Abstract
Dental plaque is a complex multispecies biofilm, and is a direct precursor of periodontal disease. The virulence of periodontal pathogens, such as Porphyromonas gingivalis, is expressed in the context of this polymicrobial community. Previously, we reported an antagonistic relationship between Streptococcus cristatus and P. gingivalis, and identified arginine deiminase (ArcA) of S. cristatus as the signaling molecule to which P. gingivalis responds by repressing the expression and production of FimA protein. Here we demonstrate that direct interaction between P. gingivalis and S. cristatus is necessary for the cell-cell communication. Two surface proteins of P. gingivalis, PGN_0294 and PGN_0806, were found to interact with S. cristatus ArcA. Using a peptide array analysis, we identified several P. gingivalis-binding sites of ArcA, which led to the discovery of an 11-mer peptide with the native sequence of ArcA that repressed expression of fimbriae and of gingipains. These data indicate that a functional motif of ArcA is sufficient to selectively alter virulence gene expression in P. gingivalis, and PGN_0294 and PGN_0806 may serve as receptors for ArcA. Our findings provide a molecular basis for future rational design of agents that interfere with the initiation and formation of a P. gingivalis-induced pathogenic community.
Introduction
Periodontitis is the 6th most common infection worldwide1 and an estimated 5–20% of the population suffers from generalized chronic periodontitis2, 3. In the US, around half of the adult population suffers from some form of periodontal disease, which constitutes a significant economic burden4. Periodontal diseases and periodontal bacteria are also physically and epidemiologically associated with severe systemic conditions such as coronary artery disease, rheumatoid arthritis, and diabetes5–7. Chronic periodontitis is the result of a breakdown of periodontal tissue-microbe homeostasis, which then leads to uncontrolled inflammation and tissue destruction8. Loss of tissue homeostasis, called dysbiosis, is initiated by communities of organisms colonizing the subgingival area. In most instances these microbial communities are associated with health, and the transition to pathogenesis requires colonization by specific pathogens such as Porphyromonas gingivalis. It has long been recognized that the presence of P. gingivalis alone is insufficient for the disease to occur in a susceptible host, and current models hold that P. gingivalis is a keystone pathogen in that it elevates the virulence of the entire community9–12. Evidence for this comes from murine models in which low levels of P. gingivalis can initiate disease, but only in the context of a microbial community9, and from primate studies where a gingipain-based vaccine caused a reduction both in P. gingivalis numbers and in the total subgingival bacterial load, as well as in inhibiting bone loss13.
Bacteria within the oral microbial community can exhibit polymicrobial synergy whereby interspecies communication enhances colonization and pathogenic potential. Conversely, oral organisms also engage in antagonistic interactions, whereby one organism inhibits the colonization or growth of another. For example, communities of P. gingivalis and Streptococcus gordonii are synergistically pathogenic in a murine model of alveolar bone loss14, whereas Streptococcus cristatus interferes with the colonization and pathogenesis of P. gingivalis in mice15. We, and others, have reported previously that arginine deiminase (ArcA) of S. cristatus represses expression of the major fimbrial adhesin of P. gingivalis, FimA16, 17. ArcA catalyzes the hydrolysis of L-arginine to L-citrulline and ammonia, and the latter is believed to be important for oral biofilm pH homeostasis and the prevention of caries18, 19. We also found that the expression of arcA was significantly higher in S. cristatus than in S. gordonii 20, suggesting that ArcA activity could define the differing roles of these two streptococcal species in the highly orchestrated formation of dental plaque. Moreover, clinical studies revealed an inverse relationship between the numbers of S. cristatus compared to P. gingivalis cells in dental plaque isolated from periodontitis subjects, suggesting that S. cristatus may be beneficial to the host by antagonizing the colonization and accumulation of P. gingivalis 21.
In this study, we investigated the components of the antagonistic communication system between P. gingivalis and S. cristatus including functional motifs of ArcA and receptor(s) of P. gingivalis. We found that direct contact is required in intergeneric communication between P. gingivalis and S. cristatus. Our results also identified a short linear domain of ArcA as the binding site for P. gingivalis. Two surface proteins of P. gingivalis, PGN_0294 (RagB) and PGN_0806, likely serve as receptors in this bacterial cell-cell communication. Equally important, we were able to show that the short ArcA-derived peptide represses expression of several established virulence genes of P. gingivalis including fimA, mfa1, rgpA, rgpB, and kgp. Exploitation of this antagonistic relationship may lead to the discovery of pharmaceutical agents to inhibit P. gingivalis colonization and pathogenicity.
Results
Direct contact is required for P. gingivalis-S. cristatus communication
To test if P. gingivalis-S. cristatus communication occurs through direct cell-cell contact, P. gingivalis 33277 and S. cristatus CC5A or its arcA mutant were separated using a transwell system with a membrane of pore size either 0.4 µm or 8 µm. After 16 h, bacteria in each the lower well were collected, and numbers of P. gingivalis 33277 and S. cristatus CC5A were determined using qPCR. 7.8 × 104 from an input of 1 × 107 CC5A cells migrated to the lower well from the Transwell insert through 8 µm pores, whereas less than 1.5 S. cristatus cells were detected in the lower well when using the membrane with 0.4 µm pore (Fig. 1a). P. gingivalis RNA was then purified and expression of the fimA gene measured using qRT-PCR. Levels of fimA expression were reduced about 2.5 fold when the 8-µm pore transwell was used (Fig. 1b). Inhibition of fimA expression by S. cristatus was not observed when P. gingivalis-S. cristatus contact was blocked by the 0.4 µm pore membrane, suggesting that direct contact is required for cell-cell communication between P. gingivalis and S. cristatus.
Figure 1.
