Abstract
In plants, leaf is crucial for photosynthesis and respiration. Leaf area and quantity are important for leaf vegetables to increase biomass. The process of leaf development involves coordinated regulation among small RNAs, transcription factors and hormones. Here, we found leaf size were regulated by transcription factors NF-YA2 and NF-YA10 in Arabidopsis. NF-YA2 and NF-YA10 overexpression increased biomass accumulation through promoting leaf growth and cell expansion. NF-YA2 and NF-YA10 were expressed in SAM and leaf vasculature. Endogenous IAA content reduced by 20% and 24% in transgenic Arabidopsis plants overexpressing NF-YA2 and NF-YA10 compared to wild-type plants. Chromatin immunoprecipitation assays revealed that NF-YA2 and NF-YA10 bound directly to the cis-element CCAAT in the promoter of the YUC2, and decreased the expression of YUC2, a YUCCA family gene. The auxin transporter gene PIN1 and auxin response factor1 and 2 (ARF1 and ARF2) genes, transcriptional repressors, were downregulated. These findings showed leaf development was regulated by NF-YA2 and NF-YA10 through the auxin-signaling pathway and may provide a new insight into the genetic engineering of vegetables biomass and crop productivity.
Introduction
Leaves are photosynthetic tissues and very important for the success of plants. The process of leaf development is composed of primordia initiation, lamina expansion and margin formation, involves coordinated regulation among small RNAs, transcription factors and hormones1. Genetic studies showed that many key factors involved in leaf development. MiR156/SPL regulation module has been reported to interact with TCP4 and this complex promoted CUC-controlled acquisition of leaf complexity in Arabidopsis 2. In relation to primordia initiation, miR160 targets ARF10, ARF16, and ARF17, three members of a divergent class of ARF genes that share high amino acid sequence similarity and present overlapping expression patterns3–5. The ARF genes regulated by miR160s are necessary for proper phyllotaxis in the rosette. Besides, MiR164 regulates organ boundary size through its modulation of the CUC1 (CUP-SHAPED COTYLEDON1) and CUC2 genes5–7. MiR319, also called miRJAW in Arabidopsis, is involved in the coordination of cell division and growth during leaf development by targeting a subset of the TCP (TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR) genes that are homologues to the Antirrhinum CIN (CINCINNATA) gene8 and the tomato LA (LANCEOLATE) gene9. AT the gene level, Class III HD-ZIP, KANADI and YABBY gene families are involved in the establishment of polarity10–12. In addition, PIN and CUC genes play crucial role in leaf margin patterning by controlling auxin-maxima formation13, 14. Furthermore, CIN (CINCINNATA) gene limits excess cell proliferation and maintains the flatness of the leaf surface by directly modulating the hormone pathways involved in patterning cell proliferation and differentiation during leaf growth15.
Auxin is a key hormone that is responsible for modulating many aspects of plant growth, including root and leaf architecture, organ patterning, and vascular development16. Current models propose that members of the PIN protein family of auxin efflux regulators represent an important part of a network for auxin distribution throughout the plant17 and mediate auxin efflux from cells and thus directional cell-to-cell transport. YUC (YUCCA) family genes of Arabidopsis encode flavin monooxygenase-like enzymes that catalyze the rate-limiting step in Trp-dependent auxin biosynthesis18. YUC genes had been proved functions are important in leaf margin development and blade outgrowth19.
