Abstract
ChHP1BP3, a chromatin complex-related protein known with dynamic features, was identified as a ChHsp70 promoter-associated factor in Crassostrea hongkongensis by DNA-affinity purification and mass spectrometry analysis. Direct interaction between purified ChHP1BP3 and the ChHsp70 promoter region was demonstrated using EMSA. ChHp1bp3 depletion led to clear enhancements in ChHsp70 mRNA expression in C. hongkongensis hemocytes. However, ChHp1bp3 overexpression in heterologous HEK293T cells correlated with fluctuations in ChHsp70 transcription. Quantitative RT-PCR analysis showed that both ChHsp70 and ChHp1bp3 transcription were responsive to external physical/chemical stresses by heat, CdCl2 and NP. This indicated a plausible correlation between ChHsp70 and ChHp1bp3 in the stress-induced genetic regulatory pathway. While, the distinctive ChHp1bp3 expression patterns upon physical and chemical stresses suggest that the mechanisms that mediate ChHp1bp3 induction might be stress-specific. This study discovered a novel role for HP1BP3 as a negative regulator in controlling Hsp70 transcription in C. hongkongensis, and contributed to better understanding the complex regulatory mechanisms governing Hsp70 transcription.
Introduction
Hsp70s are a group of stress-inducible heat shock proteins that are approximately 70 kDa1. They are highly conserved molecular chaperones that are universally present in all cellular species and play critical roles in assisting the three-dimensional folding of newly synthesized proteins and restoring disordered proteins caused by a variety of intense stresses, including heat shock, heavy metals, and toxic chemicals2, 3. Hsp70 is the largest and currently the most extensively studied stress protein family. Owing to its chaperone activity as well as its importance for allowing cells to cope with stress insults, Hsp70 has been used for many applications in various disciplines, including protecting against and therapeutic treatments for cardiovascular diseases, cancer prognosis, aiding in transmembrane protein transport, inhibiting apoptosis, and biomonitoring4–6. To further explore potential applications of Hsp70s, a comprehensive understanding of the mechanisms governing their response to various stresses is demanded.
Hsp70 activiation has been primarily investigated at the transcriptional level. Numerous studies have shown that Hsp70 transcription undergoes integrated regulation from both general and gene-specific regulators, including GAGA factor, TBP (TATA-binding protein), STAT (signal transducer and activator of transcription), and HSF-1 (heat shock factor 1)7–9. Heat-shock activation of Hsp70 is also believed to be regulated by promoter-proximal pausing, a rate-limiting step prior to productive transcription10, 11. The stall is regarded as a regulatory checkpoint; the transition of promoter-proximal Pol II into productive elongation is triggered by HSF binding signals to allow rapid and synchronous activation of Hsp70 12. The overall dynamic architecture of chromatin is critical in regulating gene expression levels, since increased chromatin dynamics facilitate factor recruitment to DNA, thus promoting activation13–15. Multiple interdependent regulators such as the TFIID and GAGA factor have been shown to manipulate nucleosome pattern and subsequently affect Hsp70 promoter architecture, most likely through associations with NURF (nucleosome remodeling factor) and dFACT (facilitates chromatin transcription)10, 16, 17. Although heat shock response is regarded as the gold standard for investigating gene regulation, the full details of its induction pathway remain incomplete.
In aquatic animals, the evaluation of stress protein expression levels has been advocated as a distinctive environmental monitoring tool18. Hsp70 is thought to be favorable for biomonitoring owing to the extreme sensitivity of the Hsp70 gene per se to various aquatic pollutants19. Oysters are considered as ideal monitoring hosts due to numerous advantageous attributes of the animals, such as sessile living, tolerance to various pressures, abundance, long lifespan, and sufficient tissue mass for analysis. Hsp70 expression in oysters has been reported to be responsive to a variety of environmental stimuli, including heat shock and various inorganic (heavy metals, salinity etc.) and organic (antifouling biocides) chemicals20–24. Despite its environmental and biological importance, the mechanisms controlling Hsp70 transcription induction in oysters remain largely unknown, although eight HSF-1 isoforms have been identified to be inducible by hypoxia in Pacific oysters25. In previous studies, we have isolated ChHsp70 promoter-bound proteins from nuclear extracts prepared from Crassostrea hongkongensis either with or without stress treatment26. However since different protein patterns were shown in DNA-affinity purification upon various stressed conditions, it was proposed that the differences are caused either by the distinct regulatory proteins showing affinity to the promoter or by dynamic interactions of specific factors from the same batch of regulators. In this study, heterochromatin protein 1 binding protein 3 (HP1BP3) was isolated after a prolonged incubation by DNA-affinity purification. A specific interaction between ChHP1BP3 and the ChHsp70 promoter region was discovered in vitro and a negative regulatory function of ChHP1BP3 on ChHsp70 transcription was demonstrated in its native host. The different ChHp1bp3 induction patterns shown by RT-PCR indicated that different mechanisms govern ChHP1BP3 induction upon physical and chemical stresses.
Results
Identification of proteins showing dynamic affinity to the ChHsp70 promoter
Our previous results of DNA-affinity purification demonstrated distinctive patterns for regulators showing affinity to the ChHsp70 promoter region under stressed or non-stressed conditions. To discover the possible involvement of putative regulators bearing dynamic binding activities, DNA affinity purification was performed with either 15 min or 30 min protein-DNA incubation times. The results are shown in Supplementary Fig. 1. When comparing the banding patterns of the two groups, most of the proteins were consistently isolated in either the control or stressed sample lanes. However, there were some protein bands that specifically appeared in the 30 min incubation stressed lanes, including one that was noticeably visible right above the 45 kDa marker. The nature of the latter band was revealed by mass spectrometry as HP1BP3 (Supplementary Fig. 2). The time- and stress- dependent affinity of HP1BP3 to the ChHsp70 promoter was of interest and was thus selected for further study.