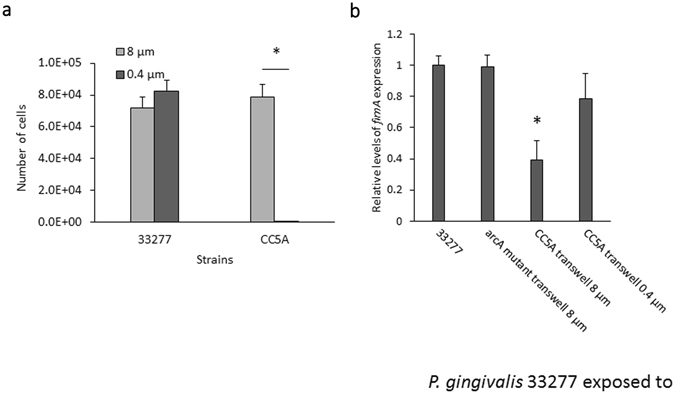
Expression of fimA in P. gingivalis in contact with S. cristatus. (a) S. cristatus CC5A migration through the Transwell insert. CC5A (1 × 107 cells) were initially incubated in the Transwell insert, and the numbers of CC5A migrated to the lower well and 33277 cells in the well were determined by qPCR. Each bar represents the number of bacteria detected in the lower well. Asterisk indicates a statistically significant difference in number of bacteria in the lower wells (n = 3; t test; p < 0.05). (b) Expression of the fimA gene in P. gingivalis in transwell chambers with S. cristatus was measured by real-time qRT-PCR. Each bar represents relative expression level of the fimA, which was normalized to that of 16 S rRNA gene. Standard deviations are indicated. Asterisk indicates a statistically significant difference in expression level of fimA compared to that in P. gingivalis without exposure to S. cristatus (n = 3; t test; p < 0.05).
Direct interaction of P. gingivalis and S. cristatus ArcA was confirmed by an immunofluorescence assay with P. gingivalis cells and purified ArcA protein. Fluorescent labeled P. gingivalis-ArcA complexes were detected by confocal microscopy. As shown in Fig. 2, ArcA had high affinity for P. gingivalis 33277, but not for AaY4, suggesting a specific interaction between ArcA and P. gingivalis surface molecules.
Figure 2.
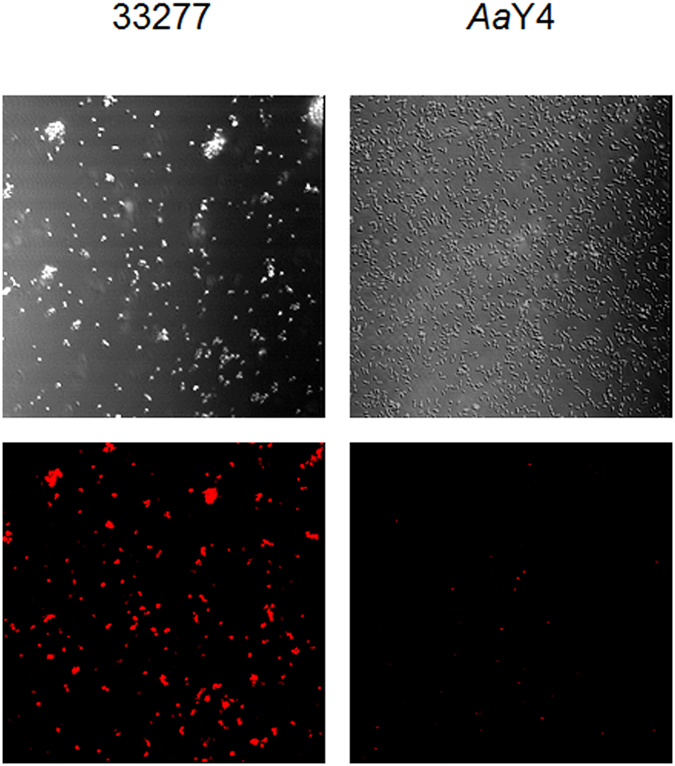
Immunofluorescence antibody images of the interaction of P. gingivalis 33277 or A. actinomycetemcomitans Y4 with ArcA. The upper panel presents differential interference contrast (DIC) images showing the location of the bacteria. The lower panels are the TRITC fluorescence labeling (red) images showing bacterial-associated ArcA. Bar is 5 µm.
Isolation of P. gingivalis surface protein(s) that interacts with ArcA of S. cristatus
To isolate and identify P. gingivalis surface molecule(s) that interact with ArcA, we performed a pull-down assay. An ArcA antibody coupled Sepharose 4B column was used to capture ArcA-interacting components from a mixture of P. gingivalis cell lysate and ArcA protein. The proteins eluted from the column were analyzed with SDS-PAGE. Three bands with molecular sizes of approximately 55, 47, and 30 kDa were detected (Fig. 3). Western blot using ArcA antibody showed that the 47 kDa protein is ArcA of S. cristatus (data not shown). The other two bands were identified by MS analysis as P. gingivalis RagB (PGN_0294) and a MotA/TolQ/ExbB proton channel family protein (PGN_0806), suggesting that these two proteins are receptors for ArcA.
Figure 3.
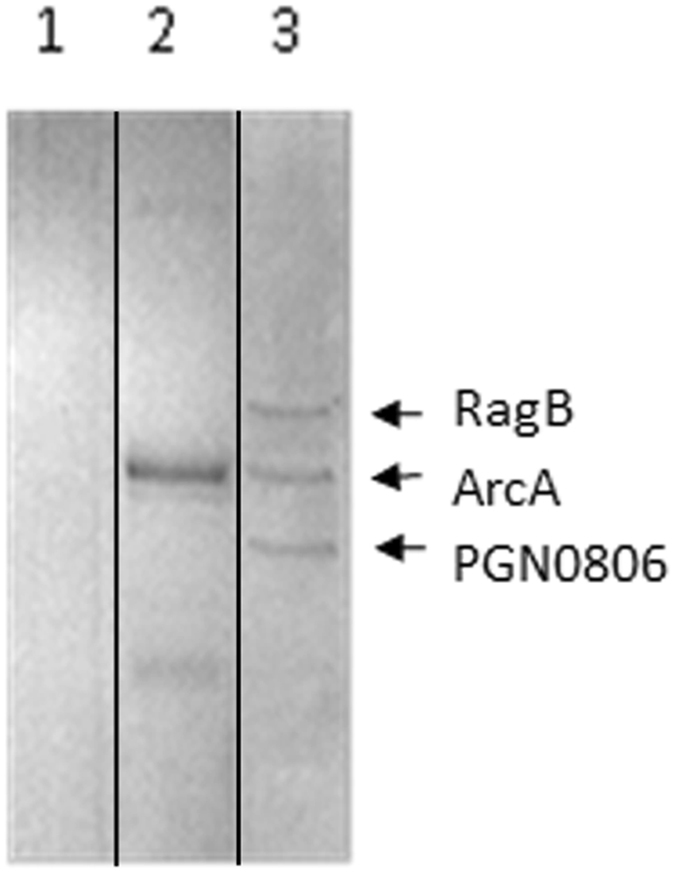
Interaction of ArcA and P. gingivalis surface proteins. SDS-PAGE analysis of proteins eluted from Sepharose 4B column. Lane 1, the proteins eluted from untreated Sepharose 4B column exposed to P. gingivalis 33277 extract; Lane 2, the proteins eluted from ArcA antibody-coupled Sepharose 4B column exposed to CC5A extract only; lane 3, the proteins eluted from ArcA antibody-coupled Sepharose 4B column exposed to P. gingivalis and CC5A extracts. Proteins were stained with Coomassie blue.