The miR169 family of Arabidopsis contains 14 genes. However, only four mature miR isoforms (a, b/c, d/e/f/g and h/i/j/k/l/m/n) are produced. The miR169 isoforms present distinct expression patterns during development20, in response to biotic21 or abiotic stresses22, 23, suggesting a functional specialization. In plants, the main targets of miR169 are genes that encode the subunit A of nuclear factor Y (NF-Y)24. This transcription factor (TF) is a heterotrimeric TF composed of NF-YA (HAP2), NF-YB (HAP3/CBF-A) and NF-YC (HAP5/CBF-C) subunits. In plants, NF-Y TFs have been linked to development25–27, signalization28 and responses to stresses23, 29–31. NF-YB and NF-YC subunits contain a histone fold domain very similar to H2A and H2B core histones32, 33 and these two subunits must form a heterodimer for stable interaction with NF-YA. The NF-Y genes present differential expression patterns during development34–37, or in response to environmental conditions38, 39, suggesting that, in different organs or under certain stimuli, only some combinations of subunits can be assembled to form the trimeric functional NF-Y factor. Arabidopsis miR169d/NF-YA2 (10) modules had been clearly shown that it plays a crucial role in stress-induced early flowering40 and root architecture in Arabidopsis 41.
Here, we showed that NF-YA2 (10) plays important roles in leaf development. Our data suggested that NF-YA2/10 can directly interact with YUC2 promoter, and decreased YUC2 expression, which in turn regulates the synthesis of auxin.
Results
NF-YA2 and NF-YA10 overexpression promote leaf initiation and development
We observed that overexpression of NY-YA2 was not only regulated Arabidopsis flowering time, but also affected leaf development. To illustrate the potential role of NY-YA2 and NY-YA10 in leaf development, we constructed NF-YA10 overexpression vector and obtained the transgenic plants. In contrast to NT, NF-YA2 and NF-YA10 overexpression plants showed larger rosettes (Fig. 1a). The rosette diameter of NF-YA2 OE and NF-YA10 OE plants were 6.8 and 6.5 cm, respectively, larger than NT (5.03 cm) (Fig. 1d). Moreover, NF-YA2 and NF-YA10 overexpression plants can generate new rosette leaves incessantly even after seeds harvest, whereas the leaves of NT plants generally decayed after harvest (Fig. 1b). Thereof NF-YA2 OE and NF-YA10 OE plants can generated more leaves than that in NT. The 40-days rosette numbers of NT, NF-YA2 OE and NF-YA10 OE were 16.6, 20.7 and 20, respectively (Fig. 1c). The biomass of NF-YA2 OE and NF-YA10 OE were increased by 24% and 28% compared to NT (Fig. 1e).
Figure 1.

Rosette phenotype of NF-YA2 and NF-YA10 overexpression lines. (a) 20-days-seedling; (b) plants at bolting stage; (c) number and diameter of rosettes for NF-YA2 and NF-YA10 overexpression lines. Thirty plants were measured for each line.
NF-YA2 and NF-YA10 overexpression expand cell size of leaves
The leaf size is determined generally by cell number and cell size. To uncover what reason result in larger leaf in NY-YA2 OE and NF-YA10 OE plants, we investigated their cell size and numbers using scanning electron (SE) microscopy. The epidermal cells of the leaves in NF-YA2 and NF-YA10 OE plants were larger than those in NT (Fig. 2a–c). These results indicate that NF-YA2 and NF-YA10 regulate leaf size by controlling cell size.
Figure 2.
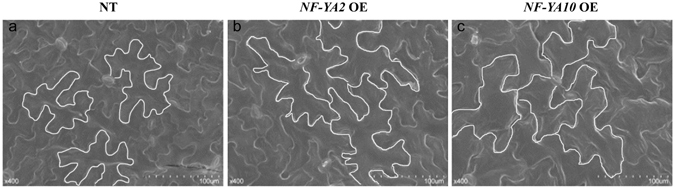
SE micrograph of a leaf of 8-week-old plant. (a) Negative transgenic plant; (b) NF-YA2 OE plant; (c) NF-YA10 OE plant.
