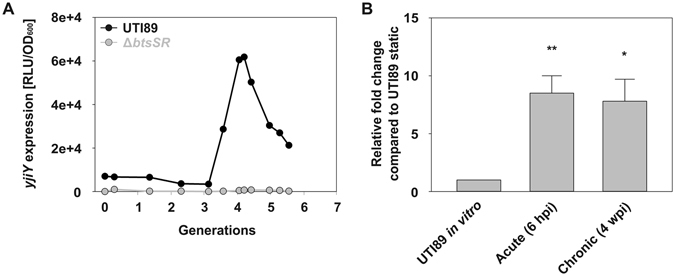
BtsS/BtsR importance during urinary tract infection. (A) The plot indicates the activity of the pBBR yjiY-lux reporter fusion in WT UTI89 and the isogenic btsS/btsR mutant during growth in LB, and depicts a representative of at least 6 biological replicates. (B) Change in the level of the yjiY transcript during acute and chronic phases of UTI relative to that measured in samples incubated in vitro without agitation. Data shown are the mean values of samples extracted from three different mice per time point. RNA was extracted from dissected bladders for each time point and enriched for bacterial RNA by depleting ribosomal and mammalian RNA as described in Materials and Methods. Profiling was performed using an yjiY-specific probe. Expression was normalized to that of the housekeeping gene gyrB and compared to expression in cDNA samples corresponding to the starting inoculum (static in vitro culture, prepared as described in Materials and methods). Relative fold change was measured using the method described by Pfaffl et al.54 (hpi, hours post infection; wpi, weeks post infection).
Mice were infected with the cystitis isolate UTI89 and RNA samples were extracted from bladder homogenates at 6 h post infection (coinciding with the acute stage of UTI), and at 2 weeks post infection from mice that went on to develop chronic cystitis. At this stage, the majority of bacteria are found on the bladder epithelial surface in the form of an extracellular biofilm17. Subsequent TaqMan-based qPCR analysis compared the expression of yjiY in the corresponding cDNA samples to the expression of yjiY in the starting bacterial inoculum. Our results demonstrated robust expression of yjiY in both the acute and chronic stages of infection from at least three separate mice per time-point (Fig. 6B). Taken together, these data suggest that, in the murine UTI model BtsS/BtsR responds to serine/pyruvate fluctuations and could play a role in promoting the infection process.
Discussion
Although numerous studies continue to demonstrate the importance of histidine kinase-mediated signal transduction in bacterial physiology, the natural ligands have been identified for very few histidine kinases24–26. This study presents compelling evidence demonstrating that the histidine kinase BtsS is a high-affinity receptor for extracellular pyruvate. Induction of the BtsS/BtsR target gene yjiY, which codes for a “carbon starvation” CstA-like transporter, correlates with the depletion of serine in complex medium, explaining the observation that BtsS/BtsR senses low serine. At the same time, while serine is being depleted from LB medium, E. coli excretes a substantial amount of pyruvate from overflow metabolism into the medium. Shortly afterwards, the concentration of extracellular pyruvate returns to its basal level (Fig. 3 and ref. 11). In previous studies, we demonstrated activation of a different system, YpdA/YpdB in response to high levels of extracellular pyruvate11 and showed that induction of YpdA/YpdB enhances BtsS/BtsR activation1. The current work provides a comprehensive characterization of how BtsS/BtsR directly responds to extracellular pyruvate levels and fine-tunes the metabolic fitness of E. coli under low-nutrient conditions. Sequence comparison between YpdA and YehU did not identify putative amino acids involved in pyruvate binding. In non-pathogenic E. coli, each of these signaling systems induces the expression of exactly one gene, which codes for a transporter. One (YhjX) belongs to the major facilitator superfamily, and is assumed to be a low-affinity carboxylate antiporter27. The other one (YjiY) belongs to the CstA-like transporters, which are characterized by an unusually large number of transmembrane helices (17 in the case of YjiY). Neither transporter has been characterized thus far. It is hypothesized that both are involved in nutrient uptake, but with different affinities. The interconnectivity between the high-affinity pyruvate signaling system BtsS/BtsR with the putative low-affinity pyruvate signaling system YpdA/YpdB by a positive and a negative feedback loop1 would provide E. coli with a network that could tailor pyruvate uptake in each individual cell according to its availability. Switching between low- and high-affinity phosphate transporters based on the needs of the individual cell was recently demonstrated for S. cerevisiae 28, and seems to be a widely distributed strategy for the maintenance of nutrient homeostasis as stocks of essential nutrients decline29.