Identification of the key functional motif of ArcA
S. cristatus ArcA is a 47 kDa protein with 409 amino acids16. We sought to identify key amino acids and the motif(s) of ArcA responsible for its inhibitory activity toward fimA expression. A peptide microarray was first performed to detect binding sites of ArcA for P. gingivalis. The arrays were incubated with surface extracts of P. gingivalis 33277, or the ΔragB or Δ0806 mutants, and binding was detected with P. gingivalis antibodies. Although the absolute binding capacities (fluorescence intensity) of these strains were significantly varied, likely due to protein degradation of surface extract in some strains, the overall patterns were consistent. Of several peaks observed (Fig. 4), a peptide with the sequence NIFKKNVGFKK (peak 4) and spanning amino acid residues 249–259, was found to have the highest binding affinity to P. gingivalis 33277 proteins, evident as the highest peak. This peak was no longer the highest when the arrays were incubated with surface extracts isolated from the Δ0806 or ΔragB mutants, corroborating the involvement of P. gingivalis proteins PGN_0806 and RagB in recognition of ArcA.
Figure 4.
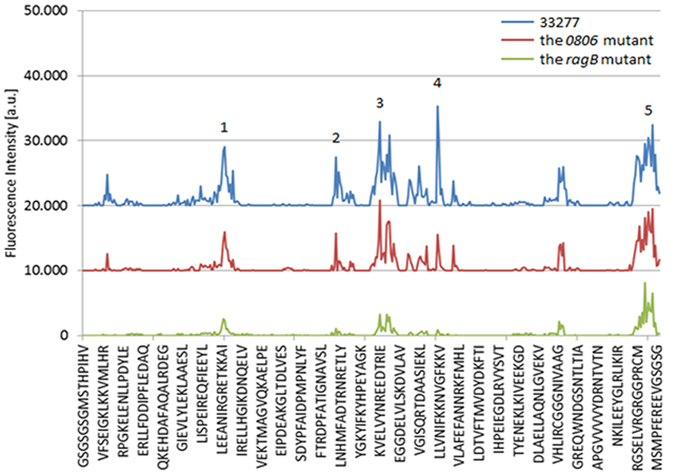
Identification of a binding region of ArcA interacting with P. gingivalis. A peptide array of ArcA was exposed to P. gingivalis 33277 and the ragB and pgn0806 mutants. The intensity plot of the peptide array signals shows as peaks with corresponding regions of ArcA. The five of the highest peaks are numbered.
Five peptides (1–5 in Table 1) were synthesized based on ArcA array peaks, and the effect of each peptide on gene expression was determined by inclusion of the peptides in the P. gingivalis growth media. As shown in Table 1, the 11 residue peptide4 from the C-terminal region of ArcA repressed expression of fimA, mfa1, kgp, rgpA/B (encoding catalytic regions of rgpA/B), and rgpA (encoding adhesin domains of RgpA) genes by at least 2 fold, at a concentration of 16 µM. Expression of pgn_0128 encoding immunoreactive 53 kDa antigen was not modulated in response to the presence of peptide4, indicating specificity for a subset of virulence-associated genes. Increased inhibitory activity (60–70%) was observed at a concentration of 64 µM (not shown), suggesting that this region is likely a key active motif of ArcA. These results also establish that the effects of ArcA on P. gingivalis virulence extend beyond repression of fimA and include genes for the gingipains and for the minor fimbrial subunit, as also shown by others17. The half inhibitory concentration (IC50) was determined by constructing a dose-response curve (0, 4, 16, and 64 µM) to measure the effectiveness of peptide4 in repressing expression of these genes. As shown in Fig. 5, the highest efficiency of peptide4 was found in inhibition of rgpA (amplified with primers corresponding to the region encoding the binding domain of RgpA, HGP44), rgpA/B (amplified with primers corresponding to the region encoding catalytic regions of RgpA and B), and fimA with IC50s of 11.2 ± 2.1, 11.7 ± 2.3 or µM, 12.4 ± 3.4, respectively. Peptide4 also showed a significantly lower IC50 for mfa1 (19.2 ± 3.3 µM) compared to that of kgp (64.3 ± 3.8 µM). It should be pointed out that peptide1, 2, 3 and 5 (Table 1) also exhibited some inhibitory activity, although at a lower efficiency. These regions along with peptide4 may be involved in formation of a structural motif that may have a higher binding capacity than peptide4 alone. These findings provide a molecular basis for the future design of inhibitors of P. gingivalis.
Table 1.
Differential expression of virulence genes in P. gingivalis in the presence of ArcA peptides.
| Peptide | Peptide sequence and residue position | Relative expression levela | |||||
|---|---|---|---|---|---|---|---|
| fimA | mfa1 | rgpA/B | rgpA | kgp | pgn0128 | ||
| P1 | I97RGRETKK | 0.88 ± 0.08 | 0.85 ± 0.05 | 0.89 ± 0.05 | 0.86 ± 0.07 | 1.06 ± 0.12 | 0.96 ± 0.06 |
| P2 | N177HMFADTRNRE | 0.80 ± 0.03 | 0.80 ± 0.10 | 0.78 ± 0.07 | 0.81 ± 0.03 | 0.90 ± 0.05 | 0.88 ± 0.03 |
| P3 | V208YNREEDTRIEGGDEL | 0.87 ± 0.10 | 0.82 ± 0.07 | 0.98 ± 0.06 | 0.84 ± 0.06 | 0.99 ± 0.12 | 0.91 ± 0.07 |
| P4 | N249IFKKNVGFKK | 0.40 ± 0.06* | 0.51 ± 0.05* | 0.38 ± 0.04* | 0.39 ± 0.04* | 0.47 ± 0.06* | 0.94 ± 0.06 |
| P5 | E389LVRGRGGPRCMSMPF | 0.97 ± 0.05 | 0.83 ± 0.04 | 0.72 ± 0.05 | 0.73 ± 0.06 | 0.96 ± 0.12 | 0.91 ± 0.06 |
a P. gingivalis 33277 was grown TSB in the presence or absence of peptide at a concentration 16 µM. Transcript levels were measured by real-time PCR. The mRNA levels of genes are indicated relative to the expression level in the absence of peptides as 1 unit. Results shown are means and standard deviations from three independent experiments. Asterisks indicate the statistical significance of expression levels at least two fold in P. gingivalis grown in TSB with/without peptides (P < 0.05; t test).
