Abstract
Gastric cancer (GC) is often diagnosed in the advanced stages and is associated with a poor prognosis. Obtaining an in depth understanding of the molecular mechanisms of GC has lagged behind compared with other cancers. This study aimed to identify candidate biomarkers for GC. An integrated analysis of microarray datasets was performed to identify differentially expressed genes (DEGs) between GC and normal tissues. Gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were then performed to identify the functions of the DEGs. Furthermore, a protein-protein interaction (PPI) network of the DEGs was constructed. The expression levels of the DEGs were validated in human GC tissues using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). A set of 689 DEGs were identified in GC tissues, as compared with normal tissues, including 202 upregulated DEGs and 487 downregulated DEGs. The KEGG pathway analysis suggested that various pathways may play important roles in the pathology of GC, including pathways related to protein digestion and absorption, extracellular matrix-receptor interaction, and the metabolism of xenobiotics by cytochrome P450. The PPI network analysis indicated that the significant hub proteins consisted of SPP1, TOP2A and ARPC1B. RT-qPCR validation indicated that the expression levels of the top 10 most significantly dysexpressed genes were consistent with the illustration of the integrated analysis. The present study yielded a reference list of reliable DEGs, which represents a robust pool of candidates for further evaluation of GC pathogenesis and treatment.
Keywords: gastric cancer, differentially expressed genes, integrated analysis, expression profile, reverse transcription-quantitative polymerase chain reaction
Introduction
Gastric cancer (GC) is one of the most prevalent cancers and the second most common cause of cancer-associated mortality worldwide (1). Almost one million people are diagnosed with GC each year. Despite decades of a steady decline in the incidence of GC, the GC fatality rate remains paradoxically high in most countries, particularly in those of East Asia (2). GC is a heterogeneous disease with numerous etiologies and potential pathways of carcinogenesis (3,4), resulting in a variation in the incidence rates of GC among different geographies, ethnicities and genders (5). One of the main etiological risk factors for GC is Helicobacter pylori infection, although only a small proportion of individuals infected with H. pylori develop GC (6,7).
Traditional methods for the treatment of GC include surgery, chemotherapy, radiation therapy and combination therapy, which is also known as multimodality therapy. However, GC is often asymptomatic during its early stage, which results in the advanced stage being generally refractory to those therapies (8). Even following radical gastrectomy, many patients experience disease recurrence and succumb to the disease within a few months to years; the 5-year survival rate of GC is ≤2/3 (9,10). Therefore, an early and effective detection method that improves the chance of treating GC is imperative.
Microarrays are good tools for investigating the pathogenesis of various diseases (11–14). Compared with traditional methods, next-generation sequencing-based microarrays have the advantages of being unbiased, as they are not limited to previously known or annotated transcripts, and allowing more accurate quantification of genes with very low or high expression levels (15). In addition, transcriptome data detects other types of transcriptional signals, including alternative splicing, transcriptional starts/stops, gene fusions and expressed alleles (16). Studies based on microarrays have provided significant insights into the molecular basis of GC and novel therapeutic targets. However, microarrays have predominantly been used to characterize the genomic alterations of GC patients, while the validation of potential target genes for GC has been rare (10,17,18), restricting the application of microarrays to clinical practice.
The present study employed an integrated analysis of microarray data from the Gene Expression Omnibus (GEO) to identify the differentially expressed genes (DEGs) between GC and normal control (NC) tissues, which were then used to construct a protein-protein interaction (PPI) network. Furthermore, the significantly enriched functions of these genes were screened and analyzed to discover the biological processes and signaling pathways associated with GC. Finally, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) of clinical samples was performed to validate the integrated analysis approach. This study may improve the methods used to elucidate the dysexpression of various genes in GC and be of some value for the future diagnosis of GC in the clinic.
Materials and methods
Microarray data and data preprocessing
Eligible GC gene expression datasets were identified by searching the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Data were included if they met the following criteria: i) The expression profile of whole genome sequencing; ii) data from the tissues of GC patients in the clinic; and iii) raw or standardized data. Raw data were normalized using the Z-score transformation method (19) to make data from different platforms comparable. Matrix Laboratory software (version 2013a; MathWorks, Natick, MA, USA) was used to identify differentially expressed probe sets between tumor and adjacent tissues. A gene-specific t-test was performed, after which P-values and the effect size of individual microarray studies were calculated. The genes with a false discovery rate of ≤0.05 were selected as the significantly DEGs. DEGs between tumor and adjacent tissues were subsequently determined. Heat map analysis was conducted using the ‘heatmap.2’ function of the R/Bioconductor package ‘gplots’ (20).