Characterization of the ChHp1bp3 cDNA
The ChHp1bp3 cDNA is 1920 bp in length and contains a 1434 bp coding region as well as 177 bp and 309 bp 5′ and 3′ untranslated regions, respectively (Fig. 1A). The peptide sequence is composed of 477 amino acids, with a molecular weight of 53.7 kDa and an isoelectronic point of 9.19. A globular domain, which has been previously reported to be structurally related to that of histone H1 and interact with nucleosomal DNA27, was discovered at the C-terminal region of the ChHP1BP3 peptide sequence (Fig. 1A). The amino acid sequence of the globular domain was then subjected to three dimensional structure prediction using the online SWISS-MODEL software (http://swissmodel.expasy.org/), and the deduced protein model revealed the presence of three α–helices and two β–sheets (Fig. 1B).
Figure 1.
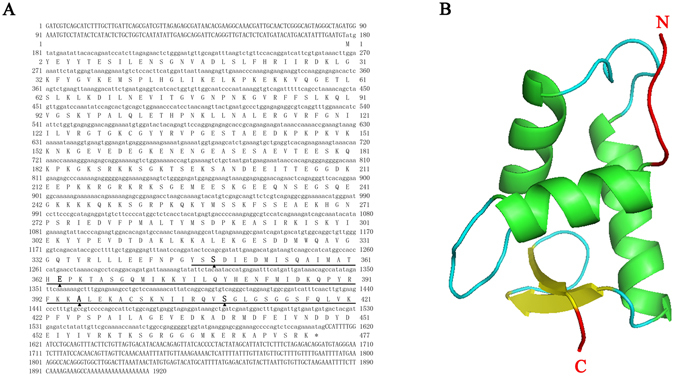
(A) The cDNA sequence and deduced amino acid sequence of ChHP1BP3 in C. hongkongensis. The nucleotides and amino acids are numbered along both margins. The region of globular domain is underlined. The putative DNA binding sites are marked by triangles. The translation stop codon is indicated with an asterisk (*). (B) Three dimensional structure of the globular domain of ChHP1BP3.
ChHP1BP3 directly interacts with the ChHsp70 promoter region in vitro
The ability of ChHP1BP3 to directly interact with the ChHsp70 promoter region was tested via EMSA using a FITC-labeled ChHsp70 promoter region and His6-ChHP1BP3 purified from Escherichia coli. The results in Fig. 2 clearly demonstrated that the addition of His6-ChHP1BP3 delayed the electrophoretic mobility of the ChHsp70 promoter region in a concentration dependent manner (Fig. 2, lanes 2 to 6). A competition assay was performed by introducing in excess amounts of unlabeled ChHsp70 promoter to the reaction mixture, which successfully out-competed the specific interactions, thus eliminating the presence of the shifted bands (Fig. 2, lanes 7, 9 and 10). However, when an irrelevant DNA fragment (+370 to +763 relative to the translational start on the ChHp1bp3 coding sequence) was introduced, the competition was largely compromised (Fig. 2, lane 8). These results indicated a specific interaction between His6-ChHP1BP3 and the ChHsp70 promoter region.
Figure 2.
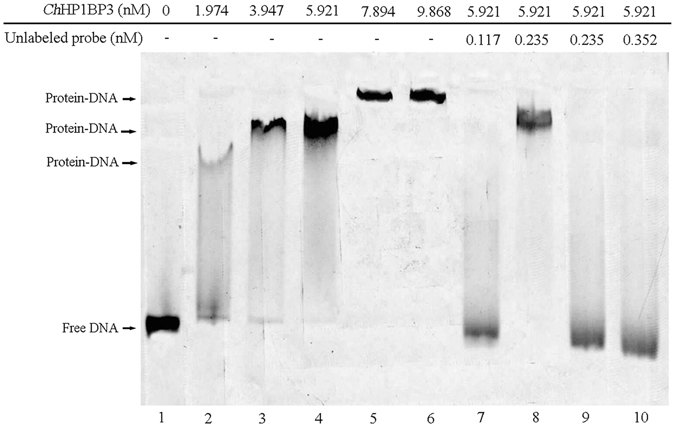
Electrophoretic mobility shift assay of the ChHsp70 promoter with purified His6-ChHP1BP3. Fifty picomoles of FITC-labeled ChHsp70 promoter was added to all the lanes. The band positions of the shifted DNA-protein complexes and free DNA are indicated by arrows.
ChHp1bp3 knockdown enhanced ChHsp70 transcription in C. hongkongensis hemocytes
To discover the regulatory functions of ChHP1BP3 on ChHsp70 transcription in either basal or heat shocked condition, ChHsp70 mRNA expression was measured by RT-RCR in both wild-type and siRNA-treated hemocytes of C. hongkongensis as described in the Materials and methods. The results are shown in Fig. 3. ChHp1Bp3 mRNA expression was significantly depleted at all the tested time points either with or without heat shock treatment when the cells were treated with siRNA, validating the successful knockdown of ChHp1bp3 expression. ChHsp70 basal transcription was clearly enhanced particularly at 24 and 48 h upon ChHp1bp3 depletion. Heat induced ChHsp70 expression was shown to be more sensitive to ChHp1bp3 levels since more significant enhancement was observed to the already induced ChHsp70 transcription when ChHp1bp3 was depleted, and the effective impact was visible nearly at all the tested time points (3 h, 6 h, 12 h, 24 h and 48 h). These results demonstrate a negative role for ChHP1BP3 in regulating ChHsp70 transcription under either basal or heat-stressed conditions in native C. hongkongensis hosts. Noticeably, an increasing expression profile was presented by the basal ChHsp70 transcription until 24 h. This induction is likely due to the experienced stress caused by altered biological context during the in vitro culture of the hemocytes.
Figure 3.