Figure 5.
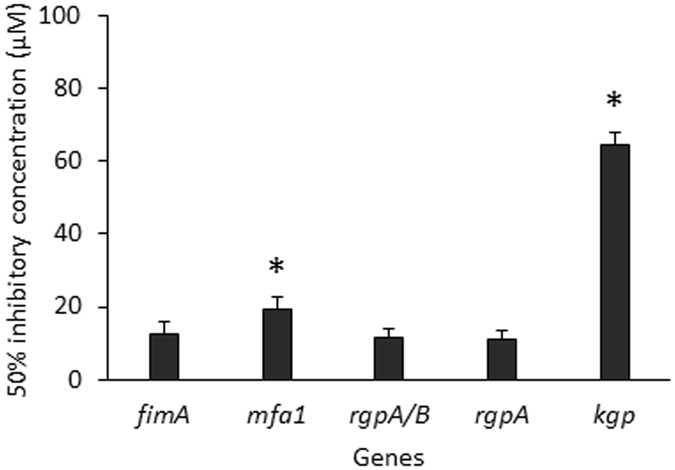
Potency of peptide4 for inhibition of virulence gene expression in P. gingivalis. The half inhibitory concentration (IC50) was measured by conducting three independent experiments to determine mRNA levels of fimA, mfa1, rgpA/B, and kgp in the presence of peptide4 at the concentrations 0, 4, 16, and 64 µM, respectively. The IC50 for each gene was established using a Microsoft Excel program with add-in for curve fitting. Asterisks indicate the statistical significances of IC50 of peptide4 for a specific gene when compared to that for the fimA gene (P < 0.05; t test).
To verify that PGN_0806 and RagB function as receptors in P. gingivalis-S. cristatus communication, we tested gene expression in the Δ0806 and ΔragB strains in the presence or absence of peptide4 and compared these to that in the wild type strain 33277. The results showed that loss of PGN_0806 prevented peptide4-dependent regulation of fimA, mfa1, rgp, and kgp (Fig. 6a). Although the ragB mutation did not completely block peptide4 activity, a significantly reduced inhibitory effect was observed toward all of the target genes. Previously, a two component regulatory system (FimS/R) was identified to be activator of the fimA expression22, 23. We thus tested the role of FimS/R in S. cristatus-P. gingivalis cell-cell communication. Although expression levels of fimA and mfa1 were repressed approximately 20 and 4 fold in the fimS and fimR mutants (data not shown), Peptide4 mediated regulation of FimA expression reminded intact in the absence of FimS and FimR (Fig. 6b), suggesting FimS/R is not involved in this bacterial cell-cell communication. These results provide strong evidence that PGN_0806 and RagB, either separately or in combination, act as receptors in the bacterial cell-cell communication between P. gingivalis and S. cristatus.
Figure 6.
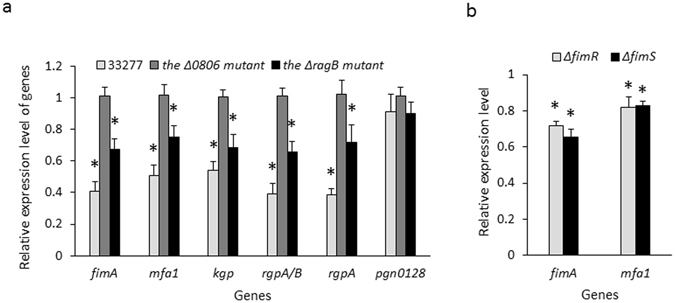
Comparison of virulence gene expression in P. gingivalis 33277 and its mutants. Expression of fimA, mfa1, rgpA + B, rgpA, and kgp was determined using qRT-PCR. P. gingivalis strains was grown TSB in the presence or absence of peptide4 at a concentration 16 µM. (a) The mRNA levels of genes in 33277, the pgn_0806, and the ragB mutants grown in the media supplemented with peptide4 are indicated relative to the expression level in P. gingivalis 33277 grown in the medium without peptide4 (1 unit). (b) The fimR (ΔfimR) and fimS (ΔfimS) mutants were grown with or without the peptide4. Each bar represents relative expression level of fimA or mfa1 in the mutants grown with peptide4 (16 µM) to those in the mutant grown in the media without peptide4 (1 unit). Results shown are means and standard deviations from three independent experiments. Asterisks indicate the statistical significance of expression levels of genes in P. gingivalis strains grown with/without peptides (P < 0.05; t test).
Expression of fimbrial proteins and gingipains at the translational level was also determined using Western blot analysis. P. gingivalis 33277 was grown with peptide4 at concentrations of 0, 3, 12, and 48 µM (0×, ¼×, 1×, and 4× IC50 of fimA expression) for 48 h. As shown in Fig. 7a,b, production of FimA, Mfa1, and HGP44 (a binding domain of RgpA) was significantly decreased in the presence of 12 and 48 µM of peptide4. However, production of immunoreactive 53 kDa antigen was not altered, consistent with the expression pattern observed at the transcriptional level. Transmission electron microscopy further showed that there were few fimbriae on the surface of P. gingivalis grown in media supplemented with peptide4 (16 µM), when compared to P. gingivalis cells grown without peptide4 (Fig. 8a,b).
Figure 7.