Functional enrichment analysis of DEGs
To determine the biological functions of DEGs, Gene Ontology (GO) enrichment analysis of biological processes, molecular functions and cellular components was performed. The online software GeneCodis3 (http://genecodis.cnb.csic.es/analysis) was used to perform this analysis (21). In addition, pathway enrichment was also performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/).
PPI network construction
A PPI network of the significantly dysexpressed genes was constructed according to data from the Biological General Repository for Interaction Datasets (http://thebiogrid.org/). Among the candidate genes, the distribution characteristics of the top 20 most significantly upregulated and downregulated DEGs were visualized using Cytoscape (22).
Collection of clinical specimens
A total of 10 patients, including 8 males and 2 females, were enrolled in the present study, among which 5 had been diagnosed with GC. The average age of the patients was 54 years (age range, 38–79 years). Frozen tissue sections were generated and examined independently by senior pathologists. Parts of each tumor tissues were frozen immediately following the operation and stored at −135°C for RNA extraction. This study was approved by the Ethics Committee of The First Affiliated Hospital of PLA General Hospital (Beijing, China).
RNA preparation and RT-qPCR
Total RNA of each sample was extracted using the RNAeasy Mini kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol. According to previous studies (23–27), 10 DEGs were retrieved from the top 20 upregulated and downregulated genes. Primers for the 10 target genes were designed using PrimerPlex 2.61 (Premier Biosoft International, Palo Alto, CA, USA) and are shown in Table I. cDNA templates were synthesized from 1–5 µg RNA using Superscript Reverse Transcriptase II (Thermo Fisher Scientific, Inc., Waltham, MA, USA). qPCR was performed on the ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR dye (Thermo Fisher Scientific, Inc.). The final reaction mixture of 12.5 µl consisted of 6.25 µl Power SYBR Green PCR Master Mix, 50 ng diluted cDNA and 1 µM of each primer. Reactions were conducted in triplicate under the following conditions: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. Melting curves (60 to 95°C) were derived for every reaction to insure a single product. Relative gene expression was evaluated using Data Assist Software, version 3.0 (Thermo Fisher Scientific, Inc.), with the human actin gene as a reference. The expression levels of each gene were determined using the 2−ΔΔCq method (28).
Table I.
Primer sequences for the target genes.
| Gene | Sequence (5′ to 3′) | Product size (bp) |
|---|---|---|
| SULF1 | F: GTAAGAAGGAAGAATCCAGCAAGAA | 187 |
| R: AGGTCACTGGGTCCTTTACACTT | ||
| SPP1 | F: ATAAGCGGAAAGCCAATGATGAGAG | 134 |
| R: TTTGGGGTCTACAACCAGCATATCT | ||
| THBS2 | F: GAACATTGGCTGGAAGGACTACAC | 126 |
| R: TCATAGATAGGTCCTGAGTCTGCCA | ||
| TOP2A | F: ATCATTGAAAATAAGCCTAAGAAAG | 197 |
| R: AGAAGATAGTTGAAGGTTGGTCC | ||
| HOXC6 | F: GGACATAACACACAGACCTCAATCG | 129 |
| R: GACCCCACTGTGCGAATTCAT | ||
| GIF | F: ATGGCATCATTGGAGACATCTACAG | 94 |
| R: TTCTTGCAGTTCCATTCCTTTTTAG | ||
| KCNE2 | F: AGAGACGGGAACACTCCAATGACC | 64 |
| R: ACTTTTCCTGCCAGTCCTCTACAATG | ||
| SST | F: AACCCAACCAGACGGAGAATGAT | 109 |
| R: GCCGGGTTTGAGTTAGCAGATCTC | ||
| GKN1 | F: AGGAAGTCATGCCCTCCATTCAATC | 149 |
| R: GTTTTTTCCGAACTTGCTCAGGTCA | ||
| LIPF | F: GAACTTTAACACGAGTCGCTTGGAT | 189 |
| R: ATGGCTGTCACATTGTAGTAGGGAG | ||
| Actin | F: ACTTAGTTGCGTTACACCCTT | 156 |
| R: GTCACCTTCACCGTTCCA |
F, forward; R, reverse.