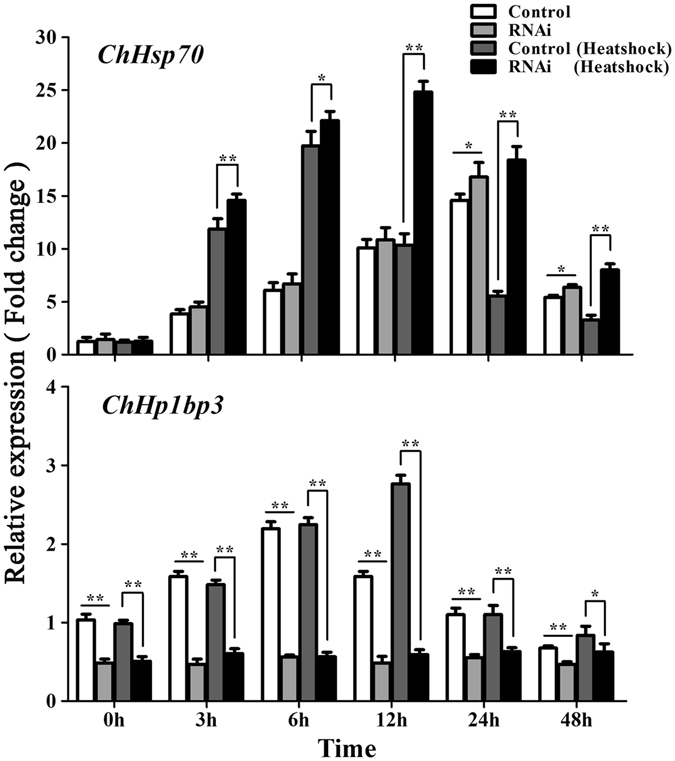
ChHsp70 and ChHp1bp3 expression levels by RT-PCR after the addition of either control RNA (Control) or siRNA (RNAi). All values are expressed as the means ± SD (n = 3). Asterisks above the bars indicate that values are significantly different from the individual controls (*P < 0.05 and **P < 0.01).
ChHP1BP3 overexpression manipulates ChHsp70 expression in heterologous host HEK293T cells
A luciferase reporter assay was conducted in HEK293 cells to discover the regulatory effects of overexpressed ChHP1BP3 on ChHsp70 expression. The results are shown in Fig. 4. ChHsp70 promoter activity was notably enhanced by overexpressed ChHP1BP3 at the first sampling point (24 h post transfection), indicating an initial positive regulatory role for ChHP1BP3 on ChHsp70 transcription. However, this activated state was soon overturned since a clear decrease in ChHsp70 promoter activity was observed at 3 h and 6 h. ChHsp70 transcription recovered from 12 h until it reached a level comparable to the control at 18 h. Interestingly, promoter activation was once again shown at 24 h, whereas the induced state was totally compromised from 48 h onwards. These results demonstrated that constitutively expressed ChHP1BP3 caused ChHsp70 expression to fluctuate, meaning that ChHP1BP3 regulated ChHsp70 expression in an oscillating manner. This suggests that a complex mechanism underlies ChHP1BP3 controlled ChHsp70 transcription in the heterologous HEK293T host.
Figure 4.
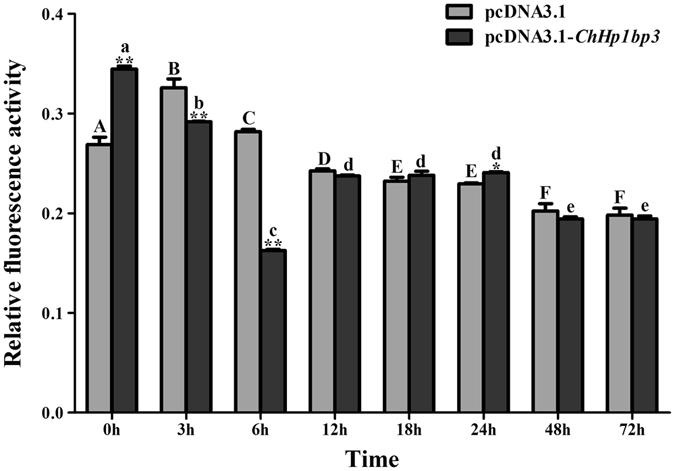
Luciferase activity driven by the ChHsp70 promoter in HEK293T cells following the addition of pcDNA3.1-ChHp1bp3 and pcDNA3.1. All values are presented as the means ± SD (n = 3). Asterisks above the bars indicate the values are significantly different from the individual controls. (*P < 0.05 and **P < 0.01). Bars on the same group with different letters are statistically significant from one another.
ChHsp70 and ChHp1bp3 transcription profiles upon various stresses
In an attempt to reveal correlations between ChHsp70 and ChHp1bp3 transcription in response to external stresses, quantitative real-time PCR was employed to determine ChHsp70 and ChHp1bp3 mRNA expression in oysters gills treated with heat, CdCl2 or 300 μg/L NP, which has been previously determined as a suitable concentration for induction26. The results are shown in Fig. 5. ChHsp70 transcription was notably induced upon stresses, with the highest peaks induced by heat, Cd and NP at 3 h, 2 d, and 4 d, respectively. After that, ChHsp70 expression declined gradually for all three treatments until it reached a level comparable to the controls. ChHp1bp3 transcription was induced by all the tested stresses. However, a different induction pattern was observed for CdCl2 and NP treatment since two different induction peaks were shown. The two peaks were accumulated at the time points of 24 h and 8 d, which either preceded or followed peak induced ChHsp70 transcription, respectively. Upon heat stress, a single ChHp1bp3 induction peak was shown at 12 h; this peak was postponed for 9 h compared to the ChHsp70 peak. These results demonstrated that both ChHsp70 and ChHp1bp3 were transcriptionally responsive to external stresses, though in distinctive patterns. This indicated a plausible correlation between the two genes in a genetic regulatory pathway induced by stresses.
Figure 5.
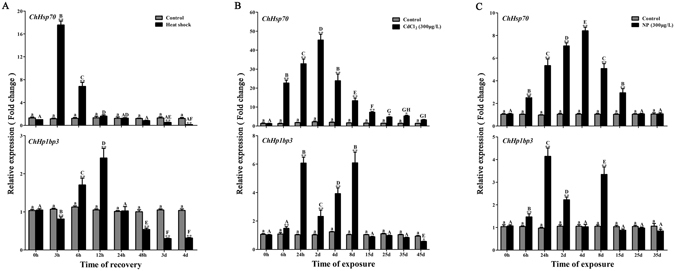
ChHsp70 and ChHp1bp3 expression levels by RT-PCR after (A) heat, (B) CdCl2 or (C) NP stress. All values are expressed as the means ± SD (n = 3). Asterisks above the bars indicate the values are significantly different from the individual controls. (*P < 0.05 and **P < 0.01). Bars on the same group with different letters are statistically significant from one another.