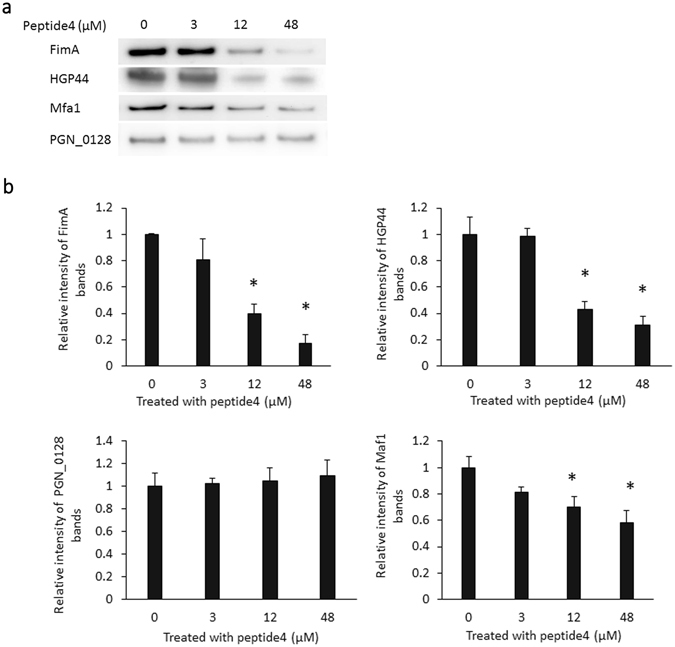
Production of fimbrial proteins and gingipains in P. gingivalis in response to peptide4. (a) Expression levels of FimA, Mfa1, Hgp44 of gingipains, and PGN_0128 (immunoreactive 53 kDa antigen) in surface extracts of P. gingivalis 33277 exposed to pwptide4 at concentrations 0, 3, 12, and 48 µM were analyzed using a Western blot analysis. (b) Semiquantitation of western blots was conducted with ImageJ software. Each bar represents the intensity of the protein band. An asterisk indicates a significant difference between the relative intensity of the protein bands in P. gingivalis exposed to peptide4 (3, 12, or 48 µM) compared to those seen in P. gingivalis not exposed (0 µM, 1 unit) (P < 0.05 by t test).
Figure 8.
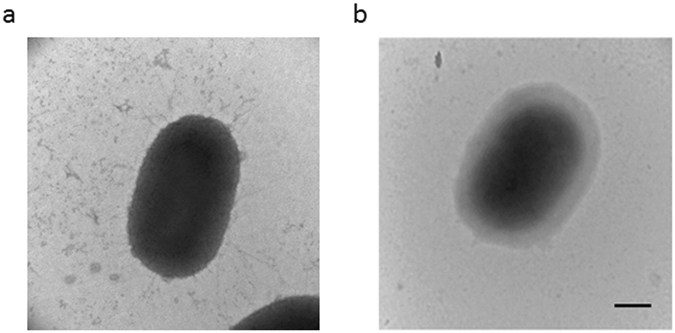
Transmission electron microscopic analysis of P. gingivalis fimbriae. Fimbrial structures were visualized using TEM. P. gingivalis strains 33277 (a) and 33277 treated with peptide4 (b) were grown on the TSB plate for 48 h and prepared by negatively staining with ammonium molybdate. Bars = 0.2 µm.
Discussion
P. gingivalis possesses a vast array of effector molecules and systems that enable the organism to colonize and survive in the oral cavity, communicate with other bacteria, and ultimately elevate the virulence of the entire microbial community24–27. Major fimbriae (long fimbriae) composed of FimA, are promiscuous adhesins and contribute to colonization, biofilm formation, cell invasion, bone resorption, and the evasion of host defense systems25, 28–37. With regard to induction of immune dysbiosis, FimA binds the CXC-chemokine receptor 4 (CXCR4) and induces crosstalk with TLR2 that inhibits the MyD88-dependent antimicrobial pathway24. Both the major and minor (Mfa1) fimbriae of P. gingivalis mediate coadhesion with S. gordonii and are thus involved in synergistic pathogenicity38–40. The majority of P. gingivalis clinical isolates are fimbriated, especially those isolated at the base of periodontal pockets41–43. Other well-known virulence factors are the gingipains which include two arginine- and one lysine-specific cysteine proteinases (RgpA, RgpB, and Kgp)44. Thus far, all tested P. gingivalis strains produce gingipains that are both membrane-associated and secreted soluble forms45. Besides their role in tissue matrix destruction due to proteolytic activity46, gingipains play an important role in biofilm formation of P. gingivalis through the C-terminal adhesive regions of RgpA and Kgp or through processing profimbrillin47, 48. Gingipains are also involved in modulating immune responses, by cleavage of secreted chemokines and intracellular immune kinases49, 50. Previously, we reported that S. cristatus ArcA represses fimA expression in P. gingivalis 16, 51. Similar results, reported by others17, 52, showed downregulation of both fimA and mfa1 fimbriae by Streptococcus intermedius ArcA. In these studies ArcA enzymatic activity is required for an effect of on biofilm formation through arginine depletion, suggesting an additional indirect role of ArcA in P. gingivalis colonization52. These observations suggest that ArcA modulates expression of fimbrial proteins in P. gingivalis both directly and indirectly. Collectively, accumulating observations suggest that ArcA modulates expression of fimbrial proteins in P. gingivalis both directly and indirectly. Here, we identified a functional motif of ArcA, located at the C-terminal and spanning amino acids 249–259, and a peptide (peptide4) derived from this region showed inhibitory activity for both mRNA and protein expression of fimbriae (FimA and Mfa1) and gingipains (RgpA/B and Kgp). Hence this peptide is a potential candidate for developing inhibitors against P. gingivalis. Based on our observation that ArcA specifically binds to the surface of P. gingivalis, it is likely that the peptide inhibitors would be specific for this organism and not have a significant inhibitory effect on early biofilm colonizers (streptococci and actinomyces). Targeting P. gingivalis alone would likely be sufficient to impede the development of a dysbiotic biofilm, as P. gingivalis is considered a keystone pathogen8, 53.
