Abstract
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) has become a frequently used strategy in gene expression studies. The relative quantification method is an important and commonly used method for the evaluation of RT-qPCR data. The key aim of this method is to identify an applicable internal reference gene. However, there are currently no data concerning the expression of reference genes for gene analysis in human tongue carcinoma cell lines and tissues. In the present study, screening was performed using 12 common reference genes, which were selected in order to provide an experimental basis for the investigation of gene expression in human tongue carcinoma. Tca-8113 and CAL-27 cell lines and a total of 8 tongue carcinoma tissue samples were investigated. The gene expression stability and the applicability of the 12 reference gene candidates were determined using the geNorm, NormFinder and BestKeeper software programs. The results from the three software programs were demonstrated to be variable following comparison. The recommended combinations were 5′-aminolevulinate synthase 1 + glucuronidase β + ribosomal protein L29 (RPL29) for the cell line + tissue group, β-2-microglobulin + RPL29 for the cell line group and peptidylprolyl isomerase A + hydroxymethylbilane synthase + RPL29 for the tissue group. These recommended internal reference genes may improve the accuracy of relative quantitation analysis of target gene expression performed by the RT-qPCR method in further gene expression research on human tongue carcinoma.
Keywords: reverse transcription quantitative polymerase chain reaction, reference gene, human tongue carcinoma
Introduction
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) is frequently used in gene expression studies and is currently considered the gold standard for accurate, sensitive and rapid measurements of gene expression. Relative quantification is an important and commonly used technique to evaluate RT-qPCR data, during which the expression levels of target genes are compared with those of a stably expressed endogenous control gene, determined simultaneously in the same biological sample. Therefore, the gene expression levels require normalization using reference genes in order to obtain reliable data. The identification of appropriate reference genes is a crucial stage involved in this approach. It is important for the ideal reference genes to be universally valid under the experimental conditions (1,2). In general, cellular maintenance genes, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB) and ribosomal RNA (18S rRNA), are selected as reference genes to examine the variability between clinical samples. However, several studies have demonstrated that the expression levels of these commonly used reference genes vary in different tissues or between treatments in the same tissue (3–6), as well as across cell types (7–9).
Tongue carcinoma is the most common malignancy of the oral cavity, accounting for 12.2% of all head and neck cancers (10,11). Tongue cancer is characterized by a high malignant degree, high local recurrence rate, high neck metastasis rate and high rate of mortality. It is the focus of oral tongue cancer surgery.
RT-qPCR is a frequently used technique to investigate differential gene expression, thus a review of the normalization standards used in quantitative gene expression studies of human tongue carcinoma was deemed necessary. To the best of our knowledge, no systematic study has previously been performed on the selection of suitable reference genes for investigating target gene profiling between human tongue carcinoma cell lines and tissues.
The present study aimed to identify the most suitable reference gene or set of genes for target gene profiling of human tongue carcinoma. The stabilities of a panel of 12 common reference genes in human tongue carcinoma cell lines and tissues were validated. The 12 candidate genes: ACTB, 5′-aminolevulinate synthase 1 (ALAS1), GAPDH, TATA-box binding protein (TBP), hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L29 (RPL29), hydroxymethylbilane synthase (HMBS), peptidylprolyl isomerase A (PPIA), pumilio RNA binding family member 1 (PUM1), glucuronidase β (GUSB), β-2-microglobulin (B2M) and 18S rRNA are frequently used as endogenous controls in the context of tongue carcinoma, but are not restricted to this. A number of these genes have been identified as optimal reference genes in certain other cancer types, including HPRT1 and ACTB (12,13). Three common software packages, geNorm (14), NormFinder (15) and Bestkeeper (16), were used to investigate these genes. The aim was to provide useful information for the selection of suitable reference genes in further gene expression studies on human tongue carcinoma.
Materials and methods
Human tongue carcinoma cell lines
The human tongue carcinoma cell line Tca-8113 was provided by Jilin Cancer Hospital (Changchun, China) and CAL-27 cells were provided by the Hospital of Stomatology, Jilin University (Changchun, China). Cells were cultivated in Iscove's modified Dulbecco's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) with 100 units of penicillin, maintained at 37°C in a 5% CO2 humidified atmosphere, according to the recommendation of the supplier.
