Abstract
Melanoma mortality rates are the highest amongst skin cancer patients. Melanoma is life threating when it grows beyond the dermis of the skin. Hence, depth is an important factor to diagnose melanoma. This paper introduces a non-invasive computerized dermoscopy system that considers the estimated depth of skin lesions for diagnosis. A 3-D skin lesion reconstruction technique using the estimated depth obtained from regular dermoscopic images is presented. On basis of the 3-D reconstruction, depth and 3-D shape features are extracted. In addition to 3-D features, regular color, texture, and 2-D shape features are also extracted. Feature extraction is critical to achieve accurate results. Apart from melanoma, in-situ melanoma the proposed system is designed to diagnose basal cell carcinoma, blue nevus, dermatofibroma, haemangioma, seborrhoeic keratosis, and normal mole lesions. For experimental evaluations, the PH2, ISIC: Melanoma Project, and ATLAS dermoscopy data sets is considered. Different feature set combinations is considered and performance is evaluated. Significant performance improvement is reported the post inclusion of estimated depth and 3-D features. The good classification scores of sensitivity = 96%, specificity = 97% on PH2 data set and sensitivity = 98%, specificity = 99% on the ATLAS data set is achieved. Experiments conducted to estimate tumor depth from 3-D lesion reconstruction is presented. Experimental results achieved prove that the proposed computerized dermoscopy system is efficient and can be used to diagnose varied skin lesion dermoscopy images.
Keywords: Melanoma in-situ, skin lesions, classification, 3D lesion reconstruction, 3D features and tumor depth estimation
This paper presents a 3D reconstruction technique for computerized dermoscopic skin lesion classification.
I. Introduction
The world health organization reports a rapid increase of skin cancer cases [1]. Skin cancer can be broadly classified as melanoma and non-melanoma type. About two to three million cases of non-melanoma cancer and 132,000 melanoma cancers are reported annually worldwide [2]. A staggering mortality rate of 75% is reported in the US alone due to skin cancer melanoma when compared to non-melanoma skin cancers [3], [4]. An average increase of 2.6% deaths caused due to melanoma has been observed annually in the past decade. Cases where early detection of skin cancer melanoma is achieved, the costs incurred towards treatments are fairly low and a five year survival rate of 95% is reported. The cost incurred towards the treatment of advance cases of melanoma is very high and the five year survival rate is only 13% [5]. Early detection of skin cancer melanoma is a challenging problem and requires attention.
To diagnose and study the skin lesions dermatologists use the dermoscopy technique
also referred to as surface skin microscopy, dermatoscopy and epiluminescence
microscopy ( [6]. The dermoscopy technique adopted is
non-invasive and is generally performed by expert dermatologists. Dermoscopy is
performed by the application of a gel on the skin lesion, then digital imaging
systems like stereomicroscope or dermatoscope are used to obtain magnified images.
Magnified skin lesion images provide additional color, structure and pattern data
not clearly visible to the naked human eye. This additional data enable the
dermatologists to identify the type of skin lesion and aid the diagnosis [7].
[6]. The dermoscopy technique adopted is
non-invasive and is generally performed by expert dermatologists. Dermoscopy is
performed by the application of a gel on the skin lesion, then digital imaging
systems like stereomicroscope or dermatoscope are used to obtain magnified images.
Magnified skin lesion images provide additional color, structure and pattern data
not clearly visible to the naked human eye. This additional data enable the
dermatologists to identify the type of skin lesion and aid the diagnosis [7].
The use of classical clinical algorithms such as ABCD (Asymmetry, Border, Color and Diameter) [8], ABCDE (Asymmetry, Border, Color, Diameter and Evolution) [9], Menzies method [10] and the seven-point checklist [11] is adopted by for the diagnosis of melanoma skin lesions. An improvement of 5–30% is achieved by using dermoscopy and classical clinical algorithms when compared to the examination carried out by the naked human eye [12]. The skill of the dermatologists is also critical to achieve accurate diagnostic performance considering dermoscopy images [13], [14]. Considering the varied type of melanoma, non-melanoma skin lesions and dependency on the skill level of dermatologist, accurate diagnosis of melanoma is still a problem.
The use of computer aided diagnosis can be used to tackle this problem. Availability of advance image processing techniques and decision making mechanisms to build computer aided diagnostic system can provide a wholistic solutions to aid early diagnosis of skin cancer melanoma. The computer aided diagnostic systems are also referred to as “Computerized dermoscopy” [22]. Computerized dermoscopy systems primarily constituted of five components A) Dermoscopy image acquisition of skin lesions, B) Region of interest identification or segmentation of skin lesion, C) Feature Extraction D) Feature selection and E) Decision making mechanisms achieved through machine learning techniques. Numerous studies published lay emphasis on the segmentation or region of interest identification [15]–[18]. Several computerized dermoscopy systems to have been developed considering all or combinations of shape, texture, color features and incorporating varied decision support mechanisms [12], [19]–[28]. To the best of our knowledge, little or no emphasis is laid so far on depth estimation and 3D reconstruction of skin lesions from 2D dermoscopic images. Considering the depth and 3D geometry of the skin lesion is critical to achieve accurate diagnosis. A detailed discussion is presented in the latter section of the paper.
Surface illumination based dermoscopy techniques are used to for image acquisition as they are inexpensive and easily available. Dermoscopy techniques like Nevoscopy [30], trans-illumination light microscopy [31], High-frequency ultrasound [32], acoustic microscopy [33] and 3D high-frequency skin ultrasound images [38], to name a few are considered by researchers to construct 3D volumes and estimate the depth of skin lesions for accurate diagnosis. The availability and the cost of these special dermoscopy imaging systems is still a problem.
A noninvasive computerized dermoscopy system to aid diagnosis of skin lesions is proposed in this paper. Special emphasis is laid to aid diagnosis of in-situ melanoma. An adaptive snake model [18] for segmentation of the 2D dermoscopic skin lesion images. To reconstruct the 3D skin lesion initially a depth map is derived from the 2D dermoscopic image. The depth map construction is adopted from the technique presented in [34]. The depth map data is fit to the 2D surface to achieve 3D skin lesion reconstruction. The 3D skin lesion is represented as structure tensors. Using the 2D skin lesion data color, texture and 2D shape features ae extracted. The 3D reconstructed skin lesion data is used to obtain the 3D shape features. The 3D shape features encompass the relative depth features estimated. To highlight and study the significance of the features, feature selection methods are considered. For decision making, three different multiclass classifiers have been considered and their performance is compared and studied. The proposed computerized dermoscopy system relies on bag-of-features (BoF), AdaBoost and Support Vector Machines (SVM) for decision making. Comparisons considering different feature combinations and classifiers is presented in the experimental study. Good classification results considering melanoma skin lesions (especially in-situ melanoma) achieved early in the research motivated authors to further expand the diagnosis on varied skin lesion types. Experimental study is conducted using dermoscopic images obtained from Atlas of Dermoscopy CD [41], PH2 dataset [42] and ISIC: Melanoma project dataset [62], [63].
The main contributions of the study is summarized as follows
-
1.
3D reconstruction from 2D dermoscopic images using depth estimation.
-
2.
3D shape features considering the 3D lesion constructed.
-
3.
Considering different algorithms for multiclass decision making.
-
4.
Comprehensive skin lesion data considered in the study namely melanoma, in-situ melanoma, atypical nevus, common nevus, basal cell carcinoma, blue nevus, dermatofibroma, haemangioma, seborrhoeic keratosis and normal mole lesions.
The remaining manuscript is organized as follows. The signifance of depth and the various stages of skin cancer melanoma is discussed in section two. A brief literature review is presented in section three. Section four discusses the proposed computerized dermoscopy system. The experimental study and discussions is in the penultimate section of the paper. Last section of the paper describes the conclusions drawn and future work.
II. Melanoma – “It is Skin Deep”
Melanoma is typically a type of skin cancer. Of all types of skin cancer known,
melanoma is the deadliest type and the highest mortality rates are reported from
patients suffering from melanoma. Melanoma cancer occurrences are predominantly
reported in the skin but occurrences in the eyes, nasal passages, throat, brain etc.
are also known. In the research presented here melanoma cancer of the skin is
considered. To diagnose melanoma of the skin a physical examination by a
dermatologist and a biopsy is generally carried out. Post confirmation, the doctors
proceed to identify the stage of the melanoma skin cancer to initiate the relevant
treatment. Stages of melanoma are described through various scales like Clarke
scale, Breslow scale, Tumor Node and Metastases (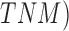 scales. The
Clarke and Breslow scale basically define the measure of the depth of the tumor i.e.
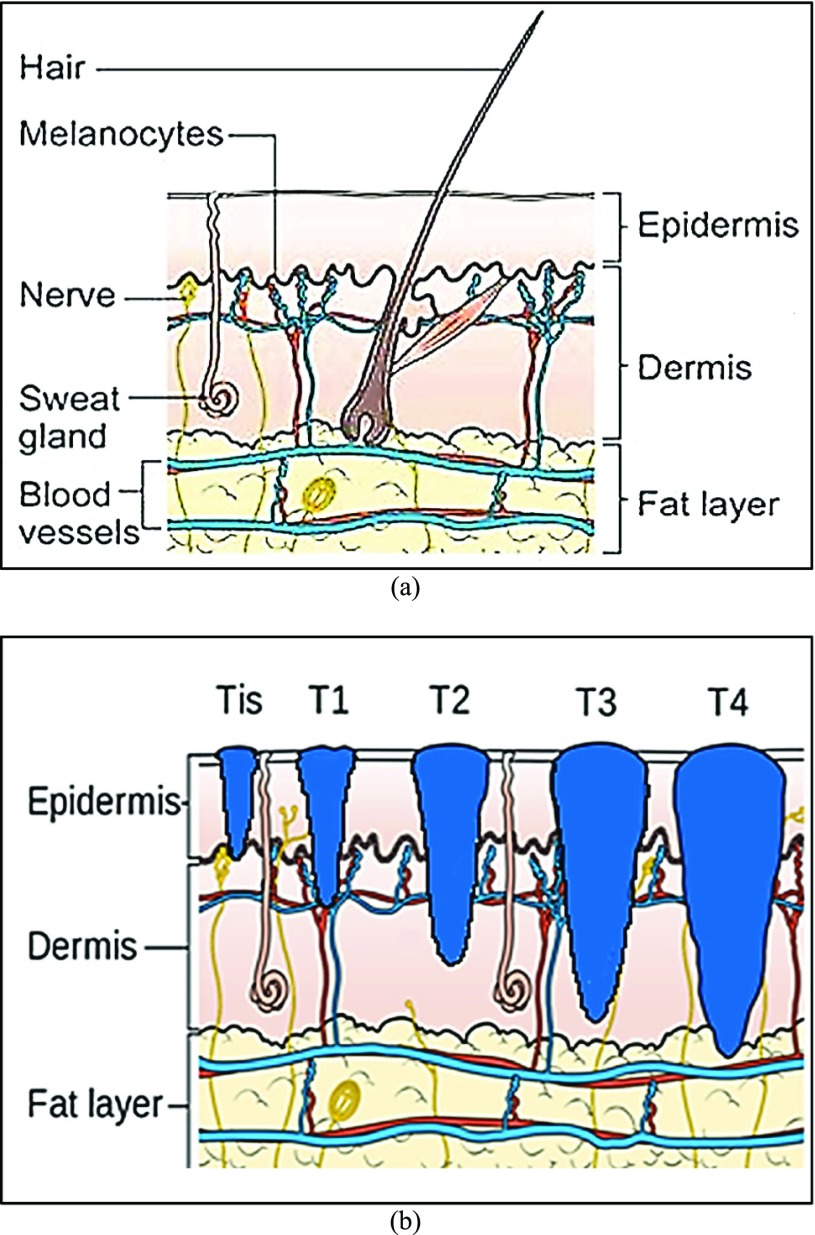
how deep the tumor has gone into the skin. Anormal skin anatomy is shown in Fig.1 (a). The T stages of melanoma defined by
Cancer Research UK [35] to measure type and
size of primary tumor and the is shown in Fig.
1.
scales. The
Clarke and Breslow scale basically define the measure of the depth of the tumor i.e.
how deep the tumor has gone into the skin. Anormal skin anatomy is shown in Fig.1 (a). The T stages of melanoma defined by
Cancer Research UK [35] to measure type and
size of primary tumor and the is shown in Fig.
1.