Cell surface receptors are important elements in signal transduction, and possess the ability to bind (sense) a specific signal, subsequently eliciting a specific cellular response. A well-known signal transduction process in bacteria involves two-component regulatory systems which involve a sensor histidine kinase and a response regulator protein54. The FimS/R two-component signal transduction in P. gingivalis predominantly regulates fimA expression55, 56 and some other genes including mfa1 22, 23. However, results from the current study showed that FimS/R was not involved in communication between P. gingivalis and S. cristatus, since expression levels of fimA and mfa1 in the fimS or fimR mutants were also modulated in response to peptide4. However, we identified two P. gingivalis surface proteins, RagB (PGN_0294), a major immunodominant antigen of P. gingivalis, and PGN_0806 annotated as a MotA/TolQ/ExbB proton channel family protein, which interact with ArcA of S. cristatus, especially with the peptide4 region of ArcA. Interestingly, the Pseudomonas aeruginosa TonB–ExbB–ExbD protein complex is reported to be involved in signal transduction57. Moreover, RagA, which is thought to associate with RagB on the P. gingivalis surface, is a TonB-dependent receptor58–60. We also demonstrate that mutation in the ragB gene partially blocks the inhibitory activity of ArcA against fimA, while the P. gingivalis strain carrying mutation in the pgn_0806 gene was completely abrogated in response to peptide4. These data corroborate the role of PGN_0806 and RagB, as receptors, in P. gingivalis-S. cristatus communication. Although we identified a signal peptide and potential receptor(s), the mechanisms of intracellular signal transduction are still unidentified. A previous study showed that expression of rgpA, but not kgp, was decreased in a prtT mutant, which indicated that expression of kgp and rgp is not coordinately regulated61. Therefore, we speculate that independent intracellular transmitters are involved in control of fimA, mfa1, kgp, and rgp.
In conclusion, we have identified a functional domain of S. cristatus ArcA that has high affinity to the surface of P. gingivalis and is able to repress expression of several well-known virulence genes involved in production of fimbriae and gingipains. We also uncovered two surface proteins of P. gingivalis, RagB and PGN_0806, which interact with ArcA and are required for bacterial cell-cell communication between P. gingivalis and S. cristatus. These results functionally characterize and molecularly dissect the antagonistic relationship between these oral bacteria that we reported earlier16, 62. Application of these findings should provide the basis for therapeutic strategies designed to reduce colonization of P. gingivalis in the oral cavity and suppress the pathogenicity of periodontitis-associated dental plaque.
Methods
Bacterial strains and growth conditions
P. gingivalis strains and A. actinomycetemcomitans Y4 were grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml), and incubated at 37 °C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). S. cristatus CC5A and isogenic ΔarcA16 were grown in Trypticase peptone broth (TPB) supplemented with 0.5% glucose at 37 °C under aerobic conditions. Erythromycin (5 μg/ml) or tetracycline (0.5 μg/ml) were added to growth media when appropriate.
Transwell co-culture assay
P. gingivalis cells (105) were inoculated in each well of a six well plate, and S. cristatus CC5A or its arcA mutant (107) was added into the transwell inserts with a polycarbonate porous membrane of pore size 0.4 µm or 8 µm. After 16 h growth, the bacterial cells in each well of the plate were collected by centrifugation. To determine the number of CC5A migrated to the lower wells, the bacteria in the lower wells were harvested by centrifugation, and DNA was released by boiling the samples for 20 mins. Numbers of bacteria were determined by qPCR using specific primers for CC5A arcA and 33277 16s-rRNA (Table S1). P. gingivalis RNA was purified using an RNeasy mini spin column which selectively lyses P. gingivalis cells and not S. cristatus cells. Expression of the fimA gene in P. gingivalis was measured using real time qRT-PCR (see below).
RNA isolation and qPCR
P. gingivalis were homogenized in Trizol Reagent (Invitrogen, Carlsbad, CA) and RNA was purified using an RNeasy mini spin column (Qiagen, Valencia, CA). RNA samples were digested on-column with RNase-free DNase, and total RNA was tested using an Agilent 2100 Bioanalyzer to ensure the quality of the samples. Primers are listed in Table S1. Amplification reactions consisted of a reverse transcription using a Bio-Rad iScript Reverse Transcription Supermix on a TC-3000 thermal cycler (Techne, Staffordshire, ST15 OSA, UK) and a real-time qRT-PCR analysis using a QuantiTect SYBR Green RT-PCR Kit (Qiagen) on an iCycler MyiQTM Real-Time PCR detection system (Bio-Rad Laboratories, Inc, Hercules, CA) according to the manufacturer’s instructions. P. gingivalis 16 S rRNA gene was used as a normalizing gene. The melting curve profile was analyzed to verify a single peak for each sample, which indicated primer specificity. The expression levels of the investigated genes for the test sample were determined relative to the untreated calibrator sample by using the comparative cycle threshold (ΔC T) method. ΔC T was calculated by subtracting the average C T value of the test sample from the average C T value of the calibrator sample, and the value used to calculate the ratio between the two by assuming 100% amplification efficiency. By loading the same amount of total RNA for any comparable samples, the ΔC T represents the difference in gene expression between the samples.
Confocal microscopy
ArcA (50 µg) was purified from S. cristatus CC5A as described22, mixed with P. gingivalis cells (108) in PBS and incubated at room temperature for 1 h. After washing three times with PBS, bacteria-protein complexes were blocked in PBS with 5% BSA for 1 h. ArcA bound to P. gingivalis was detected by staining with rabbit anti-ArcA polyclonal antibody (1:400) and tetramethyl rhodamine isothiocyanate (TRITC, 1:500)-conjugated AffiniPure Goat Anti-Rabbit IgG antibody. Visualization was with a LSM 510 inverted confocal microscope with selected filters (543 nm excitation and 560 nm emission).
Pull-down assays
To isolate and identify P. gingivalis surface molecule(s) that interacts with ArcA, we performed a pull-down assay. Surface extracts of P. gingivalis were prepared by sonication with a Sonic Dismembrator (Fisher Scientific; output control 8, 20 s × 3), and the cell debris were removed by centrifugation followed by filtration (0.2-μm pore size). Surface extracts of 33277 were mixed with purified ArcA on a rotator at room temperature for 1 h, and then added to an ArcA antibody coupled Sepharose 4B column (Sigma-Aldrich). After incubation at room temperature for 1 h, the column was washed three times with PBS containing 0.01% Tween, and proteins were eluted by 0.1 M Glycine pH 2.4. After SDS-PAGE, bands were excised and identified by liquid chromatography–mass spectrometry.
Construction of the pgn_0294 (ragB) and pgn_0806 (motA/tolQ/exbB) mutants
Insertional mutants (pgn_0294 and pgn_0806) were generated by ligation-independent cloning of PCR mediated mutagenesis (LIC-PCR)63–65. A 2.1-kb ermF-ermAM cassette was introduced into target genes by three step PCR to yield pgn_0294-erm- pgn_0294 or pgn_0806-erm-pgn_0806 DNA fragments as described previously64. The final PCR products were then introduced into P. gingivalis 33277 by electroporation. Mutants were selected on TSB plates containing erythromycin (5 µg/ml). The insertional mutation was confirmed by PCR analysis, and the mutants were designated as P. gingivalis Δ0806 or ΔragB.