Tongue carcinoma tissue samples
A total of 8 tongue carcinoma tissue samples were provided by the Tissue Bank of China-Japan Union Hospital, Jilin University (Changchun, China). The clinicopathological characteristics of the patients were summarized in Table I. The present study was approved by the Ethics Committee of the China-Japan Union Hospital, Jilin University (Changchun, China).
Table I.
Clinicopathological characteristics of patients.
| Clinicopathological characteristic | Patients with tongue carcinoma |
|---|---|
| Age (mean ± standard deviation) | 56.75±4.06 |
| Sex | |
| Male | 6 |
| Female | 2 |
| Histopathological type | |
| Squamous cell carcinomas | 8 |
| Adenocarcinoma | 0 |
| TNM stagea | |
| Stage 0 | 1 |
| Stage I | 1 |
| Stage II | 3 |
| Stage III | 2 |
| Stage IV | 1 |
According to the union for international cancer control (17).
RNA extraction and complementary DNA (cDNA) synthesis
The cell lines were recovered from liquid nitrogen and cultured for 72 h. A total of 50–100 mg tissue samples were homogenized in 1 ml TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was extracted from the cells and each tissue sample using TRIzol reagent following the manufacturer's protocol. DNase I (Sangon Biotech Co., Ltd., Shanghai, China) was used to eliminate genomic DNA contamination. The concentrations and purity of the isolated RNA were measured by NanoDrop 2000 (Thermo Fisher Scientific, Inc.).
The cDNA synthesis reaction was performed three times using the M-MuLV First Strand cDNA Synthesis kit (Sangon Biotech Co., Ltd., Shanghai, China) according to the manufacturer's protocol. The total reaction volume was 20 µl. Total RNA (1 µg), 1 µl random primer and RNase free water were mixed, incubated at 65°C for 5 min and then cooled down immediately on ice for 30 sec. The rest of the reaction reagents were added, then the mixture was incubated at 42°C for 60 min and the reaction was terminated by heating at 70°C for 10 min.
RT-qPCR
The primers of 12 putative reference genes were selected based on previous studies, and are widely used and recognized to be good reference genes (18,19). The primers were synthesized by Sangon Biotech Co., Ltd. and the sequences are listed in Table II. A Roche LightCycler 480 detection system (Roche Diagnostics GmbH, Mannheim, Germany) was used for RT-qPCR. Reactions were performed using 2xSG Fast qPCR Master Mix (Sangon Biotech Co., Ltd.) according to the manufacturer's protocol. All the samples were run in triplicate. The PCR volume was 20 µl, containing 2 µl cDNA. The following cycling conditions were used: 55°C for 5 min; 95°C for 5 min; 40 cycles of 95°C for 20 sec, 55°C for 20 sec and 72°C for 4 min. This cycle was followed by melting curve analysis, and the baseline and cycle threshold values (Cq values) were automatically determined for all the plates using Roche LightCycler 480 software v1.5.0 (Roche Diagnostics GmbH). RT-qPCR amplification products were detected by 1% agarose gel electrophoresis and melting curve to verify the specificity of the primers. A standard curve was constructed for each primer pair to determine the product specificity.
Table II.
Summary of reference genes used in the present study.
| Symbol | Official full name | Accession number | Primer sequence | Product size (bp) |
|---|---|---|---|---|
| 18S | 18S ribosomal RNA | NM_10098.1 | F:CGGCTACCACATCCAAGGAA | 186 |
| R:GCTGGAATTACCGCGGCT | ||||
| GAPDH | glyceraldehyde-3- | NM_002046.5 | F: GACAGTCAGCCGCATCTTCT | 127 |
| phosphate dehydrogenase | R: TTAAAAGCAGCCCTGGTGAC | |||
| B2M | β-2-microglobulin | NM_004048.2 | F: AGCGTACTCCAAAGATTCAGGTT | 306 |
| R: ATGATGCTGCTTACATGTCTCGAT | ||||
| ACTB | β-actin | NM_001101.3 | F: AGAAAATCTGGCACCACACC | 173 |
| R: TAGCACAGCCTGGATAGCAA | ||||
| ALAS1 | 5′-aminolevulinate synthase 1 | NM_000688.5 | F: GGCAGCACAGATGAATCAGA | 150 |
| R: CCTCCATCGGTTTTCACACT | ||||
| GUSB | glucuronidase β | NM_000181.3 | F: AGCCAGTTCCTCATCAATGG | 160 |
| R: GGTAGTGGCTGGTACGGAAA | ||||
| HPRT1 | hypoxanthine | NM_000194.2 | F: GACCAGTCAACAGGGGACAT | 132 |
| phosphoribosyltransferase 1 | R: CCTGACCAAGGAAAGCAAAG | |||