FIGURE 1.
Anatomy of the skin (a) and T Stages of melanoma (b). Based on the depth of the primary tumor the stage of melanoma is identified. (Note: Depth and dimension of tumor may vary from case to case. Figure only intends to highlight the significance of tumor depth in melanoma diagnosis) (Source: Cancer Research UK).
In Fig. 1(b), “Tis” represents an initial stage of melanoma and the tumor is on the epidermis (i.e. top layer of the skin). The primary tumor is of size T1 if the depth is less than 1 mm and is still in the epidermis. Primary tumor is of size of T2 when it has grown into the dermis of the skin and its depth ranges from 1mm to 2mm. Size of the tumor is T3 if its measured depth is 2mm to 4 mm thick and is still localized to the dermis. When the growth depth of a primary tumor is greater than 4mm and is beyond the dermis then it is said to be of T4 size.Based on how far the cancer is spread and the size of the tumor melanoma cancer is classified into five stages [35], [36].
-
Stage 0:
It is the initial stage also referred to as
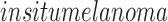 . Occurrences of abnormal melanocytes
are observed in the top layer of the skin. Melanoma detected in this
stage is 100% curable.
. Occurrences of abnormal melanocytes
are observed in the top layer of the skin. Melanoma detected in this
stage is 100% curable. -
Stage 1:
The tumor in this stage has spread into the skin but limited to the epidermis layer. No spread into the lymph or other parts of the body are detected. The tumor growth depth is between 1mm to 2mm and can exhibit ulceration (i.e. breakage of the skin). At this stage through surgical procedures the patients can be cured. Two sub classes i.e. 1A and 1B are considered based on the depth of the tumor.
-
Stage 2:
Melanoma tumor is 2mm to 4mm in size and can exhibit ulceration. No spread to lymph nodes or other parts of the body. Sub classes include 2A, 2B and 2C based on the depth and the ulceration. Cure is possible through surgical procedures.
-
Stage 3:
Tumor is more than 4mm deep and can exhibit ulceration. Cancer is spread to the lymph nodes but is still localized. Advance surgery and post-surgical care required. Survival rate is less. Sub classes include 3A, 3B and 3C.
-
Stage 4:
The tumor is more than 4mm deep and has spread to other organs and lymph nodes. Treatment at this stage is expensive and life threatening as the cancer has spread from its primary tumor site. Low survival rates amongst patients.
Based on the above discussion it is clear that the depth of the tumor is a critical parameter for diagnosis and identification of the cancer stage. Early detection of melanoma (Stage 0 and Stage 1) is the solution to reduce mortality rates amongst patients suffering from melanoma skin cancer. In the research work presented here a computerized dermoscopy system to aid early detection of melanoma is presented considering the 3D reconstruction of the lesion. The 3D reconstruction enables to estimate the relative depth of the primary tumor.
III. Literature Review
Identification of the skin lesion or region of interest in dermoscopic images is
achieved through segmentation procedures. In [15] type two fuzzy logic is used to determine the threshold for
segmentation of skin lesions. Illumination correction coupled with texture based
segmentation technique is presented in [16]. The Mimicking Expert Dermatologist’s Segmentation (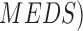 technique is
proposed in [17]. MEDS technique requires
low computation resources and provides accurate segmentation results that concur
with the segmentation carried out by dermatologists. A comparison considering
gradient vector flow, level set, expectation-maximization level set, adaptive
thresholding, fuzzy-based split-and-merge algorithm and adaptive snake segmentation
techniques is reported by Silveira et al. [18]. Of all the techniques considered the adaptive snake and the
expectation-maximization level set technique exhibit the best segmentation
performance. Adaptive snake segmentation technique is considered in the proposed
computerized dermoscopy system as low execution time and higher segmentation
accuracy is reported (when compared to the expectation-maximization level set
technique) in [18].
technique is
proposed in [17]. MEDS technique requires
low computation resources and provides accurate segmentation results that concur
with the segmentation carried out by dermatologists. A comparison considering
gradient vector flow, level set, expectation-maximization level set, adaptive
thresholding, fuzzy-based split-and-merge algorithm and adaptive snake segmentation
techniques is reported by Silveira et al. [18]. Of all the techniques considered the adaptive snake and the
expectation-maximization level set technique exhibit the best segmentation
performance. Adaptive snake segmentation technique is considered in the proposed
computerized dermoscopy system as low execution time and higher segmentation
accuracy is reported (when compared to the expectation-maximization level set
technique) in [18].
Researchers have suggested numerous works to aid early detection of melanoma considering dermoscopic images. A detailed survey can be obtained from [29].
In [21] an inspection system to identify Clark Nevi and Malignant Melanoma from pigmented skin lesions is discussed. Threshold based segmentation algorithm based on Otsu’s algorithm is considered. Shape color and texture features are extracted and binary classifiers are considered to identify the two classes of dermoscopic images considered.
In [26], using supervised mechanisms, lesion
features and maximum a posteriori (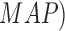 technique, a
segmentation technique is proposed. Hair removal techniques using Histograms of
Oriented Gradient (
technique, a
segmentation technique is proposed. Hair removal techniques using Histograms of
Oriented Gradient (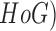 features is
also discussed. A classification mechanism is used to identify the presence of
pigment network in skin lesions. The classification is achieved considering Gaussian
and Laplacian of Gaussian (
features is
also discussed. A classification mechanism is used to identify the presence of
pigment network in skin lesions. The classification is achieved considering Gaussian
and Laplacian of Gaussian (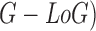 features.
features.
To identify two classes of skin lesions (malignant and benign) a computer-aided
diagnosis system is presented in [12].
Manual and automated segmentation procedures of skin lesions are discussed. Using
wavelet transforms texture features are extracted. Garnavi et al. [12] consider texture, shape and novel
boundary features in the spatial and frequency domains. Classification is achieved
using Support SVM, hidden naive Bayes (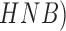 , random forest
tree (
, random forest
tree (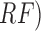 and logistic
model tree (
and logistic
model tree (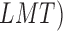 .
.
A Multi Parameter Extraction and Classification System (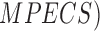 is proposed
to detect early melanoma or in situ melanoma in [24]. A six phase approach [23] is adopted to extract the color, texture
and shape features. Classification of three skin lesion types, namely
“Advanced Melanoma”, “Non-Melanoma”, “Early
Melanoma” is achieved through Neural Networks (
is proposed
to detect early melanoma or in situ melanoma in [24]. A six phase approach [23] is adopted to extract the color, texture
and shape features. Classification of three skin lesion types, namely
“Advanced Melanoma”, “Non-Melanoma”, “Early
Melanoma” is achieved through Neural Networks (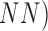 and
and 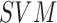 classifiers.
classifiers.
Sadeghi et al. [25] highlight the importance of detecting irregular streaks in dermoscopic images to accurately diagnose melanoma. Streak patterns, color and texture features are considered in the work presented. A simple logistic classifier is used to identify the absence/presence (regular and irregular) of streaks in dermoscopic images.
The significance of color features to classify skin lesions is put forth in [27]. A K-means clustering algorithm is incorporated to extract the color features. The Congenital nevi, combined nevi, Reed/Spitz nevi, melanomas, dysplastic nevi, blue nevi, dermal nevi, seborrheic keratosis and dermatofibroma lesion images are considered for evaluation. Using a symbolic regression algorithm the skin lesion are classified into benign or malignant types.
A. Saéz et al. [28] consider that each dermoscopic image represents a Markov model. The parameters estimated from the model are considered as the features of the skin lesion. Classification is performed to identify the globular, reticular and homogeneous patterns in the pigmented cell. Saéz et al. [28] have obtained the dermoscopic images from Interactive Atlas of Dermoscopy [37].
Based on high-level intuitive features (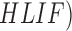 and
and 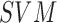 classifiers the
diagnosis of melanomas and non-melanoma skin lesions is presented in [20]. In addition to the
classifiers the
diagnosis of melanomas and non-melanoma skin lesions is presented in [20]. In addition to the 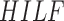 features,
low-level features and their combinations are also considered.
features,
low-level features and their combinations are also considered.
In [19] a novel equation to compute the
exposure time for skin to burn is introduced A threshold based segmentation, hair
detection and removal techniques is considered as the preprocessing steps in the
image analysis module. Shape, color and texture features are extracted to define the
skin lesion images. A two level 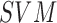 classifier is
used to identify the benign, atypical and melanoma moles from the
classifier is
used to identify the benign, atypical and melanoma moles from the 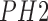 dataset [42].
dataset [42].
The importance of considering global and local features in computer aided diagnosis
methods is discussed in [22]. Use of color
and texture features (global and local) to identify melanoma and non-melanoma images
from the 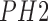 dataset is
presented. The use of,
dataset is
presented. The use of, 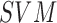 , AdaBoost and
BoF classifier is adopted for decision making.
, AdaBoost and
BoF classifier is adopted for decision making.
Based on the literature reviewed it is observed that limited work is carried out considering 3D reconstruction, depth estimation and 3D shape features of skin lesions which is critical to diagnose melanoma skin cancer. The state of art works carried out so far predominantly consider only binary decision making mechanisms. In this paper, authors consider 3D reconstruction of skin lesion images to estimate depth of the tumor and adopt multiclass decision making mechanisms.
IV. Proposed Computerized Dermoscopy System
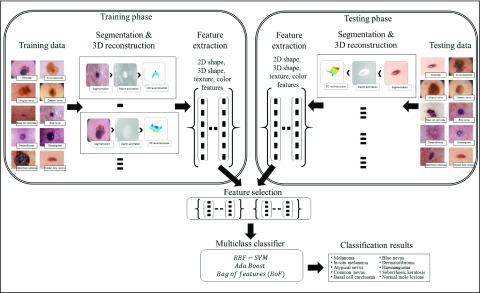
This section of paper presents the proposed computerized dermoscopy system. The main objective of the proposed system is to aid early detection of melanoma, especially in-situ melanoma. Additionally, the proposed system can also be adopted to diagnose different skin lesions types. Overview of the proposed system is shown in Fig. 2. The dermoscopic image dataset is considered to consist of training and testing data. Segmentation is performed obtain the region of interest or skin lesion to be diagnosed. A depth map is extracted from the 2D dermoscopic image. Depth map is used in constructing a 3D model corresponding to the dermoscopic image. The 3D model is represented as a structure tensor. A comprehensive feature set considering the 2D shape, 3D shape, color and texture are extracted per image. A feature selection method to understand the significance of features extracted on decision making is incorporated. For decision making, most of the related works consider binary classification mechanisms. The proposed system considers a multiclass classification mechanisms for decision making, enabling its applicability to diagnose a wide variety of skin lesion images. Dermoscopic images used for evaluation are obtained from CD-ROM of Dermoscopy [41] and PH2 dataset [42].
FIGURE 2.
Overview of the proposed computerized proposed computerized dermoscopy system for skin lesion classification.
A. Problem Formulation
Let 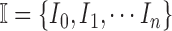 represent a set of
represent a set of 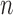 dermoscopic
images. Let
dermoscopic
images. Let 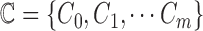 represent a set of
classes of the dermoscopic images i.e.
represent a set of
classes of the dermoscopic images i.e. 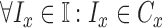 where
where 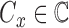 . The set
. The set 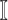 consists of images used for training and testing. Each image
consists of images used for training and testing. Each image 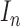 is
represented by a feature set
is
represented by a feature set 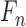 . The
training data is represented as
. The
training data is represented as 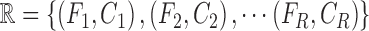 , where
, where 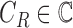 and
and 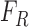 represents
the features of the
represents
the features of the 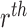 image
from the set
image
from the set 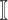 .
Similarly the testing data vector can be defined as
.
Similarly the testing data vector can be defined as 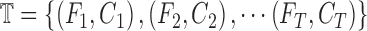 where
where 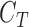 represent
the unknown classes and
represent
the unknown classes and 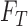 represent
a set of features extracted from the
represent
a set of features extracted from the 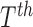 image
whose class is to be identified. Let
image
whose class is to be identified. Let 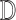 represent a decision making mechanism such that
represent a decision making mechanism such that 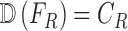 . The class identified i.e.
. The class identified i.e. 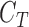 in
in 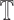 is
the diagnostic output of the proposed computerized dermoscopy system. The goal
of the proposed computerized dermoscopy system can be defined as
is
the diagnostic output of the proposed computerized dermoscopy system. The goal
of the proposed computerized dermoscopy system can be defined as
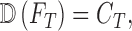 |
where 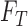 represents
the features of the image
represents
the features of the image 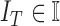 that needs to be diagnosed.
that needs to be diagnosed.