Statistical analysis
Data are expressed as the mean ± standard deviation. Comparisons of the expression levels of different genes were conducted using Student's t-test with a significance level of 0.05. Statistical analyses were conducted using SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA).
Results
DEGs in the integrated analysis of microarray datasets
Following the electronic database search, six microarray studies were obtained according to the inclusion criteria. The characteristics of the individual studies that were included in the integrated analysis are displayed in Table II. There were 53 GC patients and 259 NC patients. The integrated analysis identified a set of 689 DEGs in the GC tissues, as compared with the normal tissues, including 202 upregulated and 487 downregulated DEGs. In addition, the hierarchical clustering analysis indicated that the DEGs in GC were distinguished from those in normal tissues (Fig. 1).
Table II.
Information of the six transcriptome profiles.
| GEO ID | Samples(cancer/normal) | Platform | Country | Year |
|---|---|---|---|---|
| GSE51575 | 26/26 | GPL13607 Agilent-028004 SurePrint G3 | Korea | 2014 |
| Human GE 8×60K Microarray (Feature Number version) | ||||
| GSE29272 | 134/134 | GPL96 [HG-U133A] Affymetrix Human Genome U133A Array | USA | 2013 |
| GSE37023-GPL96 | 112/39 | GPL96 [HG-U133A] Affymetrix Human Genome U133A Array | Singapore | 2012 |
| GSE37023-GPL97 | 29/36 | GPL97 [HG-U133B] Affymetrix Human Genome U133B Array | Singapore | 2012 |
| GSE33651 | 40/12 | GPL2895 GE Healthcare/Amersham Biosciences CodeLink | Korea | 2011 |
| Human Whole Genome Bioarray | ||||
| GSE38932 | 12/12 | GPL5936 HEEBO Human oligo array | Argentina | 2012 |
GEO, Gene Expression Omnibus; HEEBO, human exonic evidence-based oligonucleotide.
Figure 1.
Heatmap visualization of the patterns of change for differentially expressed genes among samples from different datasets.
Functional enrichment analysis
GO provides a common descriptive framework and functional annotation and classification system for analyzing the gene set data. The 689 DEGs were involved in 86 signaling pathways, including the digestion and absorption of proteins, interactions between extracellular matrix (ECM) receptors, the p53 signaling pathway, the metabolism of propionate, the absorption of minerals, etc. The results of the KEGG enrichment analysis revealed that the first three most enriched pathways included protein digestion and absorption, ECM-receptor interaction and the metabolism of xenobiotics by cytochrome P450 (Tables III and IV). The detailed information on the 20 most significantly upregulated and downregulated genes is shown in Table V.
Table III.
Partial results of GO analysis.
| GO term | GO name | No. of genes | P-value |
|---|---|---|---|
| Biological processes | 22 | 7.04E-20 | |
| GO:0007586 | Digestion | 30 | 1.87E-10 |
| GO:0051301 | Cell division | 30 | 4.74E-10 |
| GO:0042493 | Response to drug | 41 | 5.44E-10 |
| GO:0007155 | Cell adhesion | 20 | 1.68E-09 |
| GO:0006805 | Xenobiotic metabolic process | 29 | 2.79E-08 |
| GO:0008285 | Negative regulation of cell proliferation | 58 | 1.05E-07 |