Discussion
Heterochromatin, a highly compacted structure of higher-order chromatin, is formed dynamically and alternately during interphase, during which most chromatin-dependent biological events occur. The non-histone proteins that temporally interact with chromatin during interphase are considered important for elucidating a variety of host cell functions28–30. HP1BP3 has been revealed to be a chromatin complex associated protein with strikingly dynamic features during the distinct stages of interphase, demonstrating that it plays critical and diverse roles in various cellular processes, including heterochromatin organization, nucleosome assembly, cell proliferation regulation, cellular response to hypoxia, and DNA-templated transcription regulation31–33. In the present paper, we reported the characterization of an HP1BP3 homologue ChHP1BP3 in C. hongkongensis. It directly interacts with the ChHsp70 promoter in vitro, and its depletion was shown to activate ChHsp70 transcription in the native host, and its overexpression caused ChHsp70 transcription to fluctuate in the heterologous host HEK293T.
Analysis of the ChHP1BP3 amino acid sequence revealed the presence of a globular “Histone H-1 like” domain (Fig. 1A), which is present in other HP1BP3 homologues27. This domain was reported to confer DNA binding characteristics34. Indeed, our EMSA results demonstrated a clear mobility shift by the ChHsp70 promoter upon the addition of ChHP1BP3. However, the shifted bands migrated more slowly when His6-ChHP1BP3 was introduced at higher concentrations (Fig. 2). This result raised the possibility of the presence of multiple ChHP1BP3 binding sites in the ChHsp70 promoter region or the formation of His6-ChHP1BP3 multimers that interact with a core binding site.
RNAi analysis demonstrated that ChHsp70 mRNA expressions was clearly induced by ChHP1BP3 depletion under either basal or heat-stressed conditions compared to the control (Fig. 3). Taken together with the above EMSA data, these results suggest that ChHP1BP3 negatively regulates ChHsp70 transcription via direct interaction with its promoter region. HP1BP3 has been previously characterized as a histone H1 related protein that associates with chromatin by binding to nucleosome surfaces and interacting with nucleosomal DNA27. These proteins are considered highly important for transcription regulation since they promote chromatin high-order structure, which has been widely shown to restrict the transcriptional machinery access to the template DNA, thus fundamentally determining host cell transcriptional activities. Indeed, some chromatin structure maintaining proteins have been shown to act as transcriptional regulators to control the expression of specific genes or gene clusters. For example, Butchko et al. reported the global regulatory role of LaeA on the expression of multiple secondary metabolite gene clusters by altering heterochromatin structure in cooperation with the velvet complex in fungi35. To confirm whether ChHP1BP3 affects C. hongkongensis chromatin structure, chromatin was extracted from both control and ChHp1bp3-depleted oyster hemocytes and subjected to partial MNase digestion using the methods described in the Supplementary data. The digestion patterns of the chromatin extracted from the hemocytes 24 h post ChHp1bp3 knockdown are shown as a representative in Supplementary Fig. 3. Compared to the control cells, the ChHp1bp3-depleted chromatin samples were always more susceptible to MNase digestion in either non-stressed or heat-stressed conditions. This suggests that ChHP1BP3 may indeed be required for the formation of higher-order chromatin structure. Thus, ChHP1BP3 depletion is expected to unpack heterochromatin and disrupt its structure, leading to the regulation of numerous DNA-dependent events including transcription. Therefore, the discovered negative regulatory role of ChHP1BP3 on ChHsp70 expression might also be a result of heterochromatin structure alterations. Taken together with the above EMSA data, it is reasonable to propose that the observed negative regulation on ChHsp70 transcription might be due to a coordinated contribution from ChHP1BP3 activities on both pathway-specific regulation and global chromatin structure change. The regulatory roles of HP1BP3 on the expression of various molecular chaperones have been previously reported in 293T by iTRAQ analysis. Though inconsistently, Hsp70 was found to be positively regulated31. This discrepancy suggests that HP1BP3 may interact in a host-dependent manner for controlling Hsp70 expression, which plausibly involves distinct regulatory mechanisms. Considering that several general (GAGA factor, TBP, etc.) and gene-specific (HSF-1, GAF, etc.) regulators have been previously discovered to control Hsp70 transcription either cooperatively or competitively, determining plausible partner regulators that interplay with ChHP1BP3 on the ChHsp70 promoter region is highly relevant to better understand mechanisms that involve ChHP1BP3 and govern ChHsp70 transcription.
Since ChHP1BP3 depletion had a positive effect on ChHsp70 expression, it was thought that conversely, ChHP1BP3 overexpression might correlate with a repressive phenotype. However, while clear repressions were indeed shown at various sampling points upon ChHP1BP3 overexpression by the luciferase reporter assay, an overall fluctuating regulatory effect was demonstrated in the heterologous HEK293T cells. It has been previously noted that the association of HP1BP3 with the chromatin complex is strikingly dynamic31. This typical feature allowed us to hypothesize that the unusual fluctuations might be due to intermittent regulation by the overexpressed ChHP1BP3 that dynamically interacted with the ChHsp70 promoter. Fortunately, this notion was supported by our DNA affinity purification result, which demonstrated a time-dependent affinity between ChHP1BP3 and the ChHsp70 promoter (Supplementary Fig. 1). The reason for the strong induction of ChHsp70 expression at the first time point was unknown, but considering that the expression of hundreds of genes could be affected by HP1BP3 depletion, it is reasonable to expect an initial emergence of any activator induced by overexpressed ChHP1BP3 in the heterologous host cells.