From (1) it is clear that a
robust feature extraction technique aiding the decision making mechanism 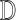 is
to be developed. The features extracted consider the 2D shape, 3D shape, color
and texture information of the skin lesions. Identification of the skin lesion
and 3D construction are also considered as the sub problems that need to be
solved.
is
to be developed. The features extracted consider the 2D shape, 3D shape, color
and texture information of the skin lesions. Identification of the skin lesion
and 3D construction are also considered as the sub problems that need to be
solved.
B. Segmentation
Dermoscopic images generally consists of normal skin and skin lesion segments.
Identification of the normal skin and skin lesion is critical to accurately
extract features. The skin lesions can be identified using segmentation
techniques. The proposed system considers an adaptive snake (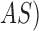 segmentation
technique to identify skin lesion regions in a set of images
segmentation
technique to identify skin lesion regions in a set of images  .
Literature presented in [18] proves the
accuracy and speed of the
.
Literature presented in [18] proves the
accuracy and speed of the 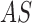 segmentation
technique. Skin texture variations, skin hair and specular reflections present
in dermoscopic images tend to induce spurious edges not belonging to the skin
lesion. Eliminating the spurious edges and accurate segmentation can be achieved
using the
segmentation
technique. Skin texture variations, skin hair and specular reflections present
in dermoscopic images tend to induce spurious edges not belonging to the skin
lesion. Eliminating the spurious edges and accurate segmentation can be achieved
using the 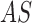 model. Based
on correlation matching in the
model. Based
on correlation matching in the 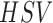 (Hue
SaturationValue) color of a skin image, intensity variations along radial
directions are identified as edges. Edges obtained are linked using a continuity
criteria to form a contour segment set. Subset of the contour segments also
known as snakes are approximated using an estimation algorithm [39] to obtain the skin lesion segment.
The regions depicting the variations of color (skin region and lesion region)
are manually selected by the user.
(Hue
SaturationValue) color of a skin image, intensity variations along radial
directions are identified as edges. Edges obtained are linked using a continuity
criteria to form a contour segment set. Subset of the contour segments also
known as snakes are approximated using an estimation algorithm [39] to obtain the skin lesion segment.
The regions depicting the variations of color (skin region and lesion region)
are manually selected by the user.
Let’s consider a boolean set 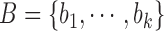 associated with
associated with 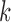 number of
contour segments identified in an image
number of
contour segments identified in an image 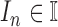 . The number of contour segment identified are
considered as features of
. The number of contour segment identified are
considered as features of 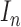 . The
contour segment set is denoted as
. The
contour segment set is denoted as 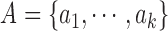 . The counter model consisting of
. The counter model consisting of 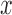 points is
defined as
points is
defined as 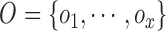 . The approximation of
. The approximation of 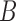 or a subset of
or a subset of 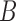 , by the model
, by the model 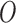 is defined as
is defined as
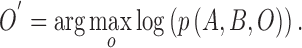 |
The approximation 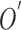 is achieved through the approximate a posteriori criterion. Equation (2) can be solved using the
is achieved through the approximate a posteriori criterion. Equation (2) can be solved using the 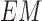 algorithm
[39]. According to the
algorithm
[39]. According to the 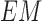 algorithm,
algorithm, 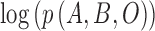 can be substituted with its
expected value with respect to the unknown variable
can be substituted with its
expected value with respect to the unknown variable  in (2).The substitution is defined
as
in (2).The substitution is defined
as
 |
where  represents
the energy of the
represents
the energy of the  contour
and the best estimate of
contour
and the best estimate of 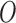 is
is 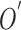 . Let
. Let 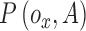 represent a potential function defined as
represent a potential function defined as
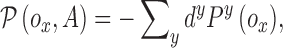 |
where 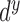 ,
, 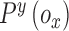 represents the confidence degree and potential
function of the
represents the confidence degree and potential
function of the 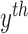 contour
segment. The confidence degree
contour
segment. The confidence degree 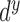 is a
probability
is a
probability 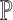 , that
the
, that
the 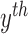 contour
segment is valid or true i.e.
contour
segment is valid or true i.e. 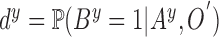
Based on the substitutions presented in [40] and (4), 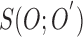 can be simplified and presented as
can be simplified and presented as
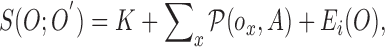 |
where 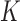 represents a
constant and
represents a
constant and 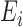 is the
internal energy.
is the
internal energy.
Potential 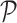 is
also referred to as an adaptive potential due to the varying nature its exhibits
during estimation. Estimation is performed based on a user specified number of
iterations. Adetailed explanation of the
is
also referred to as an adaptive potential due to the varying nature its exhibits
during estimation. Estimation is performed based on a user specified number of
iterations. Adetailed explanation of the 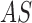 segmentation
model is found in [40].
segmentation
model is found in [40].
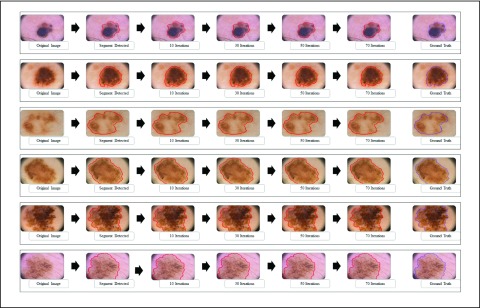
The 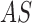 segmentation
results obtained considering dermoscopic images obtained from the PH2 dataset is
shown in Fig. 3. The number of iterations
is set to 70. A dermoscopic image is considered as the input. The normal skin
region and lesion region (center of the skin lesion) is selected manually. The
major segment (largest contour segment) detected is shown in the second column
of Fig 3. Adaption of the snake at 10,
30, 50 and 70 iterations is shown in column three to six. The ground truth
(provided in the PH2 dataset) is shown in the last column. The segmentation
accuracy is evident from the results shown in Fig 3. Results considering skin lesions extending beyond the input
image is also shown. In such cases the image boundary is considered as the
boundary of the segmented skin lesion.
segmentation
results obtained considering dermoscopic images obtained from the PH2 dataset is
shown in Fig. 3. The number of iterations
is set to 70. A dermoscopic image is considered as the input. The normal skin
region and lesion region (center of the skin lesion) is selected manually. The
major segment (largest contour segment) detected is shown in the second column
of Fig 3. Adaption of the snake at 10,
30, 50 and 70 iterations is shown in column three to six. The ground truth
(provided in the PH2 dataset) is shown in the last column. The segmentation
accuracy is evident from the results shown in Fig 3. Results considering skin lesions extending beyond the input
image is also shown. In such cases the image boundary is considered as the
boundary of the segmented skin lesion.
FIGURE 3.
The adaptive snake (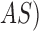 segmentation technique for 6 skin lesion images. The original image
is shown in the first column, the major segment identified is shown
in the second column. Intermediate images obtained at 10, 30 and 50
iterations are shown in column 3 to 5. The segmentation result (at
70 iterations) and the ground truth obtained is shown in the last
two columns.
segmentation technique for 6 skin lesion images. The original image
is shown in the first column, the major segment identified is shown
in the second column. Intermediate images obtained at 10, 30 and 50
iterations are shown in column 3 to 5. The segmentation result (at
70 iterations) and the ground truth obtained is shown in the last
two columns.
C. 3D Lesion Surface Reconstruction
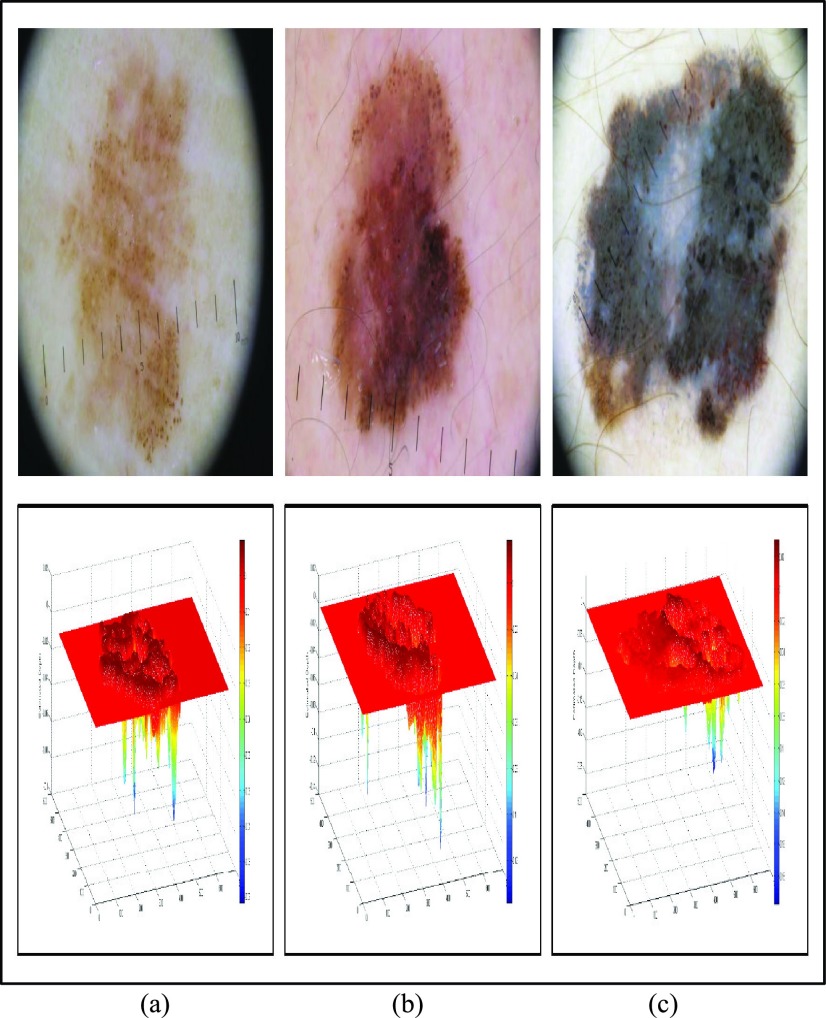
3D reconstruction is essential to estimate depth of the skin lesions. Techniques like stereo vision, structure from motion, depth from focus, depth from defocus etc. are used to estimate depth considering multiple images. Using constrained image acquisition techniques like active illumination and coded aperture method’s, depth can be estimated using single images [34]. The varying or unknown dermoscopic data acquisition parameters/settings used and the non-availability of multiple images render these mechanisms ineffective. In [34] a novel technique to estimate depth, considering a single image obtained from unconstrained image data acquisition techniques is described. The proposed computerized dermoscopy system adopts this technique to estimate the depth in dermoscopic images. Depth map obtained is fit to the underling 2D surface to enable 3D surface reconstruction. The 3D surface constructed is represented as structure tensors. The 3D surface reconstruction results considering two melanoma and one blue nevus skin lesion images is shown in Fig. 4.
FIGURE 4.
The 3D lesion surface reconstruction technique. The original image is
shown in column 1. The edge map used to compute the defocus is shown
in column 2. Sparse and the resultant depth map is shown in column
3-4. The structure tensor 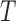 representing the 3D lesion surface is shown in the last column. (a)
3D surface reconstruction results for melanoma image obtained from
ATLAS dataset. (b) 3D surface reconstruction results for melanoma
image obtained from PH2 dataset.. (c) 3D surface reconstruction
results for blue nevus image from ATLAS dataset.
representing the 3D lesion surface is shown in the last column. (a)
3D surface reconstruction results for melanoma image obtained from
ATLAS dataset. (b) 3D surface reconstruction results for melanoma
image obtained from PH2 dataset.. (c) 3D surface reconstruction
results for blue nevus image from ATLAS dataset.
1). Depth Map Construction
The depth of the skin is computed using the estimated defocus occurrence at
edges. Input skin lesion images lesions (shown in column 1 of Fig. 4) are reblurred using a Gaussian
function. Defocus occurrences at edges (represented as edge map column 2 of
Fig.4) is obtained, as a ratio
between the gradient magnitude of the input skin lesion image and its
reblurred version. Propagating the blur observed at edges to the whole skin
lesion image, enables in computing depth maps. An ideal step edge model
considering the edge is located at 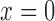 is
defined as
is
defined as
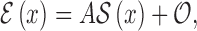 |
where 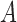 ,
, 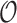 represent the amplitude and offset.
represent the amplitude and offset. 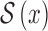 represents the step function.
represents the step function.