| GO:0007165 | Signal transduction | 31 | 3.71E-07 |
| GO:0007049 | Cell cycle | 24 | 1.22E-06 |
| GO:0005975 | Carbohydrate metabolic process | 12 | 1.31E-06 |
| GO:0043434 | Response to peptide hormone stimulus | 25 | 1.33E-06 |
| GO:0008152 | Metabolic process | 6 | 2.12E-06 |
| GO:0006600 | Creatine metabolic process | 24 | 2.35E-06 |
| GO:0000278 | Mitotic cell cycle | 10 | 2.41E-06 |
| GO:0010033 | Response to organic substance | 13 | 3.53E-06 |
| Molecular functions | |||
| GO:0005515 | Protein binding | 192 | 4.51E-23 |
| GO:0042803 | Protein homodimerization activity | 42 | 6.63E-12 |
| GO:0016491 | Oxidoreductase activity | 38 | 1.15E-11 |
| GO:0005509 | Calcium ion binding | 47 | 1.67E-11 |
| GO:0042802 | Identical protein binding | 28 | 2.71E-09 |
| GO:0005201 | Extracellular matrix structural constituent | 14 | 7.05E-09 |
| GO:0008201 | Heparin binding | 18 | 7.53E-09 |
| GO:0008009 | Chemokine activity | 10 | 2.40E-06 |
| GO:0003824 | Catalytic activity | 26 | 3.55E-06 |
| GO:0016787 | Hydrolase activity | 46 | 5.63E-06 |
| GO:0000166 | Nucleotide binding | 79 | 6.03E-06 |
| GO:0050840 | Extracellular matrix binding | 6 | 4.04E-05 |
| GO:0008233 | Peptidase activity | 28 | 5.63E-05 |
| GO:0019901 | Protein kinase binding | 18 | 8.70E-05 |
| GO:0005198 | Structural molecule activity | 17 | 0.000169 |
| Cellular components | |||
| GO:0005576 | Extracellular region | 163 | 1.54E-55 |
| GO:0005615 | Extracellular space | 93 | 6.61E-42 |
| GO:0005737 | Cytoplasm | 242 | 3.60E-36 |
| GO:0005886 | Plasma membrane | 151 | 2.66E-17 |
| GO:0005829 | Cytosol | 105 | 1.08E-15 |
| GO:0031012 | Extracellular matrix | 25 | 1.18E-14 |
| GO:0048471 | Perinuclear region of cytoplasm | 36 | 1.27E-11 |
| GO:0005887 | Integral to plasma membrane | 56 | 4.70E-10 |
| GO:0016020 | Membrane | 141 | 1.29E-09 |
| GO:0005634 | Nucleus | 173 | 4.31E-09 |
| GO:0005581 | Collagen | 15 | 1.35E-08 |
| GO:0005604 | Basement membrane | 13 | 3.41E-08 |
| GO:0005578 | Proteinaceous extracellular matrix | 21 | 4.36E-08 |
| GO:0005625 | Soluble fraction | 29 | 4.45E-08 |
| GO:0009986 | Cell surface | 24 | 2.10E-07 |
GO, gene ontology.
Table IV.
Partial results of the KEGG analysis.
| KEGG ID | KEGG term | No. of genes | P-value |
|---|---|---|---|
| hsa04974 | Protein digestion and absorption | 19 | 1.77E-13 |
| hsa04512 | ECM-receptor interaction | 16 | 9.65E-10 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 13 | 5.40E-08 |
| hsa05146 | Amoebiasis | 15 | 7.61E-08 |
| hsa04971 | Gastric acid secretion | 13 | 8.50E-08 |
| hsa04510 | Focal adhesion | 20 | 1.11E-07 |
| hsa00071 | Fatty acid metabolism | 10 | 1.70E-07 |
| hsa00982 | Drug metabolism-cytochrome P450 | 12 | 3.32E-07 |
| hsa04110 | Cell cycle | 15 | 5.78E-07 |
| hsa00010 | Glycolysis/gluconeogenesis | 11 | 7.37E-07 |
| hsa04978 | Mineral absorption | 10 | 1.03E-06 |
| hsa04115 | p53 signaling pathway | 11 | 1.42E-06 |
| hsa00640 | Propanoate metabolism | 8 | 2.52E-06 |
| hsa00330 | Arginine and proline metabolism | 9 | 1.60E-05 |
| hsa00591 | Linoleic acid metabolism | 7 | 1.71E-05 |
KEGG, kyoto encyclopedia of genes and genomes.
Table V.
Top ten significantly upregulated and downregulated DEGs.