Hsp70 has been widely acknowledged as a valid bioindicator to signify environmental stresses since its expression is extremely sensitive to various stimulants19. In our previous studies, the expression of ChHsp70 of C. hongkongensis has been demonstrated to be significantly induced by both heat and Cd stress26. Consistent with these data, similar induction profiles demonstrating induction accumulation, peaking, a gradual decline and recovery were shown by RT-PCR analysis upon the two stresses as well as by NP treatment. NP is a notorious endocrine disruptor that is frequently found but not readily biodegradable in aquatic environments, meaning that it poses continuous hazards to aquatic animals and human health36. ChHp1bp3 transcription was also induced by all three stresses, though different induction patterns were observed. The second accumulated ChHp1bp3 expression peaks induced by Cd and NP together with the single heat-induced peak appeared at points that were slightly delayed when compared with the corresponding ChHsp70 peaks. Considering the previously discovered negative regulatory function, these late accumulated ChHp1bp3 peaks might be very likely to contribute to the recovery process for promoting ChHsp70 expression back to its original level. An additional earlier ChHp1bp3 induction peak was present upon either Cd or NP stress compared with the ChHsp70 peak. It is currently unknown whether this early peak correlates directly with ChHsp70 induction based on our current data. However, its absence upon the heat stressed condition indicates that different mechanisms govern ChHp1bp3 induction following physical and chemical stress.
In summary, we demonstrated a direct interaction between ChHP1BP3 and the ChHsp70 promoter region, and a clear negative regulatory role for ChHP1BP3 on ChHsp70 transcription in C. hongkongensis. The differences in the ChHp1bp3 induction patterns due to various external stresses as well as the discrepancies in the ChHP1BP3 regulatory effects on ChHsp70 transcription in a heterologous HEK293T host cells suggest that HP1BP3 mediated regulation is rather complex and likely involves the cooperation of additional factors. This study provided novel insights into HP1BP3 mediated transcriptional regulation and presents the first evidence of a specific HP1BP3 regulatory target gene.
Materials and Methods
Oyster collection, maintenance and treatment
Oysters (C. hongkongensis) were collected from a farm located in the Yangxi suburb, in Guangdong, China. All of the sampled oysters were one-year old and had a mean shell length of 9 ± 1 cm. The oysters were transferred to Jinan University within 3 h and allowed to acclimate for 2 weeks at 24 ± 1 °C in aerated artificial seawater with 16~18‰ salinity. The oysters were fed algal slurry once a day under a natural photoperiod. The seawater was renewed every other day. After the preliminary acclimation, the oysters were randomly picked and evenly transferred to 25 L plastic tanks filled with artificial sea water supplemented with either CdCl2 or nonylphenol (NP) at 300 µg/L; the tanks without any supplements were used as the control. The seawater was renewed every other day with the initial CdCl2 or NP concentrations. For thermal stress, the acclimated oysters were transferred to a 37 °C water bath and maintained for 1 h, followed by recovery under the original conditions for 3 h.
DNA affinity purification
DNA affinity purification was performed as previously described26 but with slight modifications. Briefly, biotin-labeled ChHSP70 promoter was amplified from C. hongkongensis genomic DNA using a primer pair of P70-promoter up and P70-promoter down; the PCR product was subsequently immobilized to streptavidin-coated magnetic beads (Cat. No. 88816; Thermo) in DNA-binding buffer (10 mM Tris-HCl, 1 mM EDTA, and 2 M NaCl, pH 7.5) in accordance with the manufacturer’s instructions. After washing, the DNA coated beads were resuspended in protein-binding buffer (20 mM Tris-HCl, 4.5 mM EDTA, 60 mM NaCl, 10 mM HEPES, 5 mM CaCl2, 50 mM KCl, 9% sucrose (w/v), and 12% glycerol, pH 7.5). Then nuclear protein extracts that were previously prepared from gills of the thermal/Cd stressed and the control oysters26 were individually introduced, and the reaction mixture was incubated at room temperature for either 15 min or 30 min. A second washing step was performed using protein binding buffer, and then the proteins showing affinity to the ChHsp70 promoter were eluted with elution buffer (25 mM Tris-HCl and 200 mM NaCl, pH 7.5). The eluted proteins were then analyzed by SDS-PAGE with Coomassie Brilliant Blue staining. The individual bands of interest were identified by mass spectrometry as described in the supplementary materials.
Full-length ChHp1bp3 cDNA sequencing
Total RNA was extracted from oysters gills using the Trizol reagent (Cat. No. 15596-026; Invitrogen) following the manufacturer’s instructions. The integrity and concentration/purity of the isolated RNAs were subsequently examined by agarose gel electrophoresis and spectrometry analysis, respectively. First strand cDNA was synthesized from two micrograms of total RNA at 55 °C for 30 min with 200 U M-MLV Reverse Transcriptase (Promega) using oligo (dT) primer. The resulting cDNA pool was then applied as template to amplify the ChHp1bp3 coding sequence with the HP1BP3-up and HP1BP3-down PCR primers (Table 1). The amplified products were cloned into the pMD18-T (TaKaRa) vector, and the inserted ChHp1bp3 coding sequence was revealed by nucleotide sequencing (Liuhe BGI).
Table 1.
Synthetic oligonucleotides used in this study.