| HMBS | hydroxymethylbilane synthase | NM_000190.3 | F: AGTGTGGTGGGAACCAGC | 144 |
| R: CAGGATGATGGCACTGAACTC | ||||
| PPIA | peptidylprolyl isomerase A | NM_021130.4 | F: AGACAAGGTCCCAAAGAC | 118 |
| R: ACCACCCTGACACATAAA | ||||
| PUM1 | pumilio RNA-binding | NM_001020658.1 | F: CAGGCTGCCTACCAACTCAT | 217 |
| family member 1 | R: GTTCCCGAACCATCTCATTC | |||
| RPL29 | ribosomal protein L29 | NM_000992.2 | F: GGCGTTGTTGACCCTATTTC | 120 |
| R: GTGTGTGGTGTGGTTCTTGG | ||||
| TBP | TATA-box binding protein | NM_003194.4 | F: TGCACAGGAGCCAAGAGTGAA | 132 |
| R: CACATCACAGCTCCCCACCA |
The Cq values were identified by quantitative comparison of the amplification of the candidate genes. The Cq values were calculated to relative quantities (Q) for data analysis, in view of the PCR efficiencies of the candidate genes according to the equation: Q=2−ΔΔCq (20).
PCR efficiency
A random pool of cDNA from the samples was selected and used for 10-fold serial dilutions, ranging between 0.001 and 1X. PCR analysis was run in triplicate, as mentioned previously. PCR efficiency was calculated using the slopes of the calibration curve and by the formula: E=10−1/slope.
Statistical analysis
All the samples were divided into three groups: Cell line + tissue group, cell line group and tissue group. In order to evaluate the stability of the reference genes, three frequently used software programs, geNorm v3.5 (http://medgen.ugent.be/~jvdesomp/genorm/), NormFinder v0.953 (http://moma.dk/normfinder-software) and BestKeeper version 1 (http://www.gene-quantification.de/bestkeeper.html), were utilized. GeNorm is designed to establish reference genes for RT-qPCR and is used to analyze and determine the M-value, which refers to the stability of the reference gene expression. M is the mean pairwise variation for a given gene compared with other tested genes, following stepwise exclusion of the gene with the highest M value and calculated in order to select the most two stable genes. The default value suggested by geNorm is M=1.5. A higher M-value indicates less stable expression, and a lower M value indicates more stable expression. If M >1.5, the gene is not suitable for use as a reliable reference gene. GeNorm software was also used to analyze the pair-wise variation value of the normalization factor (V), which has a default value of 0.15. It is possible to use the value of Vn/Vn+1 to determine whether adding a novel reference gene affects the normalization factor. If the value of Vn/Vn+1 is >0.15, it is necessary to use the n+1 reference genes as internal controls. If it is <0.15, then it is not necessary to use novel reference genes. NormFinder software is a tool designed to identify the optimal reference gene among a set of candidates and it has a similar operation principle to geNorm. This program analyzes expression data, ranks the set of candidate normalization genes according to their expression stability and considers the gene with the minimum expression data as the most stable gene. It is also possible to use this software to compare the stability of inter and intragroup reference genes and propose an optimal combination of two genes. BestKeeper evaluates candidate reference gene stability based on the correlation coefficient (R-value). The genes were ranked according to their R value, with a higher R value indicating a more stable and reliable gene.
Results
Amplification specificity and efficiency of primers
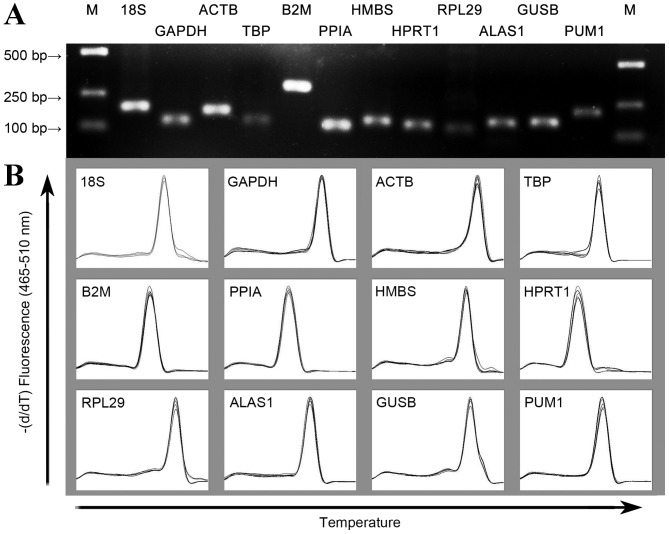
The primer sequences, corresponding length of the amplified products and PCR amplification efficiency are listed in Table II. The gel imaging system indicated that the size of the amplified fragment was consistent with the expected size, with a clear band and without primer dimers and nonspecific bands (Fig. 1A). In addition, melting curve analysis of each gene fragment amplified by qPCR revealed that all curves exhibited a single signal peak (Fig. 1B). For the candidate reference genes, the amplification efficiency range of the standard curve was 1.95–2.09 and all correlation coefficients were >0.96.