| DEG | Function | Fold-change | P-value |
|---|---|---|---|
| Upregulated | |||
| SULF1 | Exhibits arylsulfatase activity and highly specific endoglucosamine-6-sulfatase activity; diminishes heparan sulfate proteoglycan sulfation; inhibits signaling by heparin-dependent growth factors; diminishes proliferation; and facilitates apoptosis in response to exogenous stimulation | 1.9208 | 6.89E-54 |
| MEST | N/A | 1.7879 | 2.94E-50 |
| SPP1 | Probably important to cell-matrix interaction; acts as a cytokine involved in enhancing the production of interferon-γ and IL-12 and reducing the production of IL-10; and is essential in the pathway that leads to type I immunity | 2.5703 | 1.79E-47 |
| THBS2 | Mediates cell-to-cell and cell-to-matrix interactions. Ligand for cluster of differentiation 36, mediating antiangiogenic properties | 2.0385 | 9.72E-44 |
| TOP2A | Controls topological states of DNA by transient breakage and subsequent rejoining of DNA strands. | 1.9398 | 4.07E-42 |
| HOXC6 | Provides cells with specific positional identities on the anterior-posterior axis | 1.7265 | 3.34E-41 |
| HOXC10 | Provides cells with specific positional identities on the anterior-posterior axis | 1.8234 | 4.42E-38 |
| ARPC1B | Involved in regulating actin polymerization and, together with an activating nucleation-promoting factor, mediates the formation of branched actin networks | 1.4555 | 2.10E-35 |
| KIF4A | Translocates PRC1 to the plus ends of interdigitating spindle microtubules during the metaphase to anaphase transition; may play a role in mitotic chromosomal positioning and bipolar spindle stabilization | 1.4195 | 2.38E-35 |
| OLFML2B | N/A | 1.6155 | 7.38E-35 |
| Downregulated | |||
| GIF | Promotes absorption of the essential vitamin, cobalamin, in the ileum | −8.2157 | 8.04E-124 |
| ESRRG | Acts as transcription activator in the absence of bound ligand | −3.1508 | 6.98E-105 |
| KCNE2 | Assembles as a β-subunit with a voltage-gated potassium channel complex of pore-forming α-subunits; modulates the gating kinetics and enhances the stability of the channel complex; associated with KCNH2/HERG, it is proposed to form the rapidly activating component of the delayed rectifying potassium current in the heart; may associate with KCNQ2 and/or KCNQ3 and modulate the native M-type current; may associate with KCNQ1/KVLTQ1 and elicit a voltage-independent current; may associate with HCN1 and HCN2 and increase potassium current | −3.6014 | 1.29E-97 |
| SST | Somatostatin inhibits the release of somatotropin | −3.1116 | 3.67E-95 |
| GKN1 | Has mitogenic activity and may be involved in maintaining the integrity of the gastric mucosal epithelium | −8.7597 | 2.11E-91 |
| LIPF | N/A | −8.5977 | 1.14E-85 |
| CHIA | May participate in the defense against nematodes, fungi and other pathogens; plays a role in T-helper cell type 2 immune response; contributes to the response to IL-13 and inflammation in response to IL-13; stimulates chemokine production by pulmonary epithelial cells; protects lung epithelial cells against apoptosis and promotes phosphorylation of AKT1 | −4.1886 | 1.27E-80 |
| DUOX1 | Generates hydrogen peroxide, which is required for the activity of thyroid peroxidase and LPO; plays a role in thyroid hormones synthesis and LPO-mediated antimicrobial defense at the surface of mucosa; may have its own peroxidase activity through its N-terminal peroxidase-like domain | −1.9009 | 1.20E-76 |
| AKR7A3 | Can reduce the dialdehyde protein-binding form of AFB1 to the non-binding AFB1 dialcohol; may be involved in protecting the liver against the toxic and carcinogenic effects of AFB1, a potent hepatocarcinogen | −2.1215 | 1.34E-68 |
| FAM149A | N/A | −1.5039 | 1.04E-64 |
DEG, differentially expressed gene; IL, interleukin; LPO, lactoperoxidase; AFB1, aflatoxin B1; N/A, not available.
PPI network construction
A PPI network of the top 10 upregulated and downregulated DEGs is shown in Fig. 2. The network consisted of 243 edges and 251 nodes. Generally, nodes with a high degree, which measures how many neighbors a node is directly connected to, are defined as hub proteins (29,30). Three nodes, including SPP1, TOP2A and ARPC1B, showed the highest degrees.
Figure 2.
Protein-protein interaction analysis of the top 10 upregulated (red) and downregulated (blue) differentially expressed genes.
RT-qPCR validation
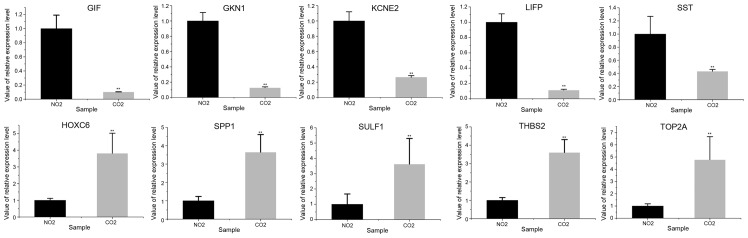
Five genes were randomly retrieved from the 10 upregulated and downregulated genes, respectively. It was shown that the expression patterns of the selected genes in GC and normal tissues in the RT-qPCR analysis were similar to those in the integrated analysis (Fig. 3; Table V), and that the difference between the GC patients and control samples were significantly different (P<0.01). The expression of SULF1, SPP1, THBS2, TOP2A and HOXC6 was upregulated in GC tissues compared with normal tissues, while the expression of GIF, KCNE2, SST, GKN1 and LIPF was downregulated in GC tissues compared with normal tissues.