| Primer | Sequence 5′ → 3′ | Position(s) | Purpose |
|---|---|---|---|
| Oligo(dT)18 | TTTTTTTTTTTTTTTTT | Reverse transcription | |
| P70-promoter up | TTCCATGCACTCTAGCTAAG | −304–285 | Amplification of ChHsp70 promoter fragment for DNA-affinity purification and EMSA |
| P70-promoter down | GAATCGTAGTCTAGTTGACTC | +40–+60 | |
| HP1BP3-up | ACATATTTGAATGTATGTATGAAT | −14–+10 | Amplification of ChHp1bp3 coding sequence |
| HP1BP3-down | ACTGTTGTGGATAAAGATTCCCT | +1528–+1550 | |
| UP | CGGCAGTGGTATCAACGCAGAGTAC | Universal primer used with 3GSP2 and 5GSP2 | |
| UP(dG) | CGGCAGTGGTATCAACGCAGAGTAC(G)10 | 5′ RACE PCR | |
| 5GSP1 | TATTTCTTCATCATTAGCA | +594–+612 | |
| 5GSP2 | TTCCCATGTTTTTCCGCCTCTGAC | +789–+812 | |
| UP (dT) | CGGCAGTGGTATCAACGCAGAGTAC(T)18 | 3′ RACE PCR | |
| 3GSP1 | TACATAGAAAAGTATTAC | +898–+915 | |
| 3GSP2 | TCAGATGACATGTGGCAGGCTGTT | +967–+990 | |
| HP1BP3-F-NdeI | GGGAATTCCATATGTATGAATAT | +1–+12 | Amplification of ChHp1bp3 for protein expression |
| HP1BP3-R-XhoI | CCGCTCGAGTTTTCTGGAGAC | +1420–+1431 | |
| HP1BP3-E-KpnI | ATAAAAGGTACCATGTATGAATATTACACAGAATCCATC | +1–+27 | Amplification of ChHp1bp3 for overexpression |
| HP1BP3-E-XhoI | ATAAAACTCGAGCTATTTTCTGGAGACTGGGGC | +1414–+1434 | |
| Actin-Up | CTGTGCTACGTTGCCCTGGACTT | +701–+723 | RT-PCR |
| Actin-Down | TGGGCACCTGAATCGCTCGTT | +829–+849 | |
| RT-HP1BP3-Up | AAACAGCCTCAGGACAGATG | +1094–+1113 | RT-PCR |
| RT-HP1BP3-Down | CAAGTTGAAATGATCCGCCAC | +1238–+1258 | |
| RT-HSP70-Up | GCCAAACTACATCAGAACGGGTC | −1860–1838 | RT-PCR |
| RT-HSP70-Down | TCCATCTCCTCTACAGTCGGTCC | −2461–2439 |
Two micrograms of total RNA was used for reverse transcription with 2 pmol of UP (dT) and 200 U M-MLV Reverse Transcriptase; the amplified products were subsequently purified using a QIA quick column (Qiagen). A homopolymeric C tail was added to the 3′-end of the cDNA using terminal deoxynucleotidyl transferase (TaKaRa) according to the manufacturer’s instructions. Then, the sample was purified again by QIA quick column (Qiagen). The resultant poly (dC)-tailed cDNA was then applied as a template for the PCR reaction using an UP (dG) primer (10 homopolymeric G) and the inner-gene-specific primer 5GSP1 (Table 1). An extra round of PCR was conducted with a 1000-fold dilution of the original PCR product as the template using the UP primer and a nested 5GSP2 primer (Table 1) to get a pure and specific DNA product. The final product was subjected to nucleotide sequencing (BGI) using the primer 5GSP2 to identify the ChHp1bp3 5′ cDNA sequence.
To determine the 3′ end sequence, the previously obtained first strand cDNA pool that was reverse transcribed with the UP (dT) primer was applied as a template for PCR amplification using a UP primer and the gene-specific primer 3GSP1. The PCR product was diluted 1000-fold and used for an additional round of PCR using a poly(dA) primer and a nested 3GSP2 primer. The obtained specific DNA product was analyzed with nucleotide sequencing using the primer 3GSP2.
Purification of Histidine-tagged ChHP1BP3 in E. coli
The ChHP1BP3 coding sequence was amplified from the C. hongkongensis cDNA pool via PCR with HP1BP3-F-NdeI and HP1BP3-R-XhoI primers. The PCR product was cloned into the pET22b(+) vector that had been digested by Nde I and Xho I to yield pET22b(+)-ChHp1bp3. The latter was transformed into E. coli BL21, which is a suitable host for protein expression. The resulting transformants were cultivated at 37 °C in LB medium supplemented with 100 μg/mL ampicillin until the OD600 reached 0.8. IPTG (isopropyl-β-D-thiogalactopyranoside) was introduced at a final concentration of 0.2 mM for induction, and the cells were incubated for an additional 6 h at 28 °C. The cells were collected by centrifugation and the cell pellet was resuspended in lysis buffer (50 mM Na2HPO4, pH 8.0, and 500 mM NaCl) and sonicated on ice to obtain a homologous solution. The cell debris was removed by centrifugation at 14,000 g at 4 °C for 20 min. The resulting cell extract was passed through a Ni-NTA Agarose column (Cat. No. 30210; Qiagen) column on a Biologic LP apparatus (Bio-Rad). The 6-histidine-tagged ChHP1BP3 (His6-ChHP1BP3) was purified to near homogeneity according to the manufacturer’s instructions.
Electrophoretic mobility shift assay (EMSA)
The 5′ FITC-labeled ChHsp70 promoter region (−304 to +62 relative to the predicted transcriptional start as +1) was amplified by PCR as described previously26. The labeled fragment was incubated with purified His6-ChHP1BP3 at various concentrations for 25 min at room temperature in binding buffer (10 mM Tris-HCl, 50 mM KCl, and 1 mM DTT, pH 7.5). Extra reactions were completed using excess amounts of specific competitors (unlabeled ChHsp70 promoter) as well as non-specific competitors (unlabeled irrelevant DNA probes). After incubation, the complexes were resolved on a 5% native polyacrylamide gel that was pre-run at 100 V for 30 min with running buffer (45 mM Tris-HCl, pH 8.3, 45 mM boric acid, and 10 mM EDTA) and then subsequently run for 90 min at 100 V. The FITC-labeled DNA bands were visualized by fluorescence imaging using an UVI Alliance 4.7 imager (UK).
ChHP1BP3 knockdown
Approximately 100 ml of hemolymph were aseptically withdrawn from the posterior adductor muscle of approximately one hundred oysters using 1 ml syringes equipped with 23-gauge needles. The hemocytes were collected by centrifugation at 3000 rpm for 2 min. The collected hemocytes were washed with ice-cold PBS three times and subsequently resuspended in 50 ml complete DMEM (Dulbecco’s Modified Eagle’s Medium containing 10% heat inactivated fetal bovine serum [FBS] and supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.4% NaCl). The resulting solution was plated on 24-well plates at a hemocyte density of 1 × 105 cells/well. Then, the plates were incubated at 28 °C for 24 h in a humidified incubator supplied with 5% CO2, so as to reach 30–50% cell confluence. Then, siRNA and control RNA prepared by Sangon were individually transfected into the hemocytes using LipofectamineTM 2000 (Invitrogen) according to the manufacturer’s instructions in DMEM complete medium supplemented with 0.4% NaCl and lacking antibiotics. The cells were incubated at 28 °C for 4 h, followed by washing with PBS. The medium was shifted back to original completed DMEM supplemented with 0.4% NaCl, followed by incubation of 20 h. For heat shock treatment, the samples were incubated at 37 °C for 20 min, followed by a recovery period at 28 °C for 1 h before collecting the first sample. The collected hemocytes were then subjected to RT-PCR analysis.