NF-YA2 and NF-YA10 are expressed in shoot apical meristems, internode and leaves
In order to explore the expression patterns of NF-YA2 and NFYA10, WT plants were transformed with promoter::GUS constructs that contained the 2 kb promoter region from NF-YA2 and NF-YA10 respectively. NF-YA2 and NF-YA10 have high level expression patterns in cotyledon vasculature and SAM in pNF-YA2::GUS and pNF-YA10::GUS plants, suggesting that they might have a role in leaf initiation and developing. NF-YA2 was expressed mainly in SAM, node and young leaves, and the expression level was rapidly decreased with leaf growth (Fig. 3a–c). NF-YA10 was expressed in SAM, node and leaves, and the expression level was increased with leaf growth. However the expression level of NF-YA10 was clearly weaker in SAM and node than that of NF-YA2 (Fig. 3d–f). The highest expression level of NF-YA2 and NF-YA10 was in SAM region and young leaves. Considering auxin is synthesized mostly in SAM and young leaf, we reduced that they might be involved in IAA regulation to affect leaf initiation and development.
Figure 3.

Expression profile of NF-YA2 and NF-YA10 in transgenic plants. (a–c) NF-YA2 expression pattern in 10-d-old, 13-d-old, and 16-d-old seedlings; (d,e) NF-YA10 expression pattern in 10-d-old, 13-d-old, and 16-d-old seedlings. Plants were grown on MS medium in a standard LD light regime.
NF-YA2 and NF-YA10 overexpression decreased endogenous IAA content
Auxin has been confirmed as the central regulator of organogenesis at the SAM. Considering the SAM expression profiles of NF-YA2 and NF-YA10, we investigated concentration of endogenous IAA in whole shoots including rosette and SAM. The data showed that IAA concentrations were decreased by 20% and 24% in the NF-YA2 and NF-YA10 overexpression lines, respectively, compared to the NT (Fig. 4), indicating NF-YA2 and NF-YA10 are involved in regulation of IAA biosynthesis and auxin-signaling pathway.
Figure 4.
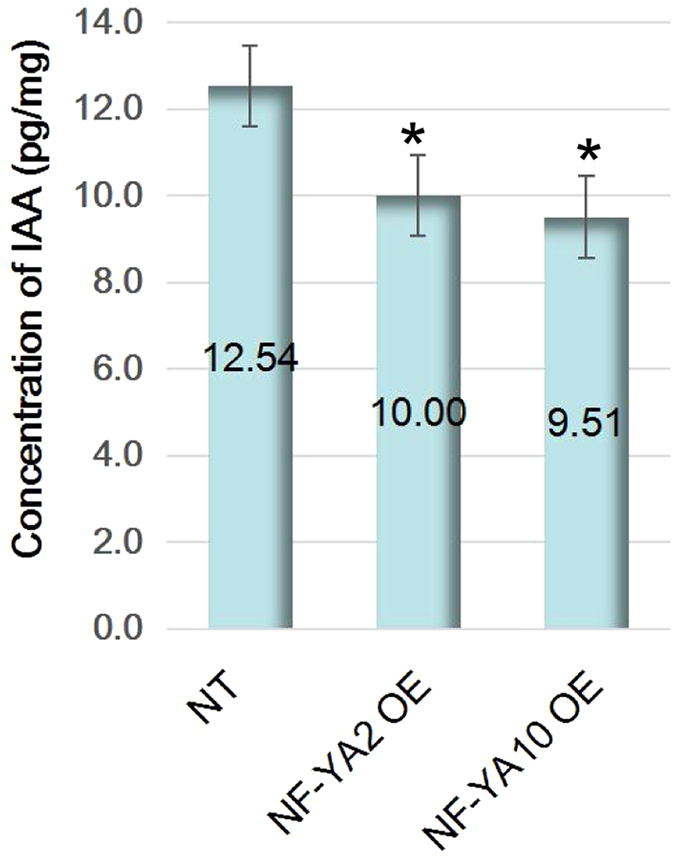
Concentrations of IAA in transgenic plants and NT.