Although nutrient sensing is crucial for host-microbe and microbe-microbe interactions, the majority of studies in non-pathogenic E. coli strains have focused on metabolic engineering, aiming to understand how processes such as elevated intracellular pyruvate levels affect metabolite distribution30, 31 or how central mutations trigger/alter metabolic fluxes32–34. In recent years, technological advances have permitted detailed analyses of in vivo metabolism35, revealing its complexity and its influence on virulence and pathogenesis36. These studies have shown pyruvate to be one of the major factors connecting cellular metabolism to cell division37. Moreover, pyruvate levels are thought to reflect the quantitative relationship between carbon and nitrogen availability in the cell, and affect amino acid biosynthesis38.
Investigations of how metabolic decisions determine pathogen fitness within host niches are increasingly uncovering opportunities for the development of robust and pathogen-specific drugs. Different E. coli pathotypes cause various clinical syndromes, depending on their genetic makeup and expression patterns. They obviously differ extensively from each other and from commensal E. coli. For example, uropathogenic and enterohemorragic E. coli differ significantly in genome content, and employ different strategies to infect their niches - the bladder and the gut, respectively. Very recent work has pointed to the need for aerobic respiration and amino acid utilization for optimal colonization of the urinary tract by UPEC18–20. Pyruvate is a crucial molecule required for fueling the aerobic arm of the tricarboxylic acid cycle, which is used by UPEC during infection18, 19, 39. Thus, it is reasonable to postulate that UPEC has the capacity to sense and respond to pyruvate fluctuations within the urinary tract. Here we demonstrate that BtsS/BtsR mediates yjiY expression during infection by a clinically relevant UPEC isolate, which strongly suggests that BtsS/BtsR constitutes part of the metabolic circuitry engaged during UTI. Thus understanding how UPEC can sense the metabolic signature of the bladder lumen, the kidney and the urothelial cell cytosol (in which they form intracellular bacterial communities) will be vital for a complete understanding of pathogen strategies and the design of more effective therapeutics. However, the study in Salmonella enterica Serovar Typhi and Typhimurium showed no detectable effect on the ability of yehUT mutant strain to invade cultured epithelial cells or induce colitis in a murine model40.
In summary, this study has uncovered a signal transduction network that responds with exquisite sensitivity to extracellular pyruvate levels and fine-tunes carbon utilization in E. coli strains. Furthermore we provide direct evidence for receptor-ligand interactions between BtsS and pyruvate, and demonstrate that BtsS/BtsR mediates yjiY expression during UTI. Future studies will focus on understanding the contribution of this system to pathogenic E. coli intra-host fitness and dissecting molecular determinants that drive the fine-tuning of bacterial fitness in response to external metabolic cues.
Methods
Strains, plasmids, and oligonucleotides
In this study we used the E. coli strains MG165541, E. coli MG1655 ΔyhjX 1, E. coli MG1655 ΔyjiY 1, BL21(DE3)42, E. coli KX146843 and the cystitis isolate UTI8944. Plasmids used in this study include the transcriptional promoter-luciferase fusion construct for PyjiY activity (pBBR yjiY-lux)3. For protein production we used the arabinose-inducible expression vectors pBAD2445, pBAD24-yehU 3 and pBAD24-yehU-TM (YehU (BtsS) amino acids 1–205). DNA fragments for plasmid construction were amplified from genomic DNA by PCR. Scarless deletion of btsS/btsR genes in UTI89 was performed using the method published by Murphy and Campellone46, and verified via PCR with oligonucleotides flanking the deleted loci. All oligonucleotide sequences are available on request.
Molecular biological techniques
Plasmid and genomic DNAs were isolated using a HiYield plasmid minikit (Suedlaborbedarf) and a DNeasy blood and tissue kit (Qiagen), respectively. DNA fragments were purified from agarose gels using a HiYield PCR cleanup and gel extraction kit (Suedlaborbedarf). Q5 DNA polymerase (New England BioLabs) was used according to the supplier’s instructions. Restriction enzymes and other DNA-modifying enzymes were also purchased from New England BioLabs and used according to the manufacturer’s directions.