Plasmid construction
The ChHp1bp3 coding sequence was amplified from the C. hongkongensis cDNA pool by PCR using the HP1BP3-E-KpnI and HP1BP3-E-XhoI primers (Table 1). The resulting PCR products were digested with the corresponding restriction enzymes and ligated into the pcDNA3.1 eukaryotic expression vector that had been cut with Kpn I and Xho I to yield pcDNA3.1-ChHp1bp3. The accuracy of fragment insertion was confirmed by sequencing (Liuhe BGI). The promoter-probe vector pGL3-ChHsp70p was constructed in our previous study26. The plasmids were kept at −20 °C until further analysis.
Dual-luciferase reporter assay
HEK293T cells were cultivated following the procedures described above for hemocytes cultivation. A confluence percentage of 90–95% was reached after plating onto 24-well plates. The dual-luciferase reporter assay was performed as described previously26. Briefly, the pGL3-ChHsp70 (200 ng/well), pRL-TK (20 ng/well, Promega), pcDNA3.1-ChHp1bp3 (200 ng/well) or empty pcDNA3.1 (200 ng/well) plasmids were co-transfected into HEK293T cells using LipofectamineTM 2000 (Invitrogen) according to the manufacturer’s instructions in the DMEM medium lacking FBS and antibiotics. After incubation at 37 °C for 4.5 h, the medium was shifted back to complete DMEM, followed by an additional incubation for 24 h prior to collecting the first sample. A Dual-luciferase Reporter Assay System (Promega) was applied to determine the luciferase activities according to the manufacturer’s instructions. Renilla luciferase activities were used to correct for the transfection efficiency.
Quantitative real-time PCR
Total RNA was extracted from hemocytes transfected with either siRNA or control RNA as well as from the gills of oysters that were either non-stressed or stressed by heat, CdCl2, or NP at various time points using the Trizol reagent (Cat. No. 15596-026; Invitrogen). Two micrograms of total RNA was used as template to synthesize the first-strand cDNA with MLV Reverse transcriptase (Promega) and oligo (dT) primer. The resulting cDNA was subsequently purified with a QIA quick column (Qiagen). Quantitative real-time PCR was performed in a 20 μL reaction mixture comprising SYBR Green Real-time PCR Master Mix (Toyobo, Japan), cDNA and one of the following gene-specific primer pairs: (1) RT-HP1BP3-Up/RT-HP1BP3-Down, (2) RT-HSP70-Up/RT-HSP70-Down or (3) Actin-Up/Actin-Down (internal control). The reaction mixtures were then subjected to the following thermal cycling conditions: (1) 95 °C for 4 min; (2) 40 cycles at 94 °C for 20 s, 62 °C for 20 s, 72 °C for 30 s; and (3) a gradient from 65 °C to 95 °C over 10 min with continuous detection. The expression of the ChHp1bp3 and ChHsp70 mRNAs was calculated relative to the amount of β-actin present. Negative controls were carried out without the addition of reverse transcriptase to confirm that the amplified products were derived from the mRNA transcripts instead of the genomic DNA. The assays were completed in triplicate for each sample.
Statistical analysis
The data were analyzed using one-way ANOVA followed by the Tukey HSD test. Differences were regarded as statistically significance at P < 0.05. IBM SPSS Statistics 19.0 was applied for the statistical analysis.
Nucleotide sequence accession number
The ChHp1bp3 cDNA sequence reported in the present study was deposited in the GenBank with accession number KX981984.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 41476100, 31170474), and the Key Natural Science Foundation of Guangdong Province (No. S2013020012720).
Author Contributions
We would like to declare that the present work is a product of the intellectual environment of the entire team, and all members have contributed in various degrees to the analytical methods used, to the data collection/interpretation, to the research concept, to the experiment design, and to the drafting/critical revision of the article. The detailed roles or tasks of each author are listed: Delin Xu Constructing idea and hypothesis for research; Planning methodology to reaching the conclusion; Logical interpretation of the results; Literature review; Construction/drafting of the whole manuscript; Critical revision of the article before submission; Final approval of version to be submitted. Qin Yang Managing experimental materials and reagents; Execution of the experiments; Data management and reporting; Presentation and interpretation of the results; Reviewing for the article before submission mainly for spelling and grammar; Final approval of version to be submitted. Miao Cui Managing experimental materials and reagents; Execution of the experiments; Data management and interpretation; Literature review; Organizing the course of the project; Reviewing the article before submission for both grammar and intellectual content; Final approval of version to be submitted. Qizhong Zhang Providing personnel, environmental and financial support and tools and instruments that are vital for the project. Constructing idea or hypothesis for research and manuscript; Supervising the course of the project or article; Reviewing the article before submission for both grammar and intellectual content; Final approval of version to be submitted.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Delin Xu and Qin Yang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01573-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miao Cui, Email: cuisanshui@163.com.
Qizhong Zhang, Email: zhangqzdr@126.com.