NF-YA2 and NF-YA10 are involved in regulation of auxin biosynthesis
To understand the molecular mechanisms and genetic regulation of leaf generation and development, we profiled the transcriptomes of developing node and leaves from NF-YA2 OE and NF-YA10 OE plants, respectively. Total of 7631 and 3607 differentially expressed genes (DEGs) were identified using cut-off values (log2FC >1 or <−1 with p-value of 0.05) in comparison with NT (Fig. 5a,b). GO enrichment analysis was performed on these two groups of DEGs to discover overrepresented functional categories (Fig. 5c). Top 10 generally changed GO terms by enrichment score (−log10P-value) were showed in Fig. 5c. The most enriched and meaningful biological process terms were related to stress responding, regulation of transcription, and plant hormone signaling, suggesting NF-YA transcription factors in plants are potentially involved in stress responding and plant development. Based on alerted endogenous IAA content in AtNF-YA2 and AtNF-YA10 OE plants, we examined differential accumulation of auxin signaling, such as auxin biosynthetic process (GO:0009851), and found that the expression of YUCCA family was clearly different between transgenic plants and NT. YUCCA family members of Arabidopsis encode Flavin monooxygenase-like enzymes that catalyze the rate-limiting step in Trp-dependent auxin biosynthesis, which play important roles in local auxin biosynthesis18. The YUC is known as a key factor in the regulatory pathway controlling leaf development42. YUC-controlled leaf developmental pathway acts synergistically with auxin polar transport43. Three genes YUC1, YUC2, and YUC6 were down-regulated in NF-YA2 OE and NF-YA10 OE lines (Supplementary Table S1). Together, functional characterization of DEGs between NF-YA2 OE line and NF-YA10 OE line indicated that NF-YA2 (10) maybe regulate auxin biosynthesis via YUCCA family genes.
Figure 5.

Up (a) and down (b) DEGs between NF-YA2 OE plant and NF-YA10 OE were analyzed using VENN and significantly enriched Gene Ontology (GO) categories with the common DEGs (c).
Based on the transcriptomes analysis, YUCCA family genes (YUC1, YUC 2, and YUC6) were potential targets regulated by NF-YA2 and NF-YA10. Thereof we investigated their expression in NT plants, NF-YA2 OE and NF-YA10 OE lines. YUC 1 and YUC6 expression in NF-YA2 OE and NF-YA10 OE plants were higher than in NT, but their differential expression were not significant. However, YUC2 was significantly down-regulated in the NF-YA2 OE and NF-YA10 OE plants (Fig. 6). These results suggest NF-YA2 and NF-YA10 can specifically regulate YUC2 expression.
Figure 6.

Yuc1, yuc2 and yuc6 expression in NT, NF-YA2 OE and NF-YA10 OE lines.
NF-YA2 and NF-YA10 specifically regulate YUC2 expression
Previous studies in human, animals and plants suggested that the CCAAT box is a binding site for NF-YA protein44, 45. Sequence analysis suggests that there are several CCAAT motifs located in the yuc2 promoter and the first intron (Fig. 7a). To investigate if NF-YAs bind to these sites, FLAG-tagged NF-YA2 and NF-YA10 transgenic Arabidopsis plants were obtained45, and ChIP was performed on the transgenic plants using anti-FLAG antibodies. Quantitative PCR (qPCR) was then performed on the YUC2 sequences using four different pairs of primers (Fig. 7a). As shown in Fig. 7b, the induced NF-YA2 and NF-YA10 transgenic plants showed a clear enrichment of the promoter (YUC2-P1and YUC2-P2) sequences in comparison with mock transgenic Arabidopsis. These results suggest that NF-YA2 and NF-YA10 regulates the expression of YUC2 by physically interacting with YUC2 promoter.
Figure 7.