Growth conditions
All strains were grown overnight in LB medium. After inoculation, bacteria were routinely grown in LB, LB supplemented with the indicated amino acids or diluted LB medium [containing 1% (w/v) NaCl] under agitation (200 rpm) at 37 °C. For solid medium, 1.5% (w/v) agar was added. Where appropriate, media were supplemented with antibiotics (ampicillin sodium salt, 100 µg/ml; kanamycin sulfate, 50 µg/ml; gentamicin sulfate, 50 µg/ml). For infection studies in mice, UTI89 was inoculated into 10 ml of LB medium and grown without shaking at 37 °C for 24 h. This culture was used to seed a fresh flask with 10 ml of a 1:1000 dilution, which was incubated for another 24 h to maximize expression of type 1 pili, as previously described47. The growth phases of E. coli marked in graphs were according to the definition in ref. 48.
In vivo expression studies
In vivo expression of yjiY was quantified by means of luciferase-based reporter-gene assays, using bacteria that had been transformed with plasmid pBBR yjiY-lux. Cells from an overnight culture were transferred to fresh medium to give a starting optical density at 600 nm (OD600) of 0.05, and incubated under aerobic growth conditions at 37 °C while OD600 and luminescence were continuously monitored. The optical density was determined in a microplate reader (Tecan Sunrise) at 600 nm. Luminescence levels were determined in a luminescence reader (Centro LB960; Berthold Technology) for 0.1 s and are reported as relative light units (RLU; counts s−1).
Identification of quorum sensing-like molecule
The cells of E. coli MG1655 ΔyjiY/pBBR yjiY-lux were cultivated in M9-minimal medium supplemented with 20 mM gluconic acid and OD600 and luminescence were constantly monitored. Cells were harvested shortly before, at and after the induction of yjiY. The supernatant was sterile filtrated (Stericup®, Filter Unit Millipore Express® PLUS(PES), 0.22 μm and separated by high pressure liquid chromatography (HPLC) using column C18-Hypersil Gold, with the gradient 1–100% (v/v) acetonitrile in 40 min. Fractions were collected, concentrated and used in reporter strain E. coli KX146843/pBBr yjiY-lux grown in M9-minimal medium supplemented with 20 mM succinate. The fraction with the highest induction potential was then analyzed via UPLC-UHR-ToF-MS using a Waters XBridge Amide column.
Production of BtsS-6His in membrane vesicles
E. coli BL21(DE3) cells were transformed with the vector pBAD24-yehU 3, which codes for BtsS-6His, and grown in LB medium at 37 °C to an OD600 of 0.5. Recombinant gene expression was induced by addition of 0.2% (w/v) arabinose. After 3 h of further incubation, cells were harvested by centrifugation, disrupted and fractionated. At each step, the pellet was resuspended in buffer consisting of MES/NaOH (pH 6), 10% (v/v) glycerol and 10 mM MgCl2. To produce BtsS-6His in right-side-out vesicles, cells were cultivated as described before. After harvesting the cells by centrifugation, lysozyme (50 µg/ml) and EDTA (10 mM) were added for 45 min at room temperature to remove the cell wall. After low-speed centrifugation, the spheroplasts were resuspended in buffer consisting of 100 mM MES/NaOH (pH 6), 30% (w/v) sucrose, 20 mM MgSO4, DNase and RNase, and then diluted 100-fold into pre-warmed buffer (100 mM MES/NaOH (pH 6)) to trigger osmotic lysis. After centrifugation, the pellet was resuspended in 100 mM MES/NaOH (pH 6), 10% (v/v) glycerol and 10 mM MgCl2. Formation of spheroplasts and right-side-out vesicles was monitored under the microscope. His-tagged BtsS was detected by Western blot analysis with an anti-His antibody (Qiagen) and an alkaline phosphatase-conjugated anti-mouse antibody as the secondary antibody.