Protein Interaction Screening
Surface extracts of P. gingivalis were collected by sonication (1 min for three times) and centrifugation (13 000 g for 30 min) followed by filtration (0.2 μm pore size). Peptide microarray analysis was performed by PEPperPRINT (Heidelberg, Germany). An ArcA microarray was generated with 409 different peptides of ArcA, and each peptide contained 15 amino acids with a peptide-peptide overlap of 14 amino acids. After blocking and washing, the array was incubated with surface extracts (100 µg/ml) isolated from P. gingivalis 33277, the 0806 mutant, or the ragB mutant. P. gingivalis surface proteins bound on the ArcA array were detected using anti-P. gingivalis antibodies and sheep anti-rabbit IgG (H + L) DyLight680. The arrays were analyzed with a LI-COR Odyssey Imaging System.
Peptide synthesis and activity
Peptides were synthesized by PEPperPRINT and Biomatik (Delaware) and purified with High Performance Liquid Chromatography (HPLC) at ≥90% purity. Peptides were resuspended in nuclease/proteinase-free PBS, aliquoted, and stored at −20 °C. Inhibitory activity of peptides were determined as described16 with slight modification. Peptides were mixed with 5 × 105 cells of P. gingivalis, spotted onto a TSB blood agar plate, and cultured for 60 h anaerobically. The expression levels of fimbrial and gingipain mRNA or proteins were measured using qRT-PCR or western blot analyses.
Western Blot Analysis
P. gingivalis cells were lysed using BugBuster® Protein Extraction Reagent (EMD Millipore, Darmstadt, Germany), and the protein concentration determined using a Bio-Rad protein assay (Bio-Rad). Lysates (0.5 µg) were separated by 12% sodium SDS-PAGE, and transferred to nitrocellulose membranes (Invitrogen, Carlsbad, CA) with a Mini Transblot Electrophoretic transfer cell (Bio-Rad) at 100 V for 1 h. The membrane was blocked with 3% BSA in PBS for 1 h and incubated with polyclonal anti-FimA, anti-Mfa1, anti-HGP44 (a C-terminal adhesin domain of gingipains), or anti-PGN-0128 antibodies diluted 1:1,000 for 1 h. After washing with PBS, the membrane was incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibodies for 1 h. The proteins were visualized using enhanced chemiluminescence (GE Healthcare Bio-Sciences Corp, Pittsburgh, PA) and measured using a semi-quantitative Western blot technique with ImageJ software (NIH, Bethesda, Maryland).
Transmission electron microscopy
P. gingivalis cells were grown on TSB blood plates for 48 h with or without peptide4. Bacterial cells were collected and resuspended in PBS. 20 µl of bacterial suspension were applied to a Formvar-coated copper grid (200 mesh, Electron Microscopy Sciences, PA) and air dried. The bacterial cells were then negatively stained with 0.5% ammonium molybdate for 4 min and observed with a transmission electron microscope (Philips CM-12, Portland, OR) operated at 80 kV.
Statistical analyses
A student’s t-test was used to determine the statistical significance of differences in gene expression profiles and growth rates of P. gingivalis strains. A p < 0.05 was considered significant.
Electronic supplementary material
Oligonucleotide primers used in this study
Acknowledgements
This work was supported by NIH grants DE022428 and 025332 (HX) and DE012505 and DE023193 (RJL) from NIDCR, and by MD007593 and MD007586 from NIMHD. The project described was also supported by NCRR grant UL1 RR024975, which is now mediated by the NCATS (2 UL1 TR000445). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank the Meharry Office for Scientific Editing and Publications for scientific editing support (S21MD000104).
Author Contributions
H.X. designed the project. M.H.H. and H.X. performed the experiments. H.X. and R.J.L. analyzed the data. H.X. and R.J.L. wrote the main manuscript text. All of the authors contributed to the discussion and provided comments on the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01551-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kassebaum NJ, et al. Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. Journal of dental research. 2014;93:1045–1053. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt B, Research S. & Therapy Committee of the American Academy of, P. Position paper: epidemiology of periodontal diseases. Journal of periodontology. 2005;76:1406–1419. doi: 10.1902/jop.2005.76.8.1406. [DOI] [PubMed] [Google Scholar]
- 3.Petersen PE, Ogawa H. The global burden of periodontal disease: towards integration with chronic disease prevention and control. Periodontology. 2012;2000(60):15–39. doi: 10.1111/j.1600-0757.2011.00425.x. [DOI] [PubMed] [Google Scholar]
- 4.Eke PI, et al. Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 to 2012. Journal of periodontology. 2015;86:611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nature reviews. Immunology. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linden GJ, Lyons A, Scannapieco FA. Periodontal systemic associations: review of the evidence. Journal of periodontology. 2013;84:S8–S19. doi: 10.1902/jop.2013.1340010. [DOI] [PubMed] [Google Scholar]
- 7.Kumar PS. Oral microbiota and systemic disease. Anaerobe. 2013;24:90–93. doi: 10.1016/j.anaerobe.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Molecular oral microbiology. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell host & microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. European journal of immunology. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nature reviews. Microbiology. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. Journal of dental research. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page RC, et al. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral microbiology and immunology. 2007;22:162–168. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 14.Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infection and immunity. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie H, Hong J, Sharma A, Wang BY. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. Journal of periodontal research. 2012;47:578–583. doi: 10.1111/j.1600-0765.2012.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie H, Lin X, Wang BY, Wu J, Lamont RJ. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology. 2007;153:3228–3234. doi: 10.1099/mic.0.2007/009050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christopher AB, Arndt A, Cugini C, Davey ME. A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology. 2010;156:3469–3477. doi: 10.1099/mic.0.042671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS microbiology letters. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 19.Curran TM, Lieou J, Marquis RE. Arginine deiminase system and acid adaptation of oral streptococci. Appl Environ Microbiol. 1995;61:4494–4496. doi: 10.1128/aem.61.12.4494-4496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin X, Lamont RJ, Wu J, Xie H. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. Journal of bacteriology. 2008;190:4367–4371. doi: 10.1128/JB.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. Journal of clinical microbiology. 2009;47:3902–3906. doi: 10.1128/JCM.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Lin X, Xie H. Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS microbiology letters. 2007;271:214–221. doi: 10.1111/j.1574-6968.2007.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo A, et al. FimR and FimS: biofilm formation and gene expression in Porphyromonas gingivalis. Journal of bacteriology. 2010;192:1332–1343. doi: 10.1128/JB.