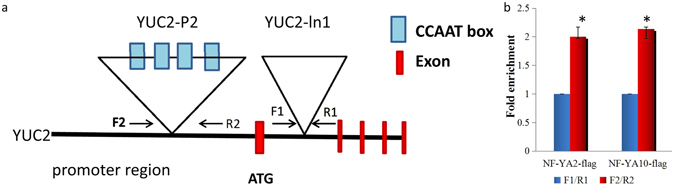
Chromatin immunoprecipitation (ChIP) assay in NF-YA2-FLAG and NF-YA10-FLAG plants. (a) Schematic structure of genomic sequences of yuc2 and the regions examined by ChIP. Two pairs of primers were used. Primer YUC2-P2 covered the promoter region containing the CCAAT cis-element. (b) Relative levels of qPCR products from the ChIP assay. Data were from one experiment with three technical replicates. Values are means ± SD, n = 3.
Effect of NF-YA2 and NF-YA10 on the expression of genes involved in IAA signaling pathway
Based on the endogenous IAA contents were decreased in the overexpression plants, the auxin transport and signaling should be changed accordingly. Thereof we examined the expression levels of the IAA efflux carrier protein gene PIN1 and auxin response factor gene ARF1. The data showed PIN1 and ARF1 expression was decreased significantly in NF-YA2OE and NF-YA10OE plants compared to NT plants (Fig. 8). These results indicated that the expression of PIN1, an IAA transporter, was decreased with lessened IAA concentration. ARF1, a repressor of auxin-induced genes, can be bound and repressed by Aux/IAA protein at low IAA concentration, delaying the process of aging (leaf increasing) in Arabidopsis. Accordingly, downregulated expression of ARFs in NF-YA2 OE and NF-YA10 OE plants also presented leaf increasing and preventing senescence. We deduced that low IAA might result in more and larger rosettes through ARF family.
Figure 8.
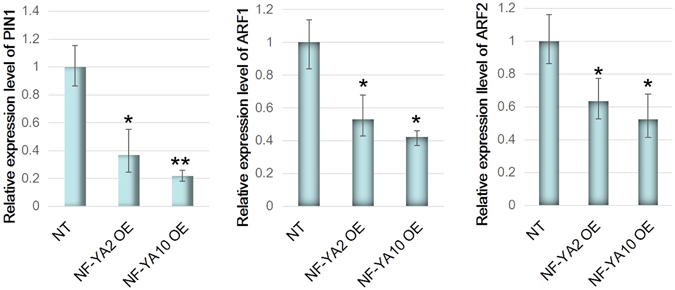
Relative expression levels of PIN1, ARF1 and ARF2 in NF-YA2 OE and NF-YA10 OE plants.
Discussion
NF-YA2 and NF-YA10 mediated leaf development
Leaf primordia of higher plants derive from the peripheral zone of the shoot apical meristem. Major outstanding questions in leaf development are initiation of the primordia, leaf patterning and ending, and how these processes are regulated accurately plays an important role in the plant life. Transcription factors are known act as regulation hub to play crucial roles in plant development processes and in response to environmental and endogenous conditions, however few of them have been linked to leaf growth. Here, we found that NF-YA2 and NF-YA10 genes were involved in leaf initiation and growth.
Members of NF-YA family can be regulated by miR169 family, which are involved in drought and nitrate responses, flowering and root architecture. Overexpression of the miR169a, which specifically targets NF-YA5, induced drought sensitivity or altered nitrogen responses23, 30 and miR169d-g mature sequence is induced by nitrate deficiency46. NF-YA2, targeted by miR169d, was involved in stress-induced flowering in Arabidopsis 40. Moreover, the regulation modules of miR169d/e/f/g isoform and the NF-YA2 target control root architecture in Arabidopsis 41. Recently, in roots, a clear upregulation of NF-YA2 and NF-YA10 has been observed in response to phosphate starvation47. Our data showed that NF-YA2 and NF-YA10 were involved in leaf development via regulating IAA biosynthesis. These results, all together, are consistent with the fact that the miR169/NF-YA module, could directly or indirectly act as a linker between plant development and responding to abiotic stresses. Indeed, auxin is a well-known operators of growth and development, which can be affected by all of these stresses in plants48.