Figure 3.
Reverse transcription-quantitative polymerase chain reaction validation of the differentially expressed genes in clinical samples. **P<0.01 vs. the control samples. NO2, control samples; CO2, gastric carcinoma samples.
Discussion
Microarrays are powerful tools for revealing the pathogenesis of human cancer and identify potential therapeutic targets (12). Microarray-based technology has been used in several studies to detect candidate genes involved in the occurrence of GC (31–34). In the present study, an integrated analysis of six transcriptome datasets was conducted and 689 DEGs were identified based on 612 samples, including 202 upregulated genes and 487 downregulated genes. The results of GO and KEGG analyses showed that the most enriched pathways included protein digestion and absorption, ECM-receptor interaction, and the metabolism of xenobiotics by cytochrome P450. The results were consistent with previous research (35,36), which identified DEGs with biological functions that were mainly involved in cell adhesion and ECM interactions.
Generally, it is accepted that, although the number of dysfunctional genes in a cancer may be limited, a large number of genes in related pathways may be affected at the expression level, and this aberrant gene transcriptional expression network is likely essential in the initiation and maintenance of the malignant phenotype (37–39). Most of the 20 most significantly DEGs have been reported to be involved in the oncogenesis and development of GC (23–26), and the functional classification of these genes was consistent with the results of GO and KEGG analyses. Three genes, including SPP1, TOP2A and ARPC1B, showed the highest connection in the PPI analysis and may have key roles in GC. In addition, SUL1, THBS2, HOXC6, SST, KCNE2, GIF, GKN1 and LIPF were also linked with more than one edge, indicating the potential role of these genes in the pathogenesis of GC.
However, the expression levels of six genes (MEST, GIF, CHIA, DUOX1, KIF4 and AKR7A3) in clinical samples were either inconsistent or ignored in previous studies (35,40). In the current study, dysregulation of these DEGs suggests that they serve roles in the oncogenesis and the development of GC. Among the six genes, the associations between GC and MEST, GIF, CHIA, and DUOX1 have never been reported. MEST encodes a member of the α/β hydrolase fold family and has the characteristic of isoform-specific imprinting. The aberrant imprinting of this gene has been linked to certain types of cancer and may be caused by promoter switching (41). GIF encodes intrinsic factor (IF), also known as gastric IF, which is a glycoprotein secreted by the parietal cells of the stomach that is necessary for the absorption of vitamin B12 (cobalamin) in the small intestine (35). CHIA may participate in the defense against nematodes, fungi and other pathogens, and play a role in the T-helper cell type 2 immune response; it is also involved in the inflammatory response and in protecting cells against apoptosis. Furthermore, CHIA is inhibited by allosamidin, suggesting that the function of this protein is dependent on carbohydrate binding (42–45). DUOX1 is the member of gp91phox homologs family and produces reactive oxygen species in various cells in response to stimuli, including growth factors, cytokines and calcium. A key role for DUOX1 in lung cancer, but not in GC, has been revealed (46). Although further studies on these genes have not been conducted, the results of the present study suggest that these genes may be considered as novel indicators for GC in the clinic.
Except for the four genes that have not previously been reported in GC, the expression levels of two genes in the current study were different from those reported in previous studies (40,47). Chromokinesin KIF4 is a member of the KIF4 subfamily and has been reported as an essential factor involved in multiple cellular process, including cell proliferation, DNA damage responses, immune cell activation, viral protein intracellular trafficking and neuronal survival in brain development (48). The overexpression of this subfamily was reported to inhibit GC cell proliferation in vitro, as well as their ability to form tumors in vivo (40). However, the expression level of KIF4 was significantly upregulated in GC samples in the current study. In addition, the expression of AKR7A3, which was reported to be upregulated in Singaporean GC patients (47), was downregulated in the RT-qPCR validation in the present study. This discrepancy in the results may be due to the heterogeneity of the GEO database, although the results still suggest complicated functions of KIF4 and AKR7A3 in the oncogenesis and development of GC.