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajishengallis G, McIntosh ML, Nishiyama SI, Yoshimura F. Mechanism and implications of CXCR4-mediated integrin activation by Porphyromonas gingivalis. Molecular oral microbiology. 2013;28:239–249. doi: 10.1111/omi.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiology and molecular biology reviews: MMBR. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral microbiology and immunology. 2000;15:341–349. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- 27.Xie H. Biogenesis and function of Porphyromonas gingivalis outer membrane vesicles. Future microbiology. 2015;10:1517–1527. doi: 10.2217/fmb.15.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulbourne PA, Ellen RP. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol. 1991;173:5266–5274. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamont RJ, Bevan CA, Gil S, Persson RE, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302X.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 30.Amano A, et al. Prophyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 31.Lin X, Wu J, Xie H. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infection and immunity. 2006;74:6011–6015. doi: 10.1128/IAI.00797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberg A, Belton CM, Park Y, Lamont RJ. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-Mediated Clearance of Porphyromonas gingivalis and Negates Its Virulence In Vivo. J Immunol. 2007;179:2359–2367. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, et al. Fimbrial proteins of porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. Journal of immunology. 2007;179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- 35.Malek R, et al. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuboniwa M, et al. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC microbiology. 2009;9:105. doi: 10.1186/1471-2180-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. Journal of oral microbiology. 2013;5:20265. doi: 10.3402/jom.v5i0.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda K, et al. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 2004;6:1163–1170. doi: 10.1016/j.micinf.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Park Y, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect Immun. 2008;76:3273–3280. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki Y, Yoshimura F, Takahashi K, Tani H, Suzuki T. Detection of fimbriae and fimbrial antigens on the oral anaerobe Bacteroides gingivalis by negative staining and serological methods. J Gen Microbiol. 1988;134:2713–2720. doi: 10.1099/00221287-134-10-2713. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki Y, Yoshimura F, Tani H, Suzuki T. Fimbriae from the oral anaerobe Bacteroides gingivalis: a screening of clinical isolates from various places. Adv Dent Res. 1988;2:301–303. doi: 10.1177/08959374880020021601. [DOI] [PubMed] [Google Scholar]
- 43.Noiri Y, Li L, Yoshimura F, Ebisu S. Localization of Porphyromonas gingivalis-carrying fimbriae in situ in human periodontal pockets. J Dent Res. 2004;83:941–945. doi: 10.1177/154405910408301210. [DOI] [PubMed] [Google Scholar]
- 44.Curtis MA, et al. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. Journal of periodontal research. 1999;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 45.Mikolajczyk-Pawlinska J, et al. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biological chemistry. 1998;379:205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- 46.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology. 2000;2000(24):153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 47.Kadowaki T, et al. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. The Journal of biological chemistry. 1998;273:29072–29076. doi: 10.1074/jbc.273.44.29072. [DOI] [PubMed] [Google Scholar]
- 48.Shi Y, et al. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. The Journal of biological chemistry. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 49.Klarstrom Engstrom K, Khalaf H, Kalvegren H, Bengtsson T. The role of Porphyromonas gingivalis gingipains in platelet activation and innate immune modulation. Molecular oral microbiology. 2015;30:62–73. doi: 10.1111/omi.12067. [DOI] [PubMed] [Google Scholar]
- 50.Barth K, Genco CA. Microbial Degradation of Cellular Kinases Impairs Innate Immune Signaling and Paracrine TNFalpha Responses. Scientific reports. 2016;6:34656. doi: 10.1038/srep34656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu J, Xie H. Role of arginine deiminase of Streptococcus cristatus in Porphyromonas gingivalis colonization. Antimicrobial agents and chemotherapy. 2010;54:4694–4698. doi: 10.1128/AAC.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cugini C, Stephens DN, Nguyen D, Kantarci A, Davey ME. Arginine deiminase inhibits Porphyromonas gingivalis surface attachment. Microbiology. 2013;159:275–285. doi: 10.1099/mic.0.062695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hajishengallis G, Lamont RJ. Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends in microbiology. 2016;24:477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zschiedrich CP, Keidel V, Szurmant H. Molecular Mechanisms of Two-Component Signal Transduction. Journal of molecular biology. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishikawa K, Yoshimura F, Duncan MJ. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Molecular microbiology. 2004;54:546–560. doi: 10.1111/j.1365-2958.2004.04291.x. [DOI] [PubMed] [Google Scholar]
- 56.Hayashi J, Nishikawa K, Hirano R, Noguchi T, Yoshimura F. Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis. Microbiology and immunology. 2000;44:279–282. doi: 10.1111/j.1348-0421.2000.tb02496.x. [DOI] [PubMed] [Google Scholar]
- 57.Wirth C, Meyer-Klaucke W, Pattus F, Cobessi D. From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor. J Mol Biol. 2007;368:398–406. doi: 10.1016/j.jmb.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 58.Goulas T, et al. Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis. Molecular oral microbiology. 2015;31:472–485. doi: 10.1111/omi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hutcherson JA, et al. Porphyromonas gingivalis RagB is a proinflammatory signal transducer and activator of transcription 4 agonist. Molecular oral microbiology. 2015;30:242–252. doi: 10.1111/omi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagano K, et al. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. Journal of medical microbiology. 2007;56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- 61.Tokuda M, Chen W, Karunakaran T, Kuramitsu HK. Regulation of protease expression in Porphyromonas gingivalis. Infection and immunity. 1998;66:5232–5237. doi: 10.1128/iai.66.11.5232-5237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie H, et al. Intergeneric communication in dental plaque biofilms. Journal of bacteriology. 2000;182:7067–7069. doi: 10.1128/JB.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J, Lin X, Xie H. OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis. FEMS microbiology letters. 2008;282:188–195. doi: 10.1111/j.1574-6968.2008.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu J, Lin X, Xie H. Regulation of hemin binding proteins by a novel transcriptional activator in Porphyromonas gingivalis. Journal of bacteriology. 2009;191:115–122. doi: 10.1128/JB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic acids research. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used in this study