Figure 1.
Specificity of RT-qPCR amplification: (A) 1% agarose gel electrophoresis of RT-qPCR amplification products. (B) Melting curve analysis. RT-qPCR, reverse transcription-quantitative polymerase chain reaction analysis.
Gene expression levels
The expression level of the candidate reference genes was determined by the Cq value, which is inversely proportional to the expression level of the gene. Higher Cq values indicated smaller expression quantities. The Cq value of all the samples ranged between 7.70 and 32.93 (Fig. 2). In all groups, 18S had the smallest mean Cq values of 9.48±1.51 (cell line + tissue group; Fig. 2A), 8.38±0.43 (cell line group; Fig. 2B) and 10.30±1.52 (tissue group; Fig. 2C) and HMBS had the greatest mean Cq values of 26.41±2.37 (cell line + tissue group; Fig. 2A), 25.06±1.70 (cell line group; Fig. 2B) and 27.43±2.37 (tissue group; Fig. 2C).
Figure 2.
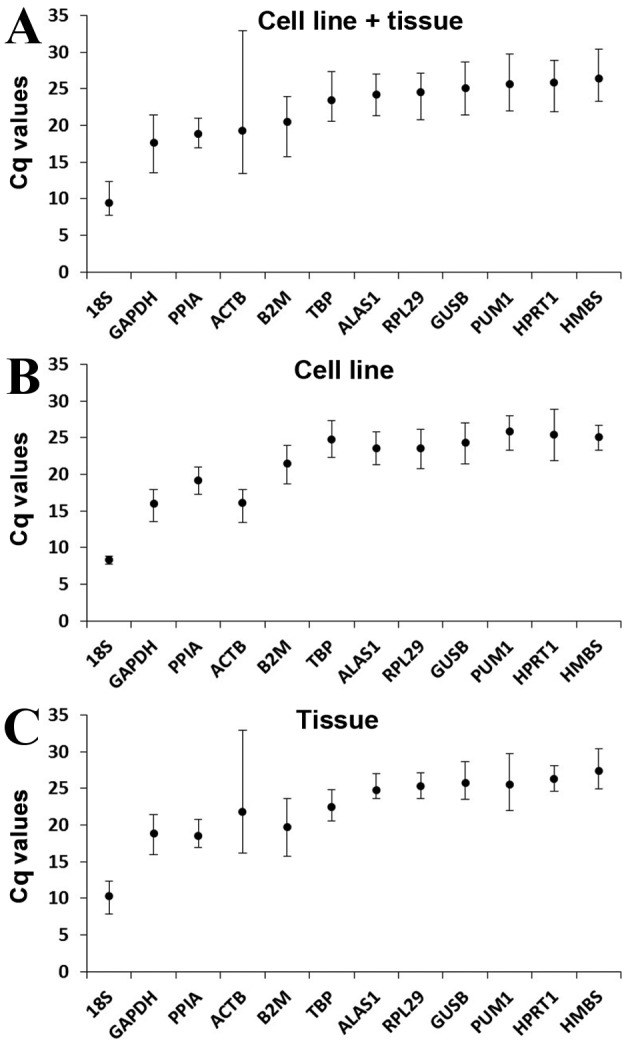
Mean Cq values of the reference genes in the experimental samples for the (A) cell line+tissue, (B) cell line and (C) tissue groups. Bars represent the mean ± standard deviation.