Based on the previous results and our finds, we put forward a model showed in Fig. 9. YUC2, a key speed-limiting gene in auxin homeostasis, acts as a direct target of NF-YA2 and NF-YA10. Overexpression of NF-YA2 and NF-YA10 decreased contents of endogenous IAA through repressing yuc2 expression. Lower IAA contents result in downregulation of PIN and ARFs family. ARF1 and ARF2, transcriptional repressors, can directly bind to promoters that contain auxin response elements (TGTCTC) to repress targets transcription, such as IAA, further influence leaf initiation and growth in Arabidopsis (Fig. 9).
Figure 9.
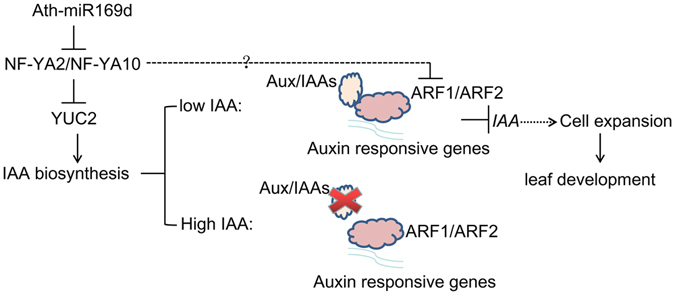
Schematic model of NF-YA2 and NF-YA10 mediated regulation of Arabidopsis leaf initiation and development through auxin signaling. IAA: indole-3-acetic acid; YUC2: YUCCA 2; ARF1: auxin response factor 1.
Over all, NF-YA family members have been proposed to control various plant responses to environmental stresses49 and development43. Our data showed that NF-YA2 and NF-YA10 were involved in leaf growth in Arabidopsis through IAA biosynthesis, providing a new insight for miR169/NF-YA module roles between abiotic stress and development.
Auxin and leaf development
All plant shoots can be described as a series of developmental modules termed phytomers, which are produced from SAM. A phytomer generally consists of a leaf, internode, and a secondary shoot meristem. Because leaf formation is part of the general lateral organ initiation program at the SAM, it is not surprising that auxin is involved. Classical micromanipulation techniques and probes that predict auxin transport pathways confirmed that dynamic auxin fluxes pattern organ initiation at the shoot apex, suggesting that auxin plays a critical role in leaf development50. Leaf initiation and leaf growth are different progress, leaf initiation requires the formation of an auxin maximum and leaf growth needs transcriptional responses mediated by ARFs51. Here concentration of endogenous IAA in whole shoots pooled rosette and SAM was decreased, which seemed to be conflicting to leaf initiation promotion. Because the amount of SAM was negligible compared to whole rosette, SAM should be separated to detect the concentration of endogenous IAA in the further experiment.
The phenotypic similarities in leaves between the NF-YA2 OE or NF-YA10 OE plants and the arf1 and arf2 mutants52 support the notion that the NF-YA genes affect an auxin-signaling process. Meanwhile certain yuc mutants were treated by combination with the auxin transport inhibitor NPA totally blocked new leaf formation, a phenotype that is not observed in the yuc mutants alone or NPA treatment alone42, suggesting that leaf development is regulated by coordinated auxin biosynthesis, transport and signalling response. So we presumed that NF-YA2 or NF-YA10 maybe target other genes in auxin-signaling pathway besides yuc2, which should be analyzed in the future.
Methods
Plant materials and culture
All experiments were performed on the Columbia ecotype of A. thaliana. Plants were grown in a controlled culture room at 22 °C with a relative humidity of 60% and 16/8 h photoperiod.
Constructs and transgenic lines
For pNF-YA2::GUS and pNF-YA10::GUS, 2000-bp region upstream of the start codon ATG of NF-YA2 and NF-YA10 was amplified from genomic DNA (all primers sequences used for cloning are listed in Table S2), respectively, to cloned into pEASY-T1 vector, which were recombined in the binary vector pCAMBIA1303 after sequencing confirmation.