Defocus blur is obtained by convolving the sharp input skin lesion image with
a point spread function. A Gaussian function 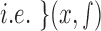 is used to approximate the point spread
function. Standard deviation
is used to approximate the point spread
function. Standard deviation 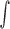 , in the Gaussian function is directly
proportional to the circle of confusion
, in the Gaussian function is directly
proportional to the circle of confusion 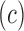 and it
is defined as
and it
is defined as  [34]. The blurred edge
[34]. The blurred edge 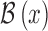 obtained using the edge model and Gaussian
function is defined as
obtained using the edge model and Gaussian
function is defined as 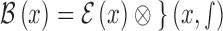 . Let
. Let 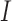 represent
an input skin lesion image. Reblurring of the input skin lesion image is
achieved using a two dimensional isotropic Gaussian function and it is
represented as
represent
an input skin lesion image. Reblurring of the input skin lesion image is
achieved using a two dimensional isotropic Gaussian function and it is
represented as 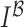 . Gradient magnitude along the
. Gradient magnitude along the 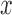 and
and 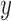 directions
of the input image is defined as
directions
of the input image is defined as
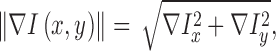 |
where 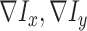 represent the gradients along
represent the gradients along 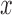 and
and 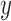 directions.
directions.
Gradient magnitude of the blurred image is computed in a similar fashion. The
gradient magnitude ratio between edge locations (in the 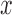 direction)
of
direction)
of  and
and 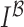 is
is 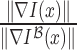 . A sparse depth map
. A sparse depth map 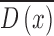 is constructed by estimating the blur scale
occurrences at each edge location. Inaccurate blur estimates at certain edge
locations are eliminated by using a joint bilateral filter and the input
skin lesion image as a reference. The resulting sparse depth map (shown in
column 3 of Fig.4.)
is constructed by estimating the blur scale
occurrences at each edge location. Inaccurate blur estimates at certain edge
locations are eliminated by using a joint bilateral filter and the input
skin lesion image as a reference. The resulting sparse depth map (shown in
column 3 of Fig.4.) 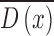 is then used to obtain the full depth map
is then used to obtain the full depth map 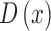 . The full depth map
. The full depth map 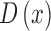 is obtained by propagating the defocus blur
estimates obtained at edge locations to the entire skin lesion image. Image
interpolation technique based on matting Laplacian technique [44] is used to obtain the full depth
map. The depth maps obtained is shown in column 4 of Fig.4. The optimal depth map
is obtained by propagating the defocus blur
estimates obtained at edge locations to the entire skin lesion image. Image
interpolation technique based on matting Laplacian technique [44] is used to obtain the full depth
map. The depth maps obtained is shown in column 4 of Fig.4. The optimal depth map 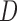 is
obtained by solving the following equation
is
obtained by solving the following equation
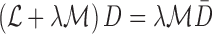 |
where 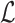 ,
, 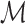 ,
, 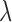 represent the matting Laplacian, diagonal matrix and scalar balance factor.
represent the matting Laplacian, diagonal matrix and scalar balance factor. 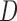 and
and 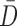 are
the vector representations of the full depth map and sparse depth maps.
are
the vector representations of the full depth map and sparse depth maps.
2). Tensor Structure Representation of Lesion Surface
Three dimensional legion surface 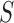 is
represented as
is
represented as 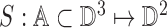 . Where
. Where 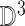 is the three dimensional space in which
is the three dimensional space in which 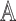 lies. A point
lies. A point 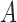 is
represented as
is
represented as 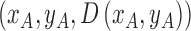 where
where 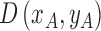 represents the depth map. The legion
reconstruction is achieved as a gradient descent of the depth map based
energy function. Nonlinear partial differential equations (
represents the depth map. The legion
reconstruction is achieved as a gradient descent of the depth map based
energy function. Nonlinear partial differential equations ( are
used to represent the gradient descent. A heat equation is used to implement
the gradient descent. Legion reconstruction using the heat equation can be
defined as
are
used to represent the gradient descent. A heat equation is used to implement
the gradient descent. Legion reconstruction using the heat equation can be
defined as
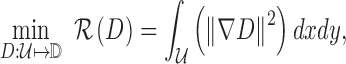 |
where 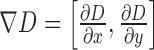 represents gradient of the depth map and
represents gradient of the depth map and 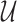 is the image domain.
is the image domain.
Perona and Malik [43] have shown
that (9) results in smooth
surfaces. To preserve lesion edges and attain smooth surfaces, Perona and
Malik [43] introduced nonlinear
anisotropic 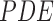 to
implement the gradient descent defined as
to
implement the gradient descent defined as
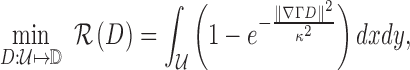 |
where 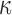 is the edge preservation constant and
is the edge preservation constant and 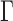 is the diffusion tensor.
is the diffusion tensor.
Using gradient decent minimization, (10) is solved in a limited number of
iterations. Let 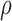 represents a tangential space obtained from the depth map. The 3D lesion
reconstructed is considered as a structure tensor
represents a tangential space obtained from the depth map. The 3D lesion
reconstructed is considered as a structure tensor 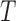 defined as
defined as
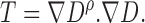 |
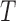 represents
a 3D lesion structure constructed, the corresponding coordinate based
eigenvectors represent minimum and maximum gradient directions. Diffusion
tensors are represented using eigenvalues and eigenvectors. Eigenvalues
represent magnitude of the gradient observed in the depth map. Eigenvectors
define direction of the gradients. The diffusion tensor
represents
a 3D lesion structure constructed, the corresponding coordinate based
eigenvectors represent minimum and maximum gradient directions. Diffusion
tensors are represented using eigenvalues and eigenvectors. Eigenvalues
represent magnitude of the gradient observed in the depth map. Eigenvectors
define direction of the gradients. The diffusion tensor 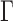 is defined as
is defined as
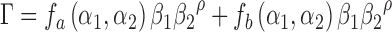 |
where 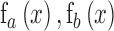 represent functions incorporated to capture the gradient deviations.
represent functions incorporated to capture the gradient deviations.
The structure tensor obtained is used to compute 3D skin lesion features. The
3D skin lesion reconstructed i.e. 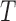 is shown
in the last column of Fig.4.
is shown
in the last column of Fig.4.
D. Feature Extraction
Characteristics of the skin lesion images are represented as features. In this paper color, texture, 2D shape and 3D shape features are considered. Accurate and robust feature representation is essential as they directly affect the performance of the skin lesion classification.
1). Color Feature Extraction
Color characteristics are often used by dermatologists to classify skin
lesions [22]. According to
dermatologists melanoma skin lesions are characterized by variegated
coloring [45]. The variegated
coloring induces high variance in the red, green and blue color space. Red,
green and blue component data of the pixels in the segmented skin lesion is
stored as vectors. The mean 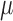 and
variance
and
variance 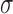 of
each channel is computed. Mean, variance computed is represented as
of
each channel is computed. Mean, variance computed is represented as 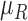 ,
, 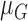 ,
, 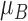 and
and 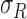 ,
, 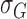 ,
, 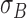 .To capture complex non-uniform color
distributions within the skin lesion, mean ratios of the mean values is
computed i.e.
.To capture complex non-uniform color
distributions within the skin lesion, mean ratios of the mean values is
computed i.e. 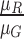 ,
, 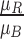 ,
, 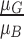 . Variations in color of the skin lesion
with respect to the surrounding skin is also considered as color features.
These features are represented as
. Variations in color of the skin lesion
with respect to the surrounding skin is also considered as color features.
These features are represented as 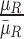 ,
, 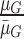 ,
, 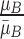 , where
, where 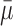 represents the mean value of surrounding/normal skin region.
represents the mean value of surrounding/normal skin region.
2). Texture Feature Extraction
To extract the texture features the segmented skin lesion image is converted
to grey scale. Haralick-features [46] are adopted to obtain the texture characteristics of the
skin lesion. Considering applicability of the proposed computerized
dermoscopy system to classify even low quality skin lesion images, Haralick
texture features is considered [21]. Texture features are computed using gray-tone
spatial-dependence matrices i.e. 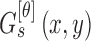 .The angle of the
spatial neighborhood
.The angle of the
spatial neighborhood 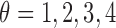 . The matrix denotes the number of the grey
tomes of
. The matrix denotes the number of the grey
tomes of 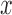 and
and 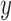 that are
spatial neighbors. The matrix
that are
spatial neighbors. The matrix 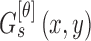 is computed at
0ř, 45ř, 90ř and 135ř degrees. The energy
feature is computed using
is computed at
0ř, 45ř, 90ř and 135ř degrees. The energy
feature is computed using
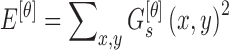 |
The homogeneity texture feature is computed using
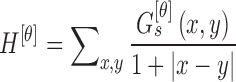 |
The contrast feature is defined as
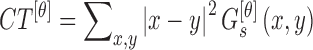 |
Mean (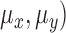 and standard deviations (
and standard deviations (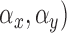 of the matrix
of the matrix  considering gray tones of
considering gray tones of 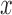 and
and 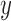 are
computed. Using mean and standard deviation the correlation feature is
computed as
are
computed. Using mean and standard deviation the correlation feature is
computed as
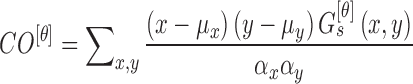 |
The mean values of 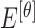 ,
, 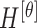 ,
, 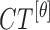 ,
, 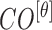 , represented as
, represented as 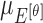 ,
, 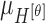 ,
, 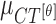 and
and 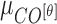 are considered as additional texture
features.
are considered as additional texture
features.
3). 2D Shape Feature Extraction
Shape, border and asymmetry features are considered as 2D shape features in
the proposed computerized dermoscopy system. A total of eleven 2D shape
features are extracted from the segmented skin lesion images. Area 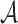 of a skin lesion is defined as the number of pixels present in the lesion.
Perimeter shape feature
of a skin lesion is defined as the number of pixels present in the lesion.
Perimeter shape feature 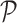 is a count of the number of pixels on the segmented skin lesion boundary.
Let
is a count of the number of pixels on the segmented skin lesion boundary.
Let 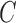 represent
the segmented skin lesion centroid. Length of a line that connects two
furthest boundary points passing through
represent
the segmented skin lesion centroid. Length of a line that connects two
furthest boundary points passing through 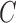 is the
greatest diameter
is the
greatest diameter 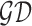 . Length of a line connecting closest lesion
boundary points and passing through
. Length of a line connecting closest lesion
boundary points and passing through 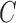 is
considered as the shortest diameter
is
considered as the shortest diameter 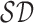 shape feature. Using area i.e.
shape feature. Using area i.e. 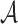 and perimeter i.e.
and perimeter i.e. 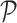 of a skin lesion, the circularity index features computed are
of a skin lesion, the circularity index features computed are 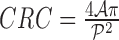 ,
, 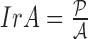 ,
, 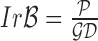 ,
, 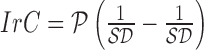 and
and 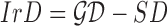 . Circularity index features computed
quantify irregularity. Major, minor axis lengths and asymmetry index
features are computed in accordance to [47].
. Circularity index features computed
quantify irregularity. Major, minor axis lengths and asymmetry index
features are computed in accordance to [47].
4). 3D Shape Feature Extraction
The maximum, minimum and average or relative depth feature is extracted from the 3D skin lesion reconstructed. In addition seven Hu invariants [48] and three affine moment invariants [49] are adopted to characterize 3D shape features of the skin lesion.
For a skin lesion image 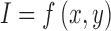 of size
of size 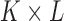 ,
the
,
the 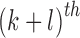 order geometric moment is defined as
order geometric moment is defined as
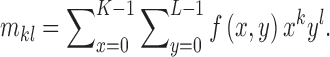 |
The 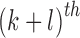 order central moment is
order central moment is
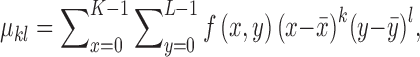 |
where 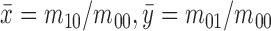 is the center of
gravity of an image
is the center of
gravity of an image 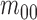 .
Considering intensity images,
.
Considering intensity images, 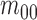 represents it quality. The moments
represents it quality. The moments 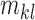 and
and 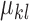 are used to represent the shape of the image.
are used to represent the shape of the image.