Stability analysis of the candidate reference gene
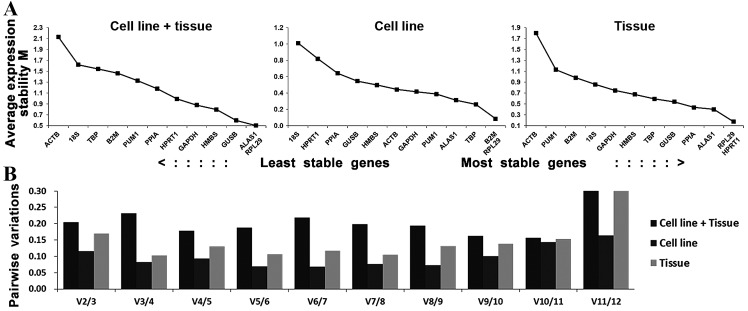
Theoretically, the 12 reference genes constituted appropriate internal controls for gene expression. In the cell line + tissue group, ALAS1 and RPL29 had the lowest M-values, followed by GUSB, which suggested that these were the most stable candidate genes for studies between human tongue carcinoma cell lines and tissue. In the cell line group, B2M and RPL29, followed by TBP, were suggested as the most stable reference genes for studies between Tca-8113 and CAL-27 cell lines. In the tissue group, RPL29 and HPRT1, followed by ALAS1, were suggested as most stable reference genes for studies on human tongue carcinoma tissue (Fig. 3A). A combination of 2 and 3 reference genes were optimal in the cell line group (V2/3=0.116) and tissue group (V3/4=0.103), respectively (Fig. 3B).
Figure 3.
GeNorm analysis of the candidate reference genes. Results are presented according to the output file of the geNorm program. (A) Stepwise exclusion of the least stable genes by calculating the M value. The x-axis from left to right indicates the ranking of the reference genes according to their expression stability and the y-axis indicates M. (B) Determination of the optimal number of reference genes for normalization.
In order to further evaluate the stability of the 12 reference genes, the present study also used the NormFinder program. PPIA + HMBS was the most stable reference gene combination in the cell line + tissue group, whilst GUSB and RPL29 were the most stably expressed genes in this group (Fig. 4A). In the cell line group, B2M + HMBS was the most stable reference gene combination, whilst PUM1 and GADPH were the most stably expressed genes (Fig. 4B). In the tissue group, PPIA + HMBS was the most stable reference gene combination, whilst HMBS and GUSB were the most stably expressed genes (Fig. 4C).
Figure 4.
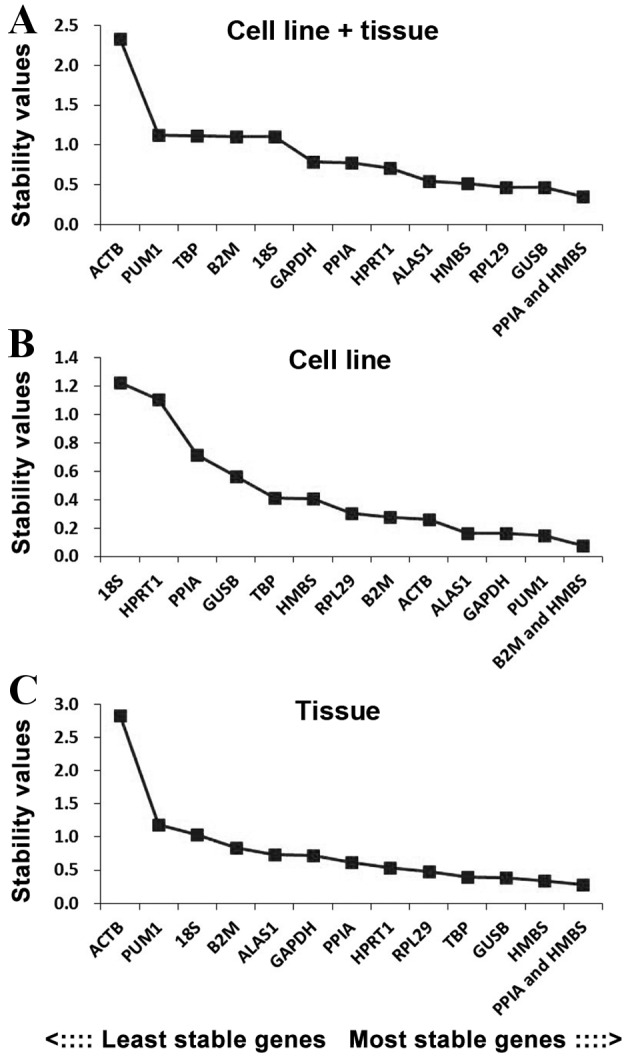
Candidate reference genes for normalization according to their expression stability for the (A) cell line+tissue, (B) cell line and (C) tissue groups, calculated using the NormFinder program. The y-axis represents the stability value. The x-axis from left to right represents the ranking of the reference genes.