The p35S::NF-YA2 overexpression lines and transgenic NF-YA2-flag plants had been generated by Xu et al.40.The p35S::NF-YA10 and NF-YA10-FLAG constructs were obtained as described40. Col 0 transformation was performed by the floral dip method53 and independent stable transgenic lines were selected.
GUS staining and microscopy
The histochemical detection of GUS activity was performed as described54 with a staining incubation overnight. Then the stained tissues were decolorized by 75% ethanol and the images were obtained using a microscope. To analyze expression in the whole organ, seedlings with different stage were obtained to detect.
RNA extraction, Real time fluorescence quantitative PCR (qRT-PCR)
Total RNA was extracted by using the Trizol procedure as described by the manufacturer (ambion) and cDNA was synthesized following the manufacturer’s instructions (5x All-In-One RT MasterMix, abm, Canada). qRT-PCR (all qPCR primers sequences used can be found in Table S2) was performed on an Applied Biosystems (http://www.AppliedBiosystems.com) Prism 7500 analyzer and SYBR Premix Ex Taq™ (CodeQPK-201, TOYOBO). For each genotype, three or four independent biological replicates, each consisting of 10 individual plants, were analysed. Sample comparisons were performed using the 2(−ΔΔCT) method55. Actin1 was reference control.
β-Oestradiol treatment of transgenic NF-YA2-FLAG and NF-YA10-FLAG plants and ChIP(Chromatin immunoprecipitation) assay were carried out as described40. All the primers used for ChIP-qPCR are listed in Supplementary Table S2.
Microarray analysis
Total RNA was extracted from seedlings of NF-YA2 OE and NF-YA10 OE lines. RNA quantity and quality were measured by Agilent 2100 Bioanalyzer. RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Agilent Arabidopsis Oligo Microarray V4.0 was adopted for detection of mRNA expression. All the microarray analysis was performed by oe Bio-tech (Shanghai, China).
Quantitative analysis of IAA
Whole shoots were harvested from plants grown under a 16:8-h photoperiod in trays (12 seedlings per tray) when the first open flower was visible. Each of the randomly arranged trays contained a single genotype and represented one replicate sample. Three replicate samples (200 mg fresh weight) were analyzed. Total IAA were detected and quantified as methyl esters by gas chromatography–mass spectroscopy (GC–MS) at Institute of Genetics and Development Biology, Chinese Academy of Sciences. (Beijing, China).
Electronic supplementary material
Acknowledgements
This project was supported by the National Key Basic Research Program (grant no. 2014CB138205), the National Key Research Program of China (grant no. 2016YFD0101002), and the National Natural Science Foundation of China (grant no. 31270318).
Author Contributions
M.Z. performed gene expression profile experiment, phenotype detection and all qRT-PCR experiment. X.H. constructed plant expression vectors, conducted Arabidopsis transformation and the ChIP experiment. M.Z. assisted with bioinformatic analysis and interpreting analysis results. M.X. designed the study and drafted the manuscript. L.W. supervised the project and edited the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01475-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miaoyun Xu, Email: xumiaoyun@caas.cn.
Lei Wang, Email: wanglei01@caas.cn.