Normalization of the central moments of higher orders using the  central moment is defined as
central moment is defined as
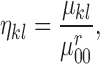 |
where  and
and  .
.
Using (19) the Hu’s
moment invariants 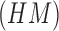 are computed as
are computed as
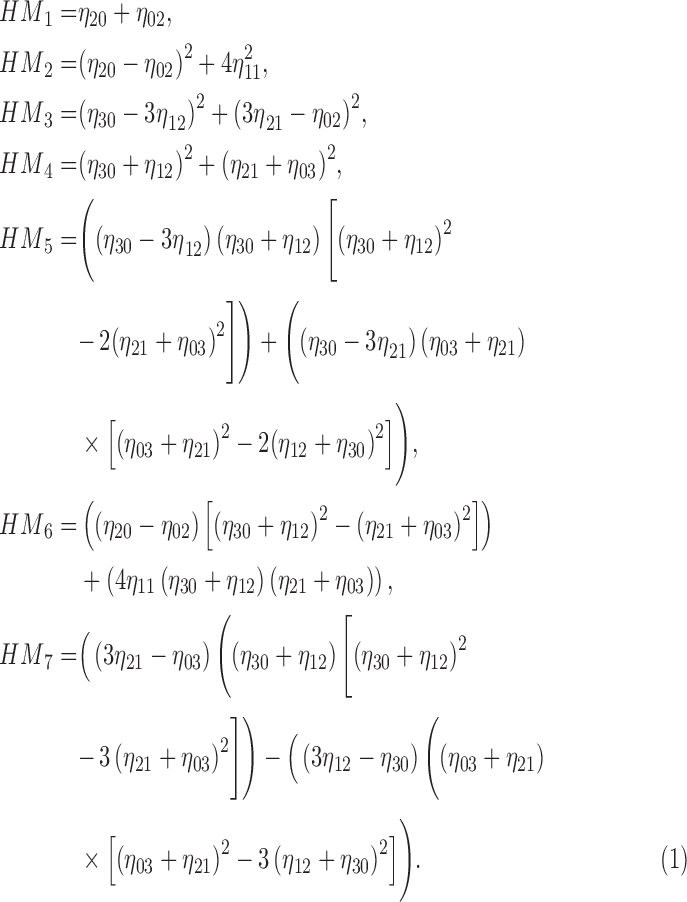 |
To provide additional 3D shape features, affine moment invariants of the first, second and third order are considered. According to [49] the features affine moment invariants are defined as
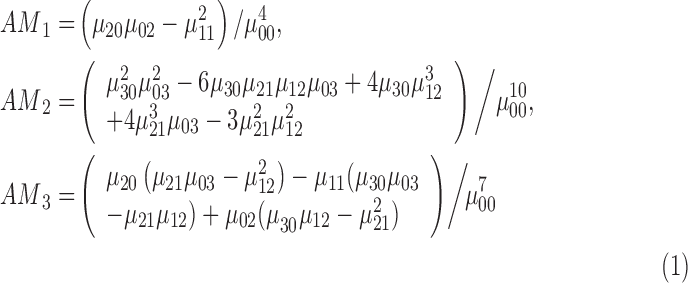 |
E. Feature Selection
Feature selection, generally is identifying an optimized subset of features
extracted that imparts highest discriminating power to the decision making
mechanism adopted. In the proposed computerized dermoscopy system color 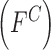 , texture
, texture 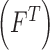 , 2D shape
, 2D shape 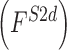 , and 3D shape
, and 3D shape 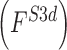 features of skin lesion images are extracted.
Apart from imparting discriminating power, feature selection is adopted to study
the impact of color features, texture features, 2D shape features, 3D shape
feature and their combinations to classification of skin lesions.
features of skin lesion images are extracted.
Apart from imparting discriminating power, feature selection is adopted to study
the impact of color features, texture features, 2D shape features, 3D shape
feature and their combinations to classification of skin lesions.
The feature set is defined as 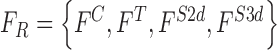 . A heuristic approach
is adopted to obtain the optimized feature set
. A heuristic approach
is adopted to obtain the optimized feature set 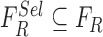 . Optimized feature set is constructed
considering different combinations of the features extracted. Resulting
performance enable in understanding the significance of features considered on
the classification system. Experimental study discussed in the subsequent
section considers four optimized feature set combining the features extracted.
The optimized feature sets considered for evaluation are defined as
. Optimized feature set is constructed
considering different combinations of the features extracted. Resulting
performance enable in understanding the significance of features considered on
the classification system. Experimental study discussed in the subsequent
section considers four optimized feature set combining the features extracted.
The optimized feature sets considered for evaluation are defined as
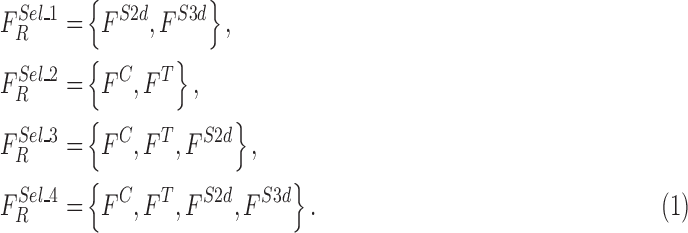 |
F. Classification
Skin lesion classification is the final step of proposed computerized dermoscopy
system. In the research work presented here, three different classes of
classifiers i.e. SVM [50], [51], AdaBoost [52] and the recently developed bag-of-features (BoF)
[53], [54] classifiers are adopted. The classifiers adopted are
also referred to as decision making mechanisms  .
Classification broadly involves two phases namely training and testing.
.
Classification broadly involves two phases namely training and testing.
In the training phase the classifiers learn from the training set  .
Feature properties with respect to the classes are derived in the training
phase. In the testing phase we wish to classify test data
.
Feature properties with respect to the classes are derived in the training
phase. In the testing phase we wish to classify test data  .
Based on the feature properties observed in training, the decision making
mechanisms
.
Based on the feature properties observed in training, the decision making
mechanisms  classifies a test image
classifies a test image 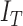 represented by feature set
represented by feature set 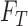 as the
resultant class
as the
resultant class 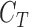 .
.
Skin lesion data is complex in nature and cannot be considered as a global model.
In the BoF decision making mechanism, skin lesion data is considered as a
combination of individual feature models rather than the complete feature set 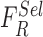 . The
BoF classifier exhibits promising results when adopted for complex image
analysis [53], [54]. Therefore, the BoF classifier was deemed applicable
to solve our skin lesion classification problem [22].
. The
BoF classifier exhibits promising results when adopted for complex image
analysis [53], [54]. Therefore, the BoF classifier was deemed applicable
to solve our skin lesion classification problem [22].
The capability to train a strong classifier from a combination of weak classifiers and appropriate feature selection capabilities exhibited by the AdaBoost algorithm motivated the authors to consider its inclusion in the proposed system [22].
SVM classifiers are robust, simple to implement and provide high degree of classification accuracy [50]. Recent works for skin lesion classification [12], [19], [20], [22] prove the applicability of SVM classifiers for decision making. A Gaussian radial basis function (RBF) kernel is considered in the proposed computerized dermoscopy system. The RBF kernel assists in deriving complex relations between the skin lesion classes and complex nonlinear skin lesion data represented as a feature vector space. A linear kernel is a special case of the RBF kernel [55], hence the authors have considered to adopt a RBF kernel in the SVM classifier.
V. Experimental Study and Discussions
In this section experimental studies conducted to evaluate performance of the proposed computerized dermoscopy system is presented. The proposed system was implemented on MATLAB. The dermoscopy data used in the experiments, experiment details, performance of the three classifiers proposed, comparisons with existing systems and the experiments based on the 3D reconstruction algorithm proposed for depth estimation is discussed.
A. Data
Data to evaluate the performance of the proposed dermoscopy system is acquired from two sources. The datasets used are summarized in Table I.
TABLE 1. Dataset Details Used for Experiments.
The PH2 database of 200 dermoscopic images [42] from Pedro Hispano hospital is considered. Four classes i.e. common nevus, atypical nevus, melanoma, in-situ melanoma (lentigo melanoma) are considered. All images in the PH2 dataset are 8-bit RGB color images. The PH2 dataset is also used to evaluate the performance in [19], [22], and [57]–[59].
The second dataset is obtained from Atlas of Dermoscopy CD published alongside [41]. Comprehensive skin lesion data of varied types with analysis from expert dermatologists is provided in the ATLAS. The authors created a custom dataset of varied skin lesion types. Atotal of 63 24-bit RGB color dermoscopic images are selected. TheATLAS dataset consists of a comprehensive set of skin lesion images rendering it more practical to evaluate the proposed computerized dermoscopy system. The ATLAS dataset created is complex and 8 type of skin lesions are considered.
B. Experiment Details
Performance of the proposed system with each classifier individually is evaluated
considering the feature selection 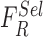 combinations defined in (22).
Training and testing data used in the experiments is obtained in accordance to
the procedure described in [22]. A
total of 4 experiments per classifier per dataset is carried out. Experiment
details and the notations used to represent them is described in Table II. Leave-one-out approach [20], [22] is adopted for testing due to the limited size of the datasets
available. To evaluate performance a cost function is derived based on the
confusion matrix obtained. Overall classification accuracy
combinations defined in (22).
Training and testing data used in the experiments is obtained in accordance to
the procedure described in [22]. A
total of 4 experiments per classifier per dataset is carried out. Experiment
details and the notations used to represent them is described in Table II. Leave-one-out approach [20], [22] is adopted for testing due to the limited size of the datasets
available. To evaluate performance a cost function is derived based on the
confusion matrix obtained. Overall classification accuracy 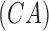 considering all skin lesion classes (i.e. 4
classes for PH2 and 8 classes for ATLAS dataset) is computed using the confusion
matrix. A tradeoff between specificity
considering all skin lesion classes (i.e. 4
classes for PH2 and 8 classes for ATLAS dataset) is computed using the confusion
matrix. A tradeoff between specificity 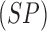 and
sensibility
and
sensibility 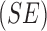 exists hence Barata [22] have introduced as cost function
exists hence Barata [22] have introduced as cost function 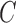 for evaluating
performance.
for evaluating
performance.
TABLE 2. Experiment Details Using Varied Feature Types and Datasets.
| Exp. No | Dataset used | Features selected | Notation |
|---|---|---|---|
| 1 | PH2 [42] | 2D shape, 3D shape | D1EX1 |
| 2 | PH2 [42] | Color, Texture | D1EX2 |
| 3 | PH2 [42] | Color, Texture, 2D shape | D1EX3 |
| 4 | PH2 [42] | Color, Texture, 2D shape, 3D shape | D1EX4 |
| 5 | ATLAS [41] | 2D shape, 3D shape | D2EX1 |
| 6 | ATLAS [41] | Color, Texture | D2EX2 |
| 7 | ATLAS [41] | Color, Texture, 2D shape | D2EX3 |
| 8 | ATLAS [41] | Color, Texture, 2D shape, 3D shape | D2EX4 |
The cost function 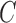 is defined as
is defined as
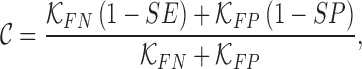 |
where 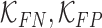 are constants and
are constants and 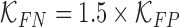 . Constants
. Constants 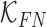 and
and 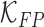 represent the false negative
represent the false negative 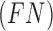 and false positive
and false positive 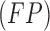 costs. In the experimental results presented
costs. In the experimental results presented 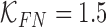 and
and 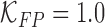 is considered.
is considered.
C. Assessment of BoF Classifier on Experiments
The BoF classifier considers a block size of 50 and the number of histogram bins
is set to 25. The k-means clustering algorithm is adopted to obtain visual
words. A total of 500 visual words is considered. Classification is achieved
using the k-Nearest Neighbor 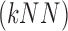 classifier. The
classifier. The  employed,
considers Euclidean distance and the number of neighbors is set to 10. Results
obtained in this study is summarized in Table III.
employed,
considers Euclidean distance and the number of neighbors is set to 10. Results
obtained in this study is summarized in Table III.
TABLE 3. Experimental Results Considering BoF Classifier.
| Exp # | 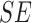 |
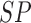 |
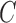 |
|---|---|---|---|
| D1EX1 | 91% | 94% | 0.079 |
| D1EX2 | 91% | 94% | 0.077 |
| D1EX3 | 93% | 96% | 0.062 |
| D1EX4 | 90% | 94% | 0.079 |
| D2EX1 | 76% | 95% | 0.164 |
| D2EX2 | 67% | 95% | 0.216 |
| D2EX3 | 77% | 96% | 0.153 |
| D2EX4 | 72% | 96% | 0.187 |
Considering PH2 dataset best performance is reported considering color, texture
and 2D shape features (D1EX3). Inclusion of 2D shape features to texture and
color exhibits a 19.4% reduction in the value of cost function 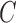 .