The BestKeeper program was also used to compare the stability of internal reference genes. As the BestKeeper program is only able to analyze 10 internal reference genes, the 2 most unstable internal reference genes indicated by the geNorm software were removed in each group. In terms of the R-value, the most stable internal reference gene in the cell line + tissue group was GUSB, followed by ALAS1 and RPL29 (Fig. 5A). In the cell line group the most stable internal reference gene was RPL29, followed by B2 M and GAPDH (Fig. 5B), and in the tissue group the most stable internal reference gene was RPL29, followed by GUSB and HPRT1 (Fig. 5C).
Figure 5.
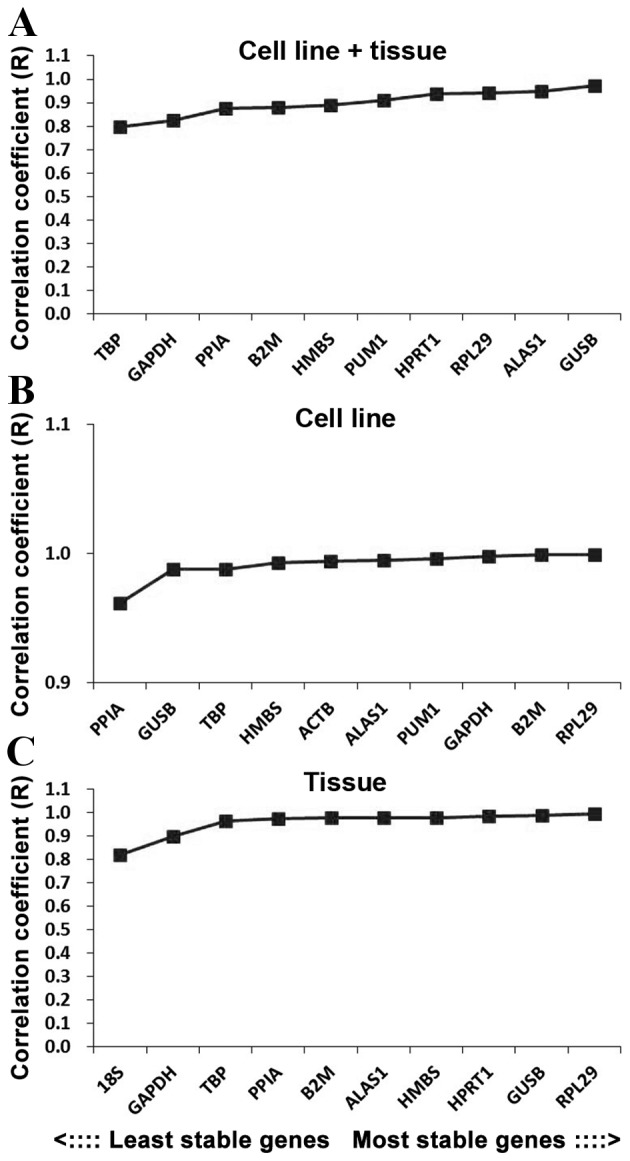
Stability values of the candidate reference genes evaluated using BestKeeper software for the (A) cell line+tissue, (B) cell line and (C) tissue groups.
Discussion
A gene with a steady expression level is required to normalize the data during detection of target gene expression, and these are known as internal reference genes. Previous studies have indicated that the majority of commonly used internal control genes have flaws. Expression levels of these genes vary significantly depending on experimental conditions, including different cell types and tissues, different individuals and different stages of cell proliferation, organ development and in vitro culture (4–6). To the best of our knowledge, the present study is first to compare the stability of commonly used internal reference genes in human tongue carcinoma cell line and tissues. As studies investigating tongue carcinoma gene profiling increase, the confirmation of stable and reliable internal control genes is required. In the present study, the reference genes commonly used in studies of gene expression in tongue carcinoma were those frequently used in studies examining molecular markers in other cancer types.