Protein-ligand interactions
Protein-ligand interactions were analyzed via DRaCALA, a method established by Roelofs et al.12. Membrane vesicles enriched for overproduced BtsS-6His were mixed with 5 µM radiolabeled (3-14C) pyruvic acid sodium salt (55 mCi mmol−1, Biotrend) or radiolabeled (3H) L-serine (11.0 Ci mmol−1, Hartmann Analytics) and incubated for 5 min at room temperature. Triplicate 5-µl aliquots were transferred to a nitrocellulose membrane, dried and visualized by a PhosphorImager. Quantification of radioactive signal was done with the software ImageQuant. The fraction bound to protein was calculated according to the signal intensities using an equation defined earlier: FB = (Iinner − Ibackground)/Itotal 12. For the evaluation the corresponding cold ligand (50 mM) was added to the reaction mixture and incubated for further 3 min. To estimate the K d value, increasing concentrations of cold pyruvate (0 µM to 2.5 mM) were used. In this case the FB value was normalized as follows: FB = (FB(NC) − FB(pyr))/FB(NC). Here, FB(NC) is calculated from the sample with pure 14C-labeled pyruvate, and FB(pyr) are all values for mixtures containing 14C-labeled pyruvate together with increasing concentrations of unlabeled pyruvate. The amount of 14C-pyruvate was kept constant in all assays.
Extraction and determination of extra- and intracellular metabolites
The reporter strain E. coli MG1655/pBBR yjiY-lux was cultivated in LB medium, and OD600 and luminescence were constantly monitored. At selected time points, cells were harvested and the supernatants were saved. Cells were washed with 10% (v/v) glycerol, and subsequently cell pellets and supernatants were analyzed via hydrophilic interaction liquid chromatography (HILIC). Acetonitrile (ACN), methanol (MeOH), ammonium acetate and ammonium hydroxide (all of LC-MS grade) were obtained from Sigma-Aldrich (Sigma-Aldrich GmbH). Water was purified using a Merck Millipore Integral water purification system to a resistance of 18 MΩ and TOC < 5 ppb. Sodium pyruvate and serine were also obtained from Sigma-Aldrich and dissolved in water at a concentration of 100 mM and further diluted with ACN. Cell pellets were extracted with 200 µl ice-cold water/MeOH (50/50, v/v). Samples were vortexed vigorously and lysed in an ice-cold sonic bath for 15 min. After centrifugation at 20,000 × g and 4 °C for 10 min, supernatants were transferred to autosampler vials.
Pyruvate and serine were quantified using a modified version of the method published by Yuan et al.49. Separation was achieved using a Waters XBridge Amide column (3.5 µm, 100 mm × 4.6 mm ID) and an ACN/water gradient using a Waters Acquity UPLC system coupled to a Bruker maXis UHR-ToF-MS (Bruker Daltonics). Eluent A consisted of 5% (v/v) ACN, 95% (v/v) water, 20 mM ammonium hydroxide, 20 mM ammonium acetate, pH 9.0 and eluent B was pure ACN.
Metabolites were eluted with the following gradient: After 3 min of 85% B, a linear decrease to 2% B over 9 min, isocratic hold for 3 min and return to initial conditions for 1 min and a 7-min re-equilibration time. Sample (5 µl) was injected via partial loop injection.
For quantification a calibration curve was generated from different concentrations of standard compounds (0 µM, 0.5 µM, 1.0 µM, 2.5 µM, 5 µM, 10 µM, 25 µM, 50 µM, 100 µM, 250 µM). If the concentration of a sample was beyond the range of the calibration curve, it was appropriately diluted with water/MeOH. Quantification was performed with Bruker Quant Analysis.
Murine infections
A cohort (15) of 7- to 8-week old female C3H/HeN mice was infected with strain UTI89 via transurethral catheterization as described in Hung et al.50. Five mice were sacrificed at 6 h after infection, marking the acute stage of UTI in this murine model51. Bladders were excised for CFU enumeration and RNA extraction. The remaining 10 mice were monitored for chronic infection using longitudinal urinalysis, as previously described52. Mice that consistently shed more than 100,000 CFUs/ml of urine were separated from the rest of the cohort, and flagged as chronically infected. These mice were sacrificed at 4 weeks post infection and bladders were obtained for CFU enumeration and RNA extraction.
To ensure the proper and humane treatment of animals, all animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and the Vanderbilt University Medical Center’s Institutional Animal Care and Use Committee (IACUC), who approved all protocols. Statistical analyses were performed using two-tailed Mann-Whitney t-test.