.
Results considering ATLAS dataset show that color and texture information alone considered in D2EX2 is insufficient to classify skin lesions. Considering shape features (2D and/or 3D) improves performance of the BoF classifier observed in D2EX1, D2EX3 and D2EX4. On the ATLAS dataset best performance is reported in D2EX3.
Classification using PH2 dataset exhibits better performance than the ATLAS dataset. This observation is due to the fact that limited training data and numerous skin lesion types are considered in the ATLAS dataset enhancing complexity. To overcome this drawback we used two additional classifiers discussed in the latter section of the paper. A noteworthy observation is that classification using the shape descriptors (i.e. D1EX1, D2EX1) exhibits similar performance to color and texture features (i.e. D1EX2, D2EX2). Inclusion of shape features to color and texture improve performance considering the BoF classifier.
D. Assessment of AdaBoost Classifier on Experiments
The AdaBoost classifier considered is built using 10 weak classifiers. Number of
bins is set to 50. This configuration is established based on a number of
iterations to obtain best performance. The classification results obtained is
shown Table IV. Considering PH2
dataset the AdaBoost classifier exhibits better results when compared to the BoF
classifier. It must be noted that in D1EX2 we report 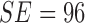 ,
, 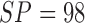 similar to
the values observed in [22] that
reports
similar to
the values observed in [22] that
reports 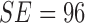 ,
, 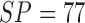 for color
and texture features considering AdaBoost classifier. In D1EX3 and D2EX4,
AdaBoost exhibits best classification results considering experiments conducted
on the datasets. Performance of the AdaBoost classifier improves on considering
the proposed 3D shape features for the ATLAS dataset. The AdaBoost classifier
exhibits better performance considering PH2 dataset when compared to ATLAS
dataset. AdaBoost classifier does not achieve acceptable performance on the
ATLAS dataset even if the number of weak classifiers are increased. A similar
observation is also reported in [22].
To overcome this drawback and improve performance across varied datasets the
authors have considered the SVM classifier.
for color
and texture features considering AdaBoost classifier. In D1EX3 and D2EX4,
AdaBoost exhibits best classification results considering experiments conducted
on the datasets. Performance of the AdaBoost classifier improves on considering
the proposed 3D shape features for the ATLAS dataset. The AdaBoost classifier
exhibits better performance considering PH2 dataset when compared to ATLAS
dataset. AdaBoost classifier does not achieve acceptable performance on the
ATLAS dataset even if the number of weak classifiers are increased. A similar
observation is also reported in [22].
To overcome this drawback and improve performance across varied datasets the
authors have considered the SVM classifier.
TABLE 4. Experimental Results Considering AdaBoost Classifier.
| Exp # | 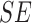 |
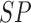 |
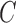 |
|---|---|---|---|
| D1EX1 | 94% | 97% | 0.049 |
| D1EX2 | 96% | 98% | 0.029 |
| D1EX3 | 96% | 98% | 0.028 |
| D1EX4 | 94% | 97% | 0.049 |
| D2EX1 | 40% | 92% | 0.394 |
| D2EX2 | 35% | 90% | 0.426 |
| D2EX3 | 44% | 92% | 0.369 |
| D2EX4 | 47% | 92% | 0.349 |
E. Assessment of SVM Classifier on Experiments
The BoF and AdaBoost classifier exhibit promising results considering PH2 dataset. On evaluation with the complex ATLAS dataset these classifiers exhibit low performance. Authors have adopted a RBF kernel in the SVM classifier to overcome this drawback and achieve acceptable performance on both PH2 and ATLAS datasets. The LIBSVM software available in [56] is used for the experimental study. Results obtained considering the RBF-SVM classifier is shown in Table V.
TABLE 5. Experimental Results Considering SVM Classifier.
| Exp # | 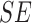 |
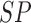 |
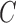 |
|---|---|---|---|
| D1EX1 | 79% | 86% | 0.184 |
| D1EX2 | 29% | 78% | 0.518 |
| D1EX3 | 49% | 86% | 0.36 |
| D1EX4 | 96% | 97% | 0.038 |
| D2EX1 | 80% | 97% | 0.136 |
| D2EX2 | 31% | 91% | 0.448 |
| D2EX3 | 59% | 94% | 0.271 |
| D2EX4 | 98% | 99% | 0.013 |
Considering PH2 dataset, the best performance is reported in D1EX4 which outperforms the results presented in [19] and [22]. A marked improvement in performance is reported on the ATLAS dataset considering the SVM classifier.
Results obtained prove that the SVM classifier exhibits better generalization performance on increasing the feature vector when compared to the other classifiers. Observe results of D1EX1, D1EX4 against D1EX2 and D1EX3 in Table V. A marked performance improvement considering the proposed 3D shape feature inclusion is reported on PH2 dataset. Similar performance improvement is reported considering ATLAS dataset. Results of D2EX1, D2EX4 against D2EX2 and D2EX3 in Table V prove the performance improvement. In [20] it is stated that performance of the SVM is directly dependent on the features extracted i.e. to project data into separable feature space. Based on the results presented it can be concluded that the 3D shape feature extracted improve classification performance (Refer D1EX1, D1EX4, D2EX1 and D2EX4 in Table V). The SVM classifier considered exhibits better performance in comparison to the AdaBoost and BoF classifier.
F. Short Note on Comparisons With Similar State of Art Systems
A tentative comparison with the other state of art systems is presented here even if we have not considered similar features and datasets.
Using the asymmetry, border, color, texture features on the Dermat dataset in [21] an accuracy of 86%, SE = 94% and SP = 68% is reported. Combining geometry, texture, border features on a custom dataset an accuracy of 91.26% is reported in [12]. Considering high level, low level features on datasets created from DermQuest, Dermatology Information System the highest accuracy of 83.59% (SE = 91.01%, SP = 73.45%) is reported by Amelard et al. [20].
Considerable amount of research work is carried out using the PH2 dataset. Using
color and texture features in [22] SE
= 98%, SP = 79% and 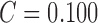 is
reported. Using color features alone the best performance of SE =
100%, SP = 75% and
is
reported. Using color features alone the best performance of SE =
100%, SP = 75% and 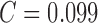 is
reported in [22]. Automated skin lesion
analysis system developed by Abuzaghleh et al. [19] reports a classification accuracy of melanoma, benign, atypical
lesions as 97.5%, 95.7%, and 96.3%. Abuzaghleh et al. [19] report a SE = 97.6%,
SP = 90.5% and average accuracy of 96.5 in [57]. Barata et al. [58] report performance measures of SE = 98%, SP
= 90% on PH2 dataset and SE = 83%, SP =
76% on EDRA datasets considering a fusion of features. A recent work
[59], introduces sparse coding of
the Scale-Invariant Feature Transform (SIFT) features for melanoma
classification. Reference [59] reports
a performance of SE = 100%, SP = 90.3% on the PH2
dataset.
is
reported in [22]. Automated skin lesion
analysis system developed by Abuzaghleh et al. [19] reports a classification accuracy of melanoma, benign, atypical
lesions as 97.5%, 95.7%, and 96.3%. Abuzaghleh et al. [19] report a SE = 97.6%,
SP = 90.5% and average accuracy of 96.5 in [57]. Barata et al. [58] report performance measures of SE = 98%, SP
= 90% on PH2 dataset and SE = 83%, SP =
76% on EDRA datasets considering a fusion of features. A recent work
[59], introduces sparse coding of
the Scale-Invariant Feature Transform (SIFT) features for melanoma
classification. Reference [59] reports
a performance of SE = 100%, SP = 90.3% on the PH2
dataset.
In comparison the proposed computerized dermoscopy system considering the PH2
dataset (and SVM classifier) reports performance results SE = 96%,
SP = 97% and 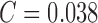 . The
classification accuracy of melanoma, common nevus, atypical nevus and in-situ
skin lesions is 100%, 93%, 90%, and 100%. The
average classification accuracy is 95.75%. Using only color features of
PH2 dataset and the AdaBoost classifier a performance of SE = 96%,
SP = 98% and
. The
classification accuracy of melanoma, common nevus, atypical nevus and in-situ
skin lesions is 100%, 93%, 90%, and 100%. The
average classification accuracy is 95.75%. Using only color features of
PH2 dataset and the AdaBoost classifier a performance of SE = 96%,
SP = 98% and 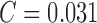 is
observed. The results considering the ATLAS dataset is SE = 98%,
SP = 99% and
is
observed. The results considering the ATLAS dataset is SE = 98%,
SP = 99% and 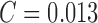 . The
classification accuracy is 96.83%. The results obtained prove that the
proposed computerized dermoscopy system is efficient and can be adopted to
diagnose skin lesions of varied types. Use of the proposed computerized
dermoscopy system to train or test new dermatologists is also mooted.
. The
classification accuracy is 96.83%. The results obtained prove that the
proposed computerized dermoscopy system is efficient and can be adopted to
diagnose skin lesions of varied types. Use of the proposed computerized
dermoscopy system to train or test new dermatologists is also mooted.
G. Assessment of Proposed 3D Skin Lesion Reconstruction Technique
A major goal of the proposed computerized dermoscopy system is to aid early detection of melanoma i.e. in-situ melanoma. Diagnosis can be efficiently achieved using the 3D reconstruction technique proposed. The 3D data / tensor provides useful insight to analyze relative depth of melanoma cancer skin lesions.