To obtain accurate experimental data and reliable conclusions, the present study used an experimental process with a number of characteristics. Cell lines and tissues are investigated in the present study. For the study of cell lines, Tca-8113 and CAL-27, which are the most commonly used tongue carcinoma cell lines for in vitro studies, were used. For the study of tissues, due to limitations for tongue carcinoma surgery, biopsy specimens were not selected by grades and stages as, according to previous research, the expression of reference genes is not directly associated with the grade or stage of a malignant tumor (12,21). The specimens were confirmed by the Pathology Department of China-Japan Union Hospital, Jilin University (Changchun, China) as malignant and the tongue cancer samples used were the most common pathological types of squamous cell carcinomas.
A total of 12 common reference genes were compared in terms of their expression stability and the geNorm, NormFinder and BestKeeper software programs, commonly used to compare stability between reference genes, were selected for data analysis.
The geNorm program was used for initial analysis. This software program is based on a pairwise-comparison statistical model. By calculating the values of M and V, the two most stable reference genes and the optimum number of reference gene combinations was determined. Following this analysis, the results suggested that ALAS1 and RPL29 in the cell line + tissue group, B2M and RPL29 in the cell line group and RPL29 and HPRT1 in the tissue group were the most stable reference genes. GUSB and the combination of PPIA and HMBS in the cell line + tissue group, PUM1 and the combination of B2M and HMBS in the cell line group, and HMBS and the combination of PPIA and HMBS in the tissue group were considered to be the most stable reference genes and the best combinations by the NormFinder software program, which is based on analysis of variance as the statistical model. Finally, in order to reduce the one-sidedness of the computing models of the aforementioned software programs, the Bestkeeper program was used for further analysis. The results suggested that GUSB, RPL29 and RPL29 were the most stable reference genes in the cell line + tissue group, cell line group and tissue group, respectively. As the rank of the candidate gene stability was slightly different, potentially caused by different calculation algorithms (22,23), no specific single reference gene was recommended as the optimal reference gene for normalizing relative quantitative investigations of tongue carcinoma. In addition, by calculating the value of V, the optimal number of reference gene combinations of the cell line + tissue group, cell line group and tissue group were 11, 2 and 3, respectively. The boundary value suggested by geNorm was 0.15, however, rather than a stringent standard consideration, which provided guidance to determine the optimal number of reference genes. Regarding the standardized principle of RT-qPCR, previous studies have recommended selecting a minimum of 3 internal control genes to perform relative quantitative investigations (24). The present study also recommended that a combination of 3 reference genes was sufficient for normalizing relative quantitative investigations. The recommended combination for the cell line + tissue group was ALAS1 + GUSB + RPL29, for the cell line group B2M + RPL29 and for the tissue group PPIA + HMBS + RPL29. These genes were ranked in the forefront of each group across the three software packages used.
The present study identified the most suitable reference genes and reference gene combinations for human tongue carcinoma cell lines and tissues for use in gene expression profile analysis. A reliable standardized method has the potential to improve understanding of the biological mechanisms underlying tongue carcinoma in the future. The relevant clarification of tumor molecular expression markers may improve the accuracy of diagnosis and estimation of prognostic factors, and provide novel treatments.
Acknowledgements
The present study was supported in part by grants from the Natural Science Foundation of China (grant no. 81503531), the Education Department of Jilin Province (grant no. 2015536) and the Science and Technology Department of Jilin Province (grant nos. 201201060 and 201215078).