RNA extraction, cDNA synthesis and qPCR
RNA extraction and bacterial RNA enrichment were performed as described by Conover et al.47. DNase treatment, reverse transcription, and real-time quantitative PCR were done as described by Guckes et al.53. qPCR analysis was carried out with three concentrations of cDNA (50 ng, 25 ng, 12.5 ng), each in triplicate for each sample, and levels of gyrB (DNA gyrase) were used for normalization. The following primers (Integrated DNA Technologies) were used for amplification: yjiY_Fwd (5′-GGCACGACGCCGAAACT-3′), yjiY_Rev (5′-GCCGTAGCCGATGAAACG-3′), gyrB_L (5′-GATGCGCGTGAAGGCCTGAATG-3′), gyrB_R (5′-CACGGGCACGGGCAGCATC-3′). The following probes (Applied Biosystems) were used for quantification; yjiY (5′FAM-TGGCTAATGAAACCGACGC-MGBNFQ-3′); gyrB (5′VIC-ACGAACTGCTG-GCGGA-MGBNFQ-3′).
Electronic supplementary material
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (Exc114/2, SPP1617, JU270/12-2 to K.J.), by Vanderbilt University Medical Center Academic Professional development funds (to M.H.) by the NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org), grant DK076169 (to M.H.), and National Science Foundation Graduate Research Fellowship Program under Grant Number 1445197 (to E.J.B.) and T32 GM07628 (to E.J.B.). We thank Lena Stelzer for excellent technical assistance.
Author Contributions
S.B., M.W., P.S.-K., M.H. and K.J. designed the experiments. S.B., I.K., M.W., E.B., A.E., C.S. performed the experiments. S.B., I.K., M.H., and K.J. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01410-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Behr S, Fried L, Jung K. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J. Bacteriol. 2014;196:2023–2029. doi: 10.1128/JB.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galperin MY. Telling bacteria: do not LytTR. Structure. 2008;16:657–659. doi: 10.1016/j.str.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraxenberger T, Fried L, Behr S, Jung K. First insights into the unexplored two-component system YehU/YehT in. Escherichia coli. J. Bacteriol. 2012;194:4272–4284. doi: 10.1128/JB.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz JE, Matin A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 1991;218:129–140. doi: 10.1016/0022-2836(91)90879-B. [DOI] [PubMed] [Google Scholar]
- 5.Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 2013;15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabnis NA, Yang H, Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 1995;270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- 7.Berzelius J. Ueber die Destillationsproducte der Traubensäure. Ann. Phys. 1835;112:1–29. doi: 10.1002/andp.18351120902. [DOI] [Google Scholar]
- 8.Prüss B, Nelms JM, Park C, Wolfe AJ. Mutations in NADH: ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selvarasu S, et al. Characterizing Escherichia coli DH5α growth and metabolism in a complex medium using genome‐scale flux analysis. Biotechnol. Bioeng. 2009;102:923–934. doi: 10.1002/bit.22119. [DOI] [PubMed] [Google Scholar]
- 10.Polzin S, et al. Growth media simulating ileal and colonic environments affect the intracellular proteome and carbon fluxes of enterohemorrhagic Escherichia coli O157: H7 strain EDL933. Appl. Environ. Microbiol. 2013;79:3703–3715. doi: 10.1128/AEM.00062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fried L, Behr S, Jung K. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J. Bacteriol. 2013;195:807–815. doi: 10.1128/JB.02051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaback H. Bacterial Membranes. Methods Enzymol. 1971;22:99–120. doi: 10.1016/0076-6879(71)22015-2. [DOI] [Google Scholar]
- 14.Foxman B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010;7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 15.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 16.Anderson GG, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 17.Hannan TJ, et al. Host–pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol. Rev. 2012;36:616–648. doi: 10.1111/j.1574-6976.2012.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadjifrangiskou M, et al. A central metabolic circuit controlled by QseC in pathogenic Escherichia coli. Mol. Microbiol. 2011;80:1516–1529. doi: 10.1111/j.1365-2958.2011.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floyd KA, et al. Adhesive fiber stratification in uropathogenic Escherichia coli biofilms unveils oxygen-mediated control of type 1 pili. PLoS Pathog. 2015;11:e1004697. doi: 10.1371/journal.ppat.1004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stec DF, et al. Alterations of urinary metabolite profile in model diabetic nephropathy. Biochem. Biophys. Res. Com. 2015;456:610–614. doi: 10.1016/j.bbrc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouatra S, et al. The human urine metabolome. PloS One. 2013;8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur P, et al. Quantitative metabolomic and lipidomic profiling reveals aberrant amino acid metabolism in type 2 diabetes. Mol. BioSyst. 2013;9:307–317. doi: 10.1039/C2MB25384D. [DOI] [PubMed] [Google Scholar]