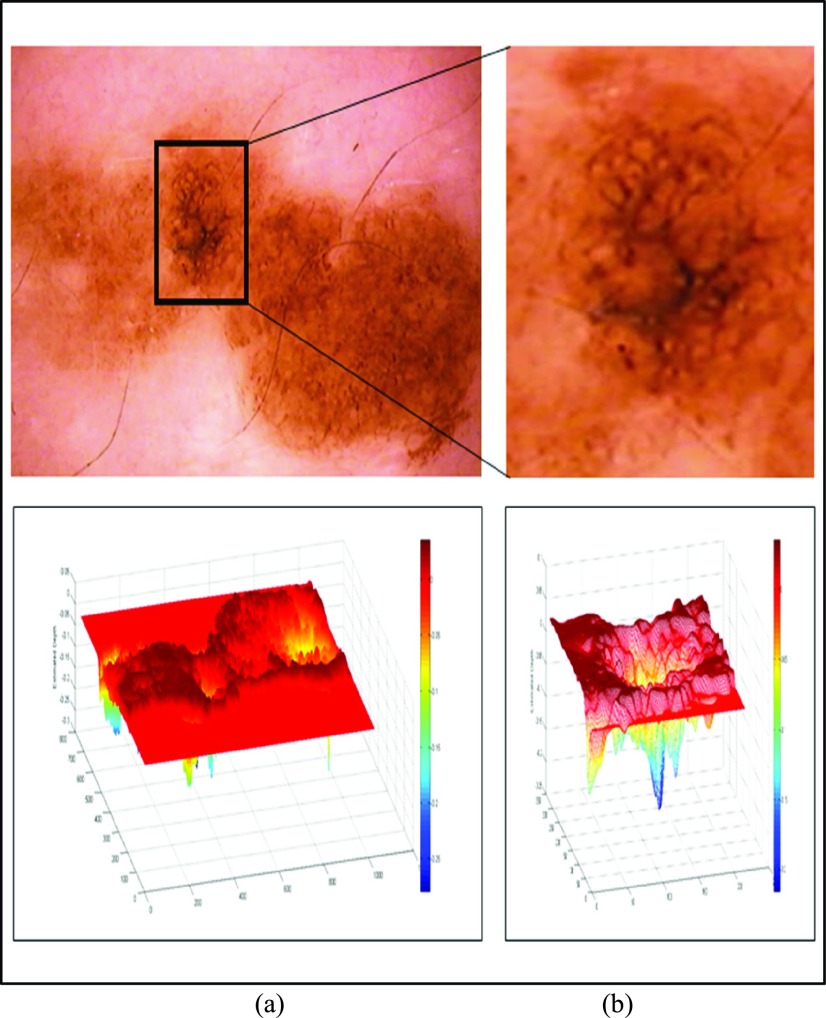
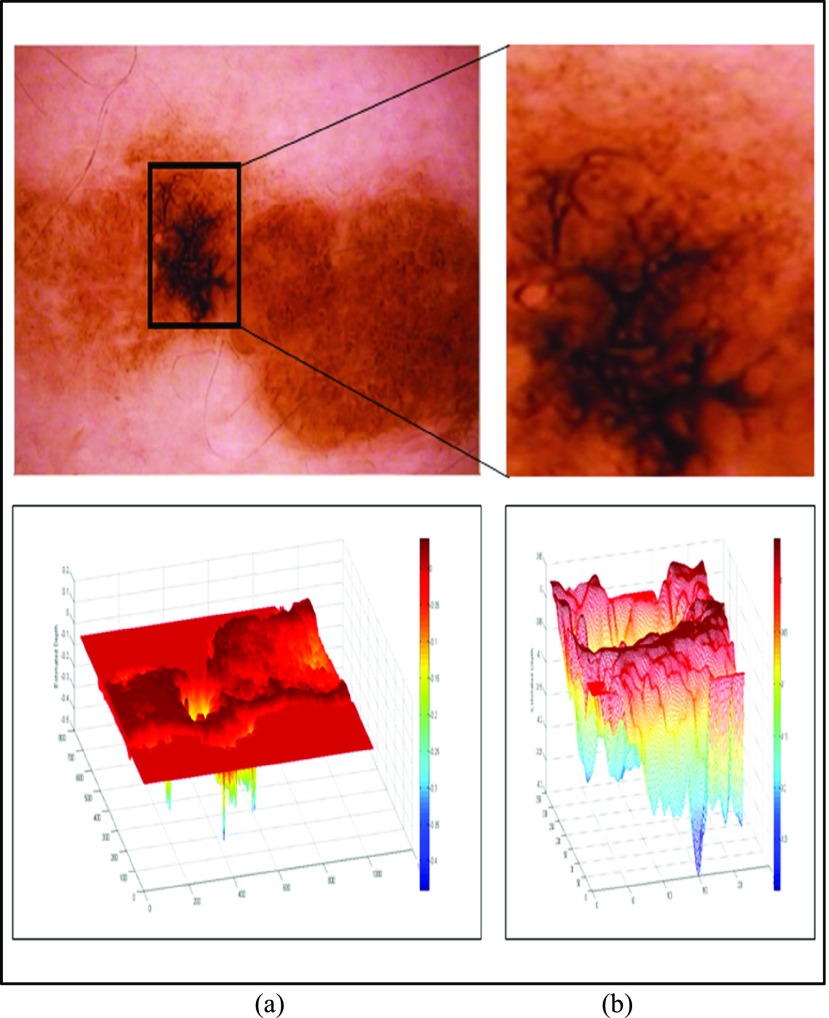
In the initial experiment we have considered an in-situ melanoma image from Chapter 16 (Follow-up of melanocytic skin lesions with digital dermoscopy) of the ATLAS CRROM [41]. Baseline image and the corresponding depth estimated is shown in Fig. 5. In Fig. 5(a) baseline image and the estimated depth is shown. Region of interest and the corresponding depth estimated is shown in Fig. 5(b). Relative depth estimated is 0.0023. Follow-up image observed after 4 months is shown in Fig. 6. Region of interest and estimated depth of the follow-up image is shown in Fig. 6(b). The relative depth estimated of the follow up image is 0.0055. Spreading of the melanoma in the region of interest is clearly evident by comparing Fig. 5(b) and Fig. 6(b). Results shown in Fig. 5, Fig. 6, and the marginal increase in relative estimated depth values validate the 3D reconstruction/estimation technique proposed in this paper.
FIGURE 5.
In-situ melanoma baseline image and 3D depth projections (a) Baseline image (top) and estimated depth (bottom). (b) Region of interest (top) and estimated depth (bottom).
FIGURE 6.
In-situ melanoma follow-up image and 3D depth projections (a) Follow-up image (top) and estimated depth (bottom). (b) Region of interest (top) and estimated depth (bottom).
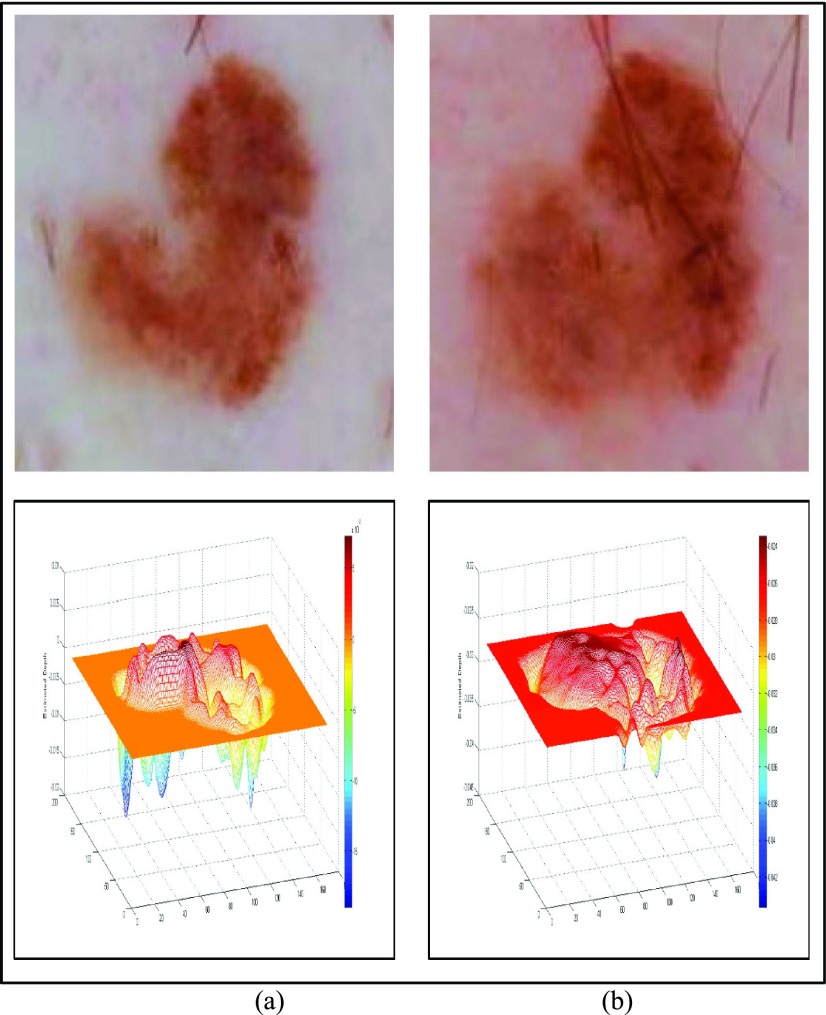
Dermoscopic images from [60] is consider to further assess performance of the proposed 3D reconstruction technique. An in-situ melanoma image (top) and the corresponding relative estimated depth (bottom) is shown Fig. 7(a). A spreading melanoma with a Breslow index of 0.5 mm and the relative depth estimated is shown in Fig. 7(b). The relative estimated depth and the spreading melanoma image with a Breslow index of 0.9mm is shown in Fig. 7(c). Relative estimated depth of the images computed using our proposed technique is reported as 0.0918, 0.1388 and 0.2437. Considering dermoscopic images shown in Fig. 7(b) and Fig. 7(c), Breslow index difference of 80% is observed. A difference of 75.58% is reported using the proposed relative depth estimation technique. The increase in the relative estimated depth and the difference measure reported validate the relative estimation accuracy.
FIGURE 7.
Melanoma images obtained from [60] and 3D depth projections (a) In-situ melanoma image (top) and estimated depth (bottom). (b) Spreading melanoma image with Breslow index 0.5mm (top) and estimated depth (bottom). (c) Spreading melanoma image with Breslow index of 0.9mm (top) and estimated depth (bottom).
To evaluate performance of the proposed 3D reconstruction technique on slow growing melanoma, dermoscopic data from [61] is considered. The baseline image and the corresponding relative estimated depth is shown in Fig. 8(a). The follow-up image after five years and the relative estimated depth is shown in Fig. 8(b). The follow-up dermoscopic skin lesion was biopsied and the melanoma was found to be 0.15mm thick. The relative estimated depth for the baseline image and follow-up image is 0.0189, 0.0436. The increasing values of the relative estimated depth prove accuracy of the proposed 3D reconstruction technique for slow growing melanoma.
FIGURE 8.
Slow growing melanoma follow-up image from [61] and 3D depth projections (a) Baseline image (top) and estimated depth (bottom). (b) Follow-up image obtained after 5 years. Biopsy reveals melanoma is 0.15mm thick (top) and estimated depth (bottom).
The International Skin Imaging Collaboration (ISIC): Melanoma Project introduced in recent times, is an academia-industry partnership providing dermoscopic data for melanoma diagnosis [62]. A large number of societies have collaborated together in the ISIC: Melanoma Project. Data provided is by far the most comprehensive set of publicly available melanoma skin lesion images [63]. A total of 4670 skin lesion images collected from various clinical trials are available in the dataset till date. Clinical/ diagnosis data corresponding to each skin lesion image is also available.
To validate 3D reconstruction technique, a set of 22 images from the ISIC archive
[63] is considered. Images whose
Breslow depth is confirmed through biopsies performed are taken for evaluation
study. Preprocessing and segmentation is performed on images. Post segmentation,
3D skin lesion construction is performed. Relative estimated depth using
proposed technique is noted. Results obtained are shown in Table VI. Large Breslow depth variations, from 0.16
to 0.9 is observed in dataset considered. Satisfactory and uniform relative
depth estimation values are reported. Minor variations (in order of 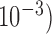 of
estimated depth values for images with similar Breslow depths is observed. Minor
variations are attributed to gradient differences observed in images. Results
presented depict that increase in estimated depth is correlated to increase in
Breslow depth, proving effectiveness of proposed 3D reconstruction technique on
large set of dermoscopic images considered.
of
estimated depth values for images with similar Breslow depths is observed. Minor
variations are attributed to gradient differences observed in images. Results
presented depict that increase in estimated depth is correlated to increase in
Breslow depth, proving effectiveness of proposed 3D reconstruction technique on
large set of dermoscopic images considered.
TABLE 6. Evaluation of 3D Skin Lesion Reconstruction Technique Using ISIC Data [63].
| Image ID from [63] | Biopsy confirmed Breslow depth | Relative estimated depth |
|---|---|---|
| ISIC_0011429 | 0.16 | 0.0448 |
| ISIC_0011430 | 0.16 | 0.0449 |
| ISIC_0011404 | 0.2 | 0.0558 |
| ISIC_0011463 | 0.22 | 0.0614 |
| ISIC_0011514 | 0.22 | 0.0610 |
| ISIC_0011428 | 0.4 | 0.1097 |
| ISIC_0011511 | 0.4 | 0.1100 |
| ISIC_0011438 | 0.48 | 0.1271 |
| ISIC_0011439 | 0.48 | 0.1291 |
| ISIC_0011458 | 0.49 | 0.1336 |
| ISIC_0011526 | 0.5 | 0.1529 |
| ISIC_0011520 | 0.62 | 0.1670 |
| ISIC_0011521 | 0.62 | 0.1693 |
| ISIC_0011405 | 0.65 | 0.1774 |
| ISIC_0000158 | 0.76 | 0.2273 |
| ISIC_0011435 | 0.8 | 0.2276 |
| ISIC_0011507 | 0.8 | 0.2203 |
| ISIC_0011503 | 0.82 | 0.2360 |
| ISIC_0011515 | 0.9 | 0.2544 |
| ISIC_0011517 | 0.9 | 0.2536 |
| ISIC_0011518 | 0.9 | 0.2531 |
| ISIC_0011519 | 0.9 | 0.2513 |
Though actual depth (currently obtained using invasive biopsy) cannot be computed, accurate estimates can be obtained using the proposed technique. The relative estimated depth is a critical feature for identification of in-situ melanoma. In addition, 3D features extracted using the 3D reconstructed skin lesion improve overall system classification performance as reported in the previous section.