In conclusion, the current study demonstrated that the analysis of expression profiles and RT-qPCR validation was able to give an explicit elucidation of the dysexpression of genes in GC. However, the results of the analysis of the expression profiles varied from study to study. Based on our results, the expression levels of six genes, including MEST, GIF, CHIA, DUOX1, KIF4 and AKR7A3, were found to be inconsistent with previous studies. These genes could potentially be valuable in the clinical treatment of GC. The present study may improve the understanding of the transcriptome status of GC and lay a foundation for further investigation of the mechanisms underlying this cancer of clinical and biological significance.
References
- 1.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhavan-Niaki H, Samadani AA. Molecular insight in gastric cancer induction: An overview of cancer stemness genes. Cell Biochem Biophys. 2014;68:463–473. doi: 10.1007/s12013-013-9749-7. [DOI] [PubMed] [Google Scholar]
- 5.Correa P, Camargo MC, Piazuelo MB, et al. The biology of gastric cancers. Springer Ebooks; 2008. [Google Scholar]
- 6.Figueiredo C, Garcia-Gonzalez MA, Machado JC. Molecular pathogenesis of gastric cancer. Helicobacter. 2013;18:S28–S33. doi: 10.1111/hel.12083. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 7.Conteduca V, Sansonno D, Lauletta G, Russi S, Ingravallo G, Dammacco F. H. pylori infection and gastric cancer: State of the art (review) Int J Oncol. 2013;42:5–18. doi: 10.3892/ijo.2012.1701. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita K, Sakuramoto S, Nemoto M, Shibata T, Mieno H, Katada N, Kikuchi S, Watanabe M. Trend in gastric cancer: 35 years of surgical experience in Japan. World J Gastroenterol. 2011;17:3390–3397. doi: 10.3748/wjg.v17.i29.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Xu J, Shi Z, Shen X, Luo T, Bi J, Nie M. Clinicopathologic characteristics and prognostic of gastric cancer in young patients. Scand J Gastroenterol. 2016;51:1043–1049. doi: 10.1080/00365521.2016.1180707. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Baek M, Yang H, Bang YJ, Kim WH, Ha JH, Kim DK, Jeoung DI. Identification of genes differentially expressed between gastric cancers and normal gastric mucosa with cDNA microarrays. Cancer Lett. 2002;184:197–206. doi: 10.1016/S0304-3835(02)00197-0. [DOI] [PubMed] [Google Scholar]
- 11.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 12.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang SN, Sun HH, Jin YM, Piao LZ, Jin DH, Lin ZH, Shen XH. Identification of differentially expressed genes in gastric cancer by high density cDNA microarray. Cancer Genet. 2012;205:147–155. doi: 10.1016/j.cancergen.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 19.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornstein P, Kyriakides TR, Yang Z, Armstrong LC, Birk DE. Thrombospondin 2 modulates collagen fibrillogenesis and angiogenesis. J Investig Dermatol Symp Proc. 2000;5:61–66. doi: 10.1046/j.1087-0024.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- 24.O'Rourke KM, Laherty CD, Dixit VM. Thrombospondin 1 and thrombospondin 2 are expressed as both homo- and heterotrimers. J Biol Chem. 1992;267:24921–24924. [PubMed] [Google Scholar]
- 25.Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 26.Acampora D, D'Esposito M, Faiella A, Pannese M, Migliaccio E, Morelli F, Stornaiuolo A, Nigro V, Simeone A, Boncinelli E. The human HOX gene family. Nucleic Acids Res. 1990;17:10385–10402. doi: 10.1093/nar/17.24.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewitt JE, Gordon MM, Taggart RT, Mohandas TK, Alpers DH. Human gastric intrinsic factor: Characterization of cDNA and genomic clones and localization to human chromosome 11. Genomics. 1991;10:432–440. doi: 10.1016/0888-7543(91)90329-D. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Fox A, Taylor D, Slonim DK. High throughput interaction data reveals degree conservation of hub proteins. Pac Symp Biocomput. 2009:391–402. doi: 10.1142/9789812836939_0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavi O, Skinner J, Gottesman MM. Network features suggest new hepatocellular carcinoma treatment strategies. BMC Syst Biol. 2014;8:88. doi: 10.1186/s12918-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay ST, Leong SH, Yu K, Aggarwal A, Tan SY, Lee CH, Wong K, Visvanathan J, Lim D, Wong WK, et al. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309–3316. [PubMed] [Google Scholar]