- 24.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 25.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudipaty SA, McEvoy MM. The histidine kinase CusS senses silver ions through direct binding by its sensor domain. Biochim. Biophys. Acta. 2014;1844:1656–1661. doi: 10.1016/j.bbapap.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pao SS, Paulsen IT, Saier MH., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wykoff DD, Rizvi AH, Raser JM, Margolin B, O’Shea EK. Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol. Cell. 2007;27:1005–1013. doi: 10.1016/j.molcel.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy S, Kafri M, Carmi M, Barkai N. The competitive advantage of a dual-transporter system. Science. 2011;334:1408–1412. doi: 10.1126/science.1207154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y-T, Bennett GN, San K-Y. The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab. Eng. 2001;3:115–123. doi: 10.1006/mben.2000.0166. [DOI] [PubMed] [Google Scholar]
- 31.Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Fischer E, Sauer U. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC‐MS. Eur. J. Biochem. 2003;270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Ou MS, Kim Y, Ingram L, Shanmugam K. Metabolic flux control at the pyruvate node in an anaerobic Escherichia coli strain with an active pyruvate dehydrogenase. Appl. Environ. Microbiol. 2010;76:2107–2114. doi: 10.1128/AEM.02545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murarka A, Clomburg JM, Gonzalez R. Metabolic flux analysis of wild-type Escherichia coli and mutants deficient in pyruvate-dissimilating enzymes during the fermentative metabolism of glucuronate. Microbiology. 2010;156:1860–1872. doi: 10.1099/mic.0.036251-0. [DOI] [PubMed] [Google Scholar]
- 35.Eylert E, et al. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 2008;69:1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x. [DOI] [PubMed] [Google Scholar]
- 36.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 37.Göhler A-K, et al. More than just a metabolic regulator - elucidation and validation of new targets of PdhR in Escherichia coli. BMC Syst. Biol. 2011;5:197. doi: 10.1186/1752-0509-5-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 39.Floyd KA, et al. The UbiI (VisC) aerobic ubiquinone synthase is required for expression of type 1 pili, biofilm formation, and pathogenesis in uropathogenic Escherichia coli. J. Bacteriol. 2016;198:2662–2672. doi: 10.1128/JB.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong VK, et al. Characterization of the yehUT Two-Component Regulatory System of Salmonella enterica Serovar Typhi and Typhimurium. PLoS One. 2013;8:e84567. doi: 10.1371/journal.pone.0084567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blattner F, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 42.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 43.Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schilling JD, Mulvey MA, Vincent CD, Lorenz RG, Hultgren SJ. Bacterial invasion augments epithelial cytokine responses to Escherichia coli through a lipopolysaccharide-dependent mechanism. J. Immunol. 2001;166:1148–1155. doi: 10.4049/jimmunol.166.2.1148. [DOI] [PubMed] [Google Scholar]
- 45.Guzman L, Belin D, Carson M, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic Escherichia coli. BMC Mol. Biol. 2003;4:1. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conover MS, et al. Metabolic Requirements of Escherichia coli in Intracellular Bacterial Communities during Urinary Tract Infection Pathogenesis. mBio. 2016;7:e00104–00116. doi: 10.1128/mBio.00104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pörtner, R. Grundlagen der Bioverfahrenstechnik. In Antranikian, G. (Ed.) Angewandte Mikrobiologie Springer-Verlag, Berlin, Heidelberg p. 240 (2006)
- 49.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung C-S, Dodson KW, Hultgren SJ. A murine model of urinary tract infection. Nat. Protoc. 2009;4:1230–1243. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Justice SS, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hannan TJ, Mysorekar IU, Hung CS, Isaacson-Schmid ML, Hultgren SJ. Early severe inflammatory responses to uropathogenic Escherichia coli predispose to chronic and recurrent urinary tract infection. PLoS Pathog. 2010;6:e1001042. doi: 10.1371/journal.ppat.1001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guckes KR, et al. Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc. Natl. Acad. Sci. USA. 2013;110:16592–16597. doi: 10.1073/pnas.1315320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45–e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