References
- 1.Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 2.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Ali H, Du Z, Li X, Yang Q, Zhang YC, Wu M, Li Y, Zhang G. Identification of suitable reference genes for gene expression studies using quantitative polymerase chain reaction in lung cancer in vitro. Mol Med Rep. 2015;11:3767–3773. doi: 10.3892/mmr.2015.3159. [DOI] [PubMed] [Google Scholar]
- 4.Ma H, Yang Q, Li D, Liu J. Validation of suitable reference genes for quantitative polymerase chain reaction analysis in rabbit bone marrow mesenchymal stem cell differentiation. Mol Med Rep. 2015;12:2961–2968. doi: 10.3892/mmr.2015.3776. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q, Ali HA, Yu S, Zhang L, Li X, Du Z, Zhang G. Evaluation and validation of the suitable control genes for quantitative PCR studies in plasma DNA for non-invasive prenatal diagnosis. Int J Mol Med. 2014;34:1681–1687. doi: 10.3892/ijmm.2014.1944. [DOI] [PubMed] [Google Scholar]
- 6.Yu S, Yang Q, Yang JH, Du Z, Zhang G. Identification of suitable reference genes for investigating gene expression in human gallbladder carcinoma using reverse transcription quantitative polymerase chain reaction. Mol Med Rep. 2015;11:2967–2974. doi: 10.3892/mmr.2014.3008. [DOI] [PubMed] [Google Scholar]
- 7.He YX, Zhang Y, Yang Q, Wang C, Su G. Selection of suitable reference genes for reverse transcription-quantitative polymerase chain reaction analysis of neuronal cells differentiated from bone mesenchymal stem cells. Mol Med Rep. 2015;12:2291–2300. doi: 10.3892/mmr.2015.3671. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Yang Q, Bai J, Xuan Y, Wang Y. Identification of appropriate reference genes for human mesenchymal stem cell analysis by quantitative real-time PCR. Biotechnol Lett. 2015;37:67–73. doi: 10.1007/s10529-014-1652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Yang Q, Bai J, Yang Y, Zhong L, Wang Y. Identification of optimal reference genes for quantitative PCR studies on human mesenchymal stem cells. Mol Med Rep. 2015;11:1304–1311. doi: 10.3892/mmr.2014.2841. [DOI] [PubMed] [Google Scholar]
- 10.Moore SR, Johnson NW, Pierce AM, Wilson DF. The epidemiology of tongue cancer: A review of global incidence. Oral Dis. 2000;6:75–84. doi: 10.1111/j.1601-0825.2000.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, Du Y, Tao J, Wu Y, Chen N. Expression of cytosolic phospholipase A2 and cyclooxygenase 2 and their significance in human oral mucosae, dysplasias and squamous cell carcinomas. ORL J Otorhinolaryngol Relat Spec. 2008;70:242–248. doi: 10.1159/000130872. [DOI] [PubMed] [Google Scholar]
- 12.Ohl F, Jung M, Xu C, Stephan C, Rabien A, Burkhardt M, Nitsche A, Kristiansen G, Loening SA, Radonić A, Jung K. Gene expression studies in prostate cancer tissue: Which reference gene should be selected for normalization? J Mol Med (Berl) 2005;83:1014–1024. doi: 10.1007/s00109-005-0703-z. [DOI] [PubMed] [Google Scholar]
- 13.Huan P, Maosheng T, Zhiqian H, Long C, Xiaojun Y. TLR4 expression in normal gallbladder, chronic cholecystitis and gallbladder carcinoma. Hepatogastroenterology. 2012;59:42–46. doi: 10.5754/hge10258. [DOI] [PubMed] [Google Scholar]
- 14.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:Research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 17.Sobin LH, Gospodarowicz MK, Wittekind C, editors. TNM Classification of Malignant Tumours. 7th. Wiley-Blackwell; Chichester, UK: 2009. International Union Against Cancer. [Google Scholar]
- 18.Battula VL, Bareiss PM, Treml S, Conrad S, Albert I, Hojak S, Abele H, Schewe B, Just L, Skutella T, Bühring HJ. Human placenta and bone marrow derived MSC cultured in serum-free, b-FGF-containing medium express cell surface frizzled-9 and SSEA-4 and give rise to multilineage differentiation. Differentiation. 2007;75:279–291. doi: 10.1111/j.1432-0436.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- 19.Mane VP, Heuer MA, Hillyer P, Navarro MB, Rabin RL. Systematic method for determining an ideal housekeeping gene for real-time PCR analysis. J Biomol Tech. 2008;19:342–347. [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399:257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, Yang W, Lan Q, Jiang Z. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using real-time PCR. PLoS One. 2012;7:e33278. doi: 10.1371/journal.pone.0033278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brugè F, Venditti E, Tiano L, Littarru GP, Damiani E. Reference gene validation for qPCR on normoxia- and hypoxia-cultured human dermal fibroblasts exposed to UVA: Is β-actin a reliable normalizer for photoaging studies? J Biotechnol. 2011;156:153–162. doi: 10.1016/j.jbiotec.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Wisnieski F, Calcagno DQ, Leal MF, dos Santos LC, Gigek Cde O, Chen ES, Pontes TB, Assumpção PP, de Assumpção MB, Demachki S, et al. Reference genes for quantitative RT-PCR data in gastric tissues and cell lines. World J Gastroenterol. 2013;19:7121–7128. doi: 10.3748/wjg.v19.i41.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]