References
- 1.Whitley D, Goldberg SP, Jordan WD. Heat shock proteins: a review of the molecular chaperones. J Vasc Surg. 1999;29:748–751. doi: 10.1016/S0741-5214(99)70329-0. [DOI] [PubMed] [Google Scholar]
- 2.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta SC, et al. Heat shock proteins in toxicology: how close and how far? Life Sci. 2010;86:377–384. doi: 10.1016/j.lfs.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Zorzi E, Bonvini P. Inducible hsp70 in the regulation of cancer cell survival: analysis of chaperone induction, expression and activity. Cancers (Basel). 2011;3:3921–3956. doi: 10.3390/cancers3043921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deniset JF, Pierce GN. Heat Shock Proteins: Mediators of Atherosclerotic Development. Curr Drug Targets. 2015;16:816–826. doi: 10.2174/1389450116666150416115423. [DOI] [PubMed] [Google Scholar]
- 6.Young JC. The role of the cytosolic HSP70 chaperone system in diseases caused by misfolding and aberrant trafficking of ion channels. Dis Model Mech. 2014;7:319–329. doi: 10.1242/dmm.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PB, Jr., Lis JT. Cooperative and competitive protein interactions at the hsp70 promoter. J Biol Chem. 1997;272:33227–33233. doi: 10.1074/jbc.272.52.33227. [DOI] [PubMed] [Google Scholar]
- 8.Stephanou A, Isenberg DA, Nakajima K, Latchman DS. Signal transducer and activator of transcription-1 and heat shock factor-1 interact and activate the transcription of the Hsp-70 and Hsp-90beta gene promoters. J Biol Chem. 1999;274:1723–1728. doi: 10.1074/jbc.274.3.1723. [DOI] [PubMed] [Google Scholar]
- 9.Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 10.Tsai SY, et al. GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. Epigenetics Chromatin. 2016;9:32. doi: 10.1186/s13072-016-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 12.Duarte FM, et al. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 2016;30:1731–1746. doi: 10.1101/gad.284430.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanctot C, et al. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 14.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 15.Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta. 2008;1783:2174–2184. doi: 10.1016/j.bbamcr.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 17.Shimojima T, et al. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders BM. Stress proteins in aquatic organisms: an environmental perspective. Crit Rev Toxicol. 1993;23:49–75. doi: 10.3109/10408449309104074. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay I, Nazir A, Saxena DK, Chowdhuri DK. Heat shock response: hsp70 in environmental monitoring. J Biochem Mol Toxicol. 2003;17:249–254. doi: 10.1002/jbt.10086. [DOI] [PubMed] [Google Scholar]
- 20.Park MS, et al. Effects of Antifouling Biocides on Molecular and Biochemical Defense System in the Gill of the Pacific Oyster Crassostrea gigas. PLoS One. 2016;11:e0168978. doi: 10.1371/journal.pone.0168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farcy E, Voiseux C, Lebel JM, Fievet B. Transcriptional expression levels of cell stress marker genes in the Pacific oyster Crassostrea gigas exposed to acute thermal stress. Cell Stress Chaperones. 2009;14:371–380. doi: 10.1007/s12192-008-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Proteomic basis of stress responses in the gills of the pacific oyster Crassostrea gigas. J Proteome Res. 2015;14:304–317. doi: 10.1021/pr500940s. [DOI] [PubMed] [Google Scholar]
- 23.Cruz-Rodriguez LA, Chu FL. Heat-shock protein (HSP70) response in the eastern oyster, Crassostrea virginica, exposed to PAHs sorbed to suspended artificial clay particles and to suspended field contaminated sediments. Aquat Toxicol. 2002;60:157–168. doi: 10.1016/S0166-445X(02)00008-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Zhang Q. Molecular cloning, characterization and expression of heat shock protein 70 gene from the oyster Crassostrea hongkongensis responding to thermal stress and exposure of Cu(2+) and malachite green. Gene. 2012;497:172–180. doi: 10.1016/j.gene.2012.01.058. [DOI] [PubMed] [Google Scholar]
- 25.Kawabe S, Yokoyama Y. Novel isoforms of heat shock transcription factor 1 are induced by hypoxia in the Pacific oyster Crassostrea gigas. J Exp Zool A Ecol Genet Physiol. 2011;315:394–407. doi: 10.1002/jez.685. [DOI] [PubMed] [Google Scholar]
- 26.Miao Z, Xu D, Cui M, Zhang Q. High mobility group protein DSP1 negatively regulates HSP70 transcription in Crassostrea hongkongensis. Biochem Biophys Res Commun. 2016;474:634–639. doi: 10.1016/j.bbrc.2016.03.163. [DOI] [PubMed] [Google Scholar]
- 27.Garfinkel BP, et al. HP1BP3 is a novel histone H1 related protein with essential roles in viability and growth. Nucleic Acids Res. 2015;43:2074–2090. doi: 10.1093/nar/gkv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalski A, Palyga J. Chromatin compaction in terminally differentiated avian blood cells: the role of linker histone H5 and non-histone protein MENT. Chromosome Res. 2011;19:579–590. doi: 10.1007/s10577-011-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartova E, et al. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56:711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masumi A. Histone acetyltransferases as regulators of nonhistone proteins: the role of interferon regulatory factor acetylation on gene transcription. J Biomed Biotechnol. 2011;2011:640610–6. doi: 10.1155/2011/640610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutta B, et al. Profiling of the Chromatin-associated Proteome Identifies HP1BP3 as a Novel Regulator of Cell Cycle Progression. Mol Cell Proteomics. 2014;13:2183–2197. doi: 10.1074/mcp.M113.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta B, et al. Quantitative profiling of chromatome dynamics reveals a novel role for HP1BP3 in hypoxia-induced oncogenesis. Mol Cell Proteomics. 2014;13:3236–3249. doi: 10.1074/mcp.M114.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garfinkel BP, et al. Proportionate Dwarfism in Mice Lacking Heterochromatin Protein 1 Binding Protein 3 (HP1BP3) Is Associated With Alterations in the Endocrine IGF-1 Pathway. Endocrinology. 2015;156:4558–4570. doi: 10.1210/en.2015-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashihara K, et al. The middle region of an HP1-binding protein, HP1-BP74, associates with linker DNA at the entry/exit site of nucleosomal DNA. J Biol Chem. 2010;285:6498–6507. doi: 10.1074/jbc.M109.092833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butchko RA, et al. Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides. Fungal Genet Biol. 2012;49:602–612. doi: 10.1016/j.fgb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Soares A, et al. Nonylphenol in the environment: a critical review on occurrence, fate, toxicity and treatment in wastewaters. Environ Int. 2008;34:1033–1049. doi: 10.1016/j.envint.2008.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