References
- 1.Moon J, Hake S. How a leaf gets its shape. Curr Opin Plant Biol. 2011;14:24–30. doi: 10.1016/j.pbi.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Rubio-Somoza I, et al. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Opin. Plant Biol. 2014;24:2714–2719. doi: 10.1016/j.cub.2014.09.058. [DOI] [PubMed] [Google Scholar]
- 3.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JW, et al. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu PP, et al. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 6.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 8.Palatnik JF, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 9.Ori N, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 2007;39:787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 10.Siegfried KR, et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- 11.Byrne ME. Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genet. 2006;2:e89. doi: 10.1371/journal.pgen.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhart BJ, et al. Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell. 2013;25:3228–3249. doi: 10.1105/tpc.113.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008;40:1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 14.Kawamura E, Horiguchi G, Tsukaya H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 2010;62:429–441. doi: 10.1111/j.1365-313X.2010.04156.x. [DOI] [PubMed] [Google Scholar]
- 15.Das Gupta M, Aggarwal P, Nath U. CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 2014;204:901–912. doi: 10.1111/nph.12963. [DOI] [PubMed] [Google Scholar]
- 16.Millner PA. The auxin signal. Curr. Opin. Cell Biol. 1995;7:224–231. doi: 10.1016/0955-0674(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 17.Friml J, Benkova E, Mayer U, Palme K, Muster G. Automated whole mount localisation techniques for plant seedlings. Plant J. 2003;34:115–124. doi: 10.1046/j.1365-313X.2003.01705.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, et al. YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 2011;157:1805–1819. doi: 10.1104/pp.111.186395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Ibeas D, et al. Analysis of the melon (Cucumis melo) small RNAome by high-throughput pyrosequencing. BMC Genomics. 2011;12:393. doi: 10.1186/1471-2164-12-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh K, Talla A, Qiu W. Small RNA profiling of virus-infected grapevines: evidences for virus infection-associated and variety-specific miRNAs. Funct. Integr. Genomics. 2012;12:659–669. doi: 10.1007/s10142-012-0292-1. [DOI] [PubMed] [Google Scholar]
- 22.Licausi F, et al. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 2011;190:442–456. doi: 10.1111/j.1469-8137.2010.03451.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M, Ding H, Zhu JK, Zhang F, Li WX. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011;190:906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhoades MW, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 25.Lotan T, et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 26.Combier JP, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenkel S, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warpeha KM, et al. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson DE, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li WX, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zemzoumi K, Frontini M, Bellorini M. & Mantovani, R. NF-Y histone fold alpha1 helices help impart CCAAT specificity. J. Mol. Biol. 1999;286:327–337. doi: 10.1006/jmbi.1998.2496. [DOI] [PubMed] [Google Scholar]
- 34.Gusmaroli G, Tonelli C, Mantovani R. Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene. 2001;264:173–185. doi: 10.1016/S0378-1119(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 35.Gusmaroli G, Tonelli C, Mantovani R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene. 2002;283:41–48. doi: 10.1016/S0378-1119(01)00833-2. [DOI] [PubMed] [Google Scholar]
- 36.Siefers N, et al. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao S, Kumimoto RW, Siriwardana CL, Risinger JR, Holt BF., 3rd Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PloS one. 2011;6:e21805. doi: 10.1371/journal.pone.0021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pant BD, et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Des Marais DL, et al. Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell. 2012;24:893–914. doi: 10.1105/tpc.112.096180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu MY, et al. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 2014;65:89–101. doi: 10.1093/jxb/ert353. [DOI] [PubMed] [Google Scholar]
- 41.Sorin C, et al. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014;202:1197–1211. doi: 10.1111/nph.12735. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pallai R, Simpkins H, Chen J, Parekh HK. The CCAAT box binding transcription factor, nuclear factor-Y (NF-Y) regulates transcription of human aldo-keto reductase 1C1 (AKR1C1) gene. Gene. 2010;459:11–23. doi: 10.1016/j.gene.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang G, He H, Yu D. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PloS one. 2012;7:e48951. doi: 10.1371/journal.pone.0048951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo J, et al. The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 2012;12:62. doi: 10.1186/1471-2229-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavy M, Estelle M. Mechanisms of auxin signaling. Development. 2016;143:3226–3229. doi: 10.1242/dev.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PloS one. 2012;7:e48138. doi: 10.1371/journal.pone.0048138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinhardt D, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 51.Sluis A, Hake S. Organogenesis in plants: initiation and elaboration of leaves. Trends in Genetics. 2015;6:300–306. doi: 10.1016/j.tig.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Ellis CM, et al. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 53.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 54.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