- 32.Wu CM, Lee YS, Wang TH, Lee LY, Kong WH, Chen ES, Wei ML, Liang Y, Hwang TL. Identification of differential gene expression between intestinal and diffuse gastric cancer using cDNA microarray. Oncol Rep. 2006;15:57–64. [PubMed] [Google Scholar]
- 33.Dang Y, Wang YC, Huang QJ. Microarray and next-generation sequencing to analyse gastric cancer. Asian Pac J Cancer Prev. 2014;15:8033–8039. doi: 10.7314/APJCP.2014.15.19.8035. [DOI] [PubMed] [Google Scholar]
- 34.Song B, Du J, Deng N, Ren JC, Shu ZB. Comparative analysis of gene expression profiles of gastric cardia adenocarcinoma and gastric non-cardia adenocarcinoma. Oncol Lett. 2016;12:3866–3874. doi: 10.3892/ol.2016.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, Ni Z, Zhang M, Kong X, Hoffman LL, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–1207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 37.Koslowski M, Sahin U, Mitnacht-Kraus R, Seitz G, Huber C, Türeci O. A placenta-specific gene ectopically activated in many human cancers is essentially involved in malignant cell processes. Cancer Res. 2007;67:9528–9534. doi: 10.1158/0008-5472.CAN-07-1350. [DOI] [PubMed] [Google Scholar]
- 38.Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, Gardner GJ, Hao K, Wong WH, Barrett JC, et al. Expression profiling of serous low malignant potential, low-grade and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 39.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, Polunovsky VA. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 40.Gao J, Sai N, Wang C, Sheng X, Shao Q, Zhou C, Shi Y, Sun S, Qu X, Zhu C. Overexpression of chromokinesin KIF4 inhibits proliferation of human gastric carcinoma cells both in vitro and in vivo. Tumor Biol. 2011;32:53–61. doi: 10.1007/s13277-010-0090-0. [DOI] [PubMed] [Google Scholar]
- 41.Nishita Y, Yoshida I, Sado T, Takagi N. Genomic imprinting and chromosomal localization of the human MEST gene. Genomics. 1996;36:539–542. doi: 10.1006/geno.1996.0502. [DOI] [PubMed] [Google Scholar]
- 42.Lunardi D, Abelli L, Panti C, Marsili L, Fossi MC, Mancia A. Transcriptomic analysis of bottlenose dolphin (Tursiops truncatus) skin biopsies to assess the effects of emerging contaminants. Mar Environ Res. 2016;114:74–79. doi: 10.1016/j.marenvres.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Hartl D, He CH, Koller B, Da Silva CA, Homer R, Lee CG, Elias JA. Acidic mammalian chitinase is secreted via an ADAM17/epidermal growth factor receptor-dependent pathway and stimulates chemokine production by pulmonary epithelial cells. J Biol Chem. 2008;283:33472–33482. doi: 10.1074/jbc.M805574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Żurawska-Płaksej E, Ługowska A, Hetmańczyk K, Knapik-Kordecka M, Piwowar A. Neutrophils as a source of chitinases and chitinase-like proteins in type 2 diabetes. PLoS One. 2015;10:e0141730. doi: 10.1371/journal.pone.0141730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gooday GW, Brydon LJ, Chappell LH. Chitinase in female onchocerca gibsoni and its inhibition by allosamidin. Mol Biochem Parasitol. 1988;29:223–225. doi: 10.1016/0166-6851(88)90077-1. [DOI] [PubMed] [Google Scholar]
- 46.Kamata T. Roles of Nox1 and other Nox isoforms in cancer development. Cancer Sci. 2009;100:1382–1388. doi: 10.1111/j.1349-7006.2009.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nair NG, Blessantoli M, Vennila JJ. Gene expression in gastric cancer for Singapore and UK population: An Insilico comparative approach. Res J Recent Sci. 2014;3 [Google Scholar]
- 48.Taniwaki M, Takano A, Ishikawa N, Yasui W, Inai K, Nishimura H, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin Cancer Res. 2007;13:6624–6631. doi: 10.1158/1078-0432.CCR-07-1328. [DOI] [PubMed] [Google Scholar]