VI. Conclusion
Amongst all skin cancers known, melanoma accounts for the majority of deaths reported. Melanoma is curable if diagnosed early. Use of non-invasive computerized dermoscopy techniques to diagnose skin lesions is commonly adopted. Identifying depth of the melanoma tumor into the skin is essential to ascertain the stage of cancer. Existing computerized dermoscopy techniques lay marginal or no emphasis on depth for diagnosis. Authors here believe “Melanoma is Skin Deep” and introduce a computerized dermoscopy system in this paper that incorporates depth estimation. A 3D skin lesion reconstruction technique using 2D dermoscopic images is proposed. Segmentation is achieved using the adaptive snake technique. The 3D reconstruction is achieved by fitting the depth map estimated to the underling 2D surface. Color, texture and 2D shape features are extracted. Based on the 3D tensor structure constructed, depth and 3D shape features are extracted. Feature selection to study the effects of features and their combinations on decision making is proposed. For decision making BoF, AdaBoost and SVM classifiers is applied. Experimental study is conducted using the PH2 and ATLAS datasets. Results considering different feature combinations and BoF, AdaBoost, SVM classifiers is presented. In view of the results, it is concluded that inclusion of 3D shape features proposed (that include the estimated depth features) enhance performance aiding accurate diagnosis of varied skin lesion types. The SVM achieves best classification scores of SE = 96% and SP = 97% on PH2 dataset. The SVM classification score of SE = 98% and SP = 99% on ATLAS dataset is reported.
The depth estimation technique proposed in this paper is naïve. Depth estimation performance is evaluated on ISIC: Melanoma project dataset and data obtained from literature published. Though good performance is reported, improvement of the estimation technique is always an open issue. Future of the work presented here is to identify depth estimation error using clinical data and devise new techniques to minimize errors. Authors welcome dermatologists/ researchers to undertake a collaborative research to develop the proposed system further considering clinical data.
Acknowledgment
Authors express their appreciation to Dr Michael A. Marchetti (Memorial Sloan Kettering Cancer Center, USA) for his invaluable support and guidance provided in this research. The authors would like to thank members of the Automatic Computer-based Diagnosis System for Dermoscopy Images (ADDI) project group in providing access to the PH2 dataset. Authors of this paper, praise the efforts involved in accumulating and provisioning of melanoma dermoscopy data by ISIC: Melanoma Project. The authors would like to thank the reviewers for their valuable suggestions that helped us improve the paper.
Biographies

T. Y. Satheesha received the master’s degree from the B.M.S. College of Engineering, Bengaluru, India. He is currently pursuing the Ph.D. degree with Jawaharlal Nehru Technological University, Anantapur, India. He is currently an Assistant Professor with ECE Department, Nagarjuna College of Engineering and Technology, Bengaluru. His current research interest is biomedical image processing.

D. Sathyanarayana received the Ph.D. degree from Jawaharlal Nehru Technological University, Hyderabad, India, in 2009. He is currently a Professor and the Head of ECE Department, Rajeev Gandhi Memorial College of Engineering and Technology, Nandyala, India. His areas of interests are signal and image processing. He is a member in professional societies like the ISTE and the IETE.

M. N. Giri Prasad received the Ph.D. degree from Jawaharlal Nehru Technological University, Hyderabad, India, in 2003. He is currently a Professor with ECE Department, Jawaharlal Nehru Technological University Anantapur, Anantapur, India. His areas of interests are wireless communication, biomedical instrumentation, digital image processing, vhdl coding, evolutionary computing, biomedical signal, and image processing. He is currently a BOS Chairman for Jawaharlal Nehru Technological University Anantapur, also he is a member in professional societies like the ISTE, the IEI, and the NAFEN.

Kashyap D. Dhruve received the bachelor’s degree from the Dayanada Sagar College of Engineering, Bengaluru, India. He is currently a Director of Research and Development Team with Planet-I Technologies, Bengaluru. He has focused on several research projects with the defense industry. His current research interests include image processing, wireless networks, gene sequencing, and cloud computing.
References
- [1].WHO Intersun, accessed on Jul. 21, 2015. [Online]. Available: http://www.who.int/uv/faq/skincancer/en/index1.html
- [2].Baldwin L. and Dunn J., “Global controversies and advances in skin cancer,” Asian–Pacific J. Cancer Prevention, vol. 14, no. 4, pp. 2155–2157, 2013. [DOI] [PubMed] [Google Scholar]
- [3].American Cancer Society, Cancer Facts&Figures 2015, American Cancer Society, 2015.
- [4].Howlader N., et al. , “SEER cancer statistics review,” Nat. Cancer Inst, Bethesda, MD, USA, Tech. Rep., Apr. 2012, pp. 1975–2010. [Google Scholar]
- [5].“U.S. Emerging melanoma therapeutics market,” Tech. Rep. a090-52, 2001.
- [6].Pehamberger H., Steiner A., and Wolff K., “In vivo epiluminescence microscopy of pigmented skin lesions-I: Pattern analysis of pigmented skin lesions,” J. Amer. Acad. Dermatol., vol. 17, pp. 571–583, Sep. 1987. [DOI] [PubMed] [Google Scholar]
- [7].Mayer J., “Systematic review of the diagnostic accuracy of dermatoscopy in detecting malignant melanoma,” Med. J. Australia, vol. 167, no. 4, pp. 206–210, Aug. 1997. [DOI] [PubMed] [Google Scholar]
- [8].Stolz W., Riemann A., and Cognetta A., “ABCD rule of dermatoscopy: A new practical method for early recognition of malignant melanoma,” Eur. J. Dermatol., vol. 4, pp. 521–527, Sep. 1994. [Google Scholar]
- [9].Blum A., Rassner G., and Garbe C., “Modified ABC-point list of dermoscopy:A simplified and highly accurate dermoscopic algorithm for the diagnosis of cutaneous melanocytic lesions,” J. Amer. Acad. Dermatol., vol. 48, no. 5, pp. 672–678, May 2003. [DOI] [PubMed] [Google Scholar]
- [10].Menzies S., Ingvar C., Crotty K., and McCarthy W., “Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features,” Arch. Dermatol., vol. 132, no. 10, pp. 1178–1182, Oct. 1996. [PubMed] [Google Scholar]
- [11].Argenziano G., Fabbrocini G., Carli P., De Giorgi V., Sammarco E., and Delfino M., “Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: Comparison of the ABCD rule of dermatoscopy and a new7-point checklist based on pattern analysis,” Arch. Dermatol., vol. 134, no. 12, pp. 1563–1570, Sep. 1998. [DOI] [PubMed] [Google Scholar]
- [12].Garnavi R., Aldeen R., Bailey M. J., “Computer-aided diagnosis of melanoma using border- and wavelet-based texture analysis,” IEEE Trans. Inf. Technol. Biomed., vol. 16, no. 6, pp. 1239–1252, Nov. 2012. [DOI] [PubMed] [Google Scholar]
- [13].Binder M., et al. , “Epiluminescence microscopy of small pigmented skin lesions: Short-term formal training improves the diagnosis performance of dermatologists,” J. Amer. Acad. Dermatol., vol. 36, no. 2, pp. 197–202, Feb. 1997. [DOI] [PubMed] [Google Scholar]
- [14].Braun R. P., Rabinovitz H., Tzu J. E., and Marghoob A. A., “Dermoscopy research—An update,” Seminars Cutaneous Med. Surgery, vol. 28, no. 3, pp. 165–171, 2009. [DOI] [PubMed] [Google Scholar]
- [15].YÜksel M. E. and Borlu M., “Accurate segmentation of dermoscopic images by image thresholding based on type-2 fuzzy logic,” IEEE Trans. Fuzzy Syst., vol. 17, no. 4, pp. 976–982, Aug. 2009. [Google Scholar]
- [16].Glaister J., Wong A., and Clausi D. A., “Segmentation of skin lesions from digital images using joint statistical texture distinctiveness,” IEEE Trans. Biomed. Eng., vol. 61, no. 4, pp. 1220–1230, Apr. 2014. [DOI] [PubMed] [Google Scholar]
- [17].Peruch F., Bogo F., Bonazza M., Cappelleri V.-M., and Peserico E., “Simpler, faster, more accurate melanocytic lesion segmentation through MEDS,” IEEE Trans. Biomed. Eng., vol. 61, no. 2, pp. 557–565, Feb. 2014. [DOI] [PubMed] [Google Scholar]
- [18].Silveira M., et al. , “Comparison of segmentation methods for melanoma diagnosis in dermoscopy images,” IEEE J. Sel. Topics Signal Process., vol. 3, no. 1, pp. 35–45, Feb. 2009. [Google Scholar]
- [19].Abuzaghleh O., Barkana B. D., and Faezipour M., “Noninvasive real-time automated skin lesion analysis system for melanoma early detection and prevention,” IEEE J. Transl. Eng. Health Med., vol. 3, 2015, Art. no. 4300212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Amelard R., Glaister J., Wong A., and Clausi D. A., “High-level intuitive features (HLIFs) for intuitive skin lesion description,” IEEE Trans. Biomed. Eng., vol. 62, no. 3, pp. 820–831, Mar. 2015. [DOI] [PubMed] [Google Scholar]
- [21].Alcon J. F., et al. , “Automatic imaging system with decision support for inspection of pigmented skin lesions and melanoma diagnosis,” IEEE J. Sel. Topics Signal Process., vol. 3, no. 1, pp. 14–25, Feb. 2009. [Google Scholar]
- [22].Barata C., Ruela M., Francisco M., Mendonça T., and Marques J. S., “Two systems for the detection of melanomas in dermoscopy images using texture and color features,” IEEE Syst. J., vol. 8, no. 3, pp. 965–979, Sep. 2014. [Google Scholar]
- [23].Fatima R., Khan M. Z. A., Govardhan A., and Dhruve K. D., “Computer aided multi-parameter extraction system to aid early detection of skin cancer melanoma,” Int. J. Comput. Sci. Netw. Secur., vol. 12, no. 10, pp. 74–86, Oct. 2012. [Google Scholar]
- [24].Fathima R., Khan M. Z. A., Govardhan A., and Dhruve K., “Detecting in-situ melanoma using multi parameter extraction and neural classification mechanisms,” Int. J. Comput. Eng. Technol., vol. 4, no. 1, pp. 16–33, 2013. [Google Scholar]
- [25].Sadeghi M., Lee T. K., McLean D., Lui H., and Atkins M. S., “Detection and analysis of irregular streaks in dermoscopic images of skin lesions,” IEEE Trans. Med. Imag., vol. 32, no. 5, pp. 849–861, May 2013. [DOI] [PubMed] [Google Scholar]
- [26].Wighton P., Lee T. K., Lui H., McLean D. I., and Atkins M. S., “Generalizing common tasks in automated skin lesion diagnosis,” IEEE Trans. Inf. Technol. Biomed., vol. 15, no. 4, pp. 622–629, Jul. 2011. [DOI] [PubMed] [Google Scholar]
- [27].Celebi M. E. and Zornberg A., “Automated quantification of clinically significant colors in dermoscopy images and its application to skin lesion classification,” IEEE Syst. J., vol. 8, no. 3, pp. 980–984, Sep. 2014. [Google Scholar]
- [28].Saéz A., Serrano C., and Acha B., “Model-based classification methods of global patterns in dermoscopic images,” IEEE Trans. Med. Imag., vol. 33, no. 5, pp. 1137–1147, May 2014. [DOI] [PubMed] [Google Scholar]
- [29].Maglogiannis I. and Doukas C. N., “Overview of advanced computer vision systems for skin lesions characterization,” IEEE Trans. Inf. Technol. Biomed., vol. 13, no. 5, pp. 721–733, Sep. 2009. [DOI] [PubMed] [Google Scholar]
- [30].Dhawan A. P., Gordon R., and Rangayyan R. M., “Nevoscopy: Three-dimensional computed tomography of nevi and melanomas in situ by transillumination,” IEEE Trans. Med. Imag., vol. 3, no. 2, pp. 54–61, Jun. 1984. [DOI] [PubMed] [Google Scholar]
- [31].D’Alessandro B. and Dhawan A. P., “3-D volume reconstruction of skin lesions for melanin and blood volume estimation and lesion severity analysis,” IEEE Trans. Med. Imag., vol. 31, no. 11, pp. 2083–2092, Nov. 2012. [DOI] [PubMed] [Google Scholar]
- [32].Vogt M. and Ermert H., “In vivo ultrasound biomicroscopy of skin: Spectral system characteristics and inverse filtering optimization,” IEEE Trans. Ultrason., Ferroelect., Freq. Control, vol. 54, no. 8, pp. 1551–1559, Aug. 2007. [DOI] [PubMed] [Google Scholar]
- [33].Tittmann B. R., Miyasaka C., Maeva E., and Shum D., “Fine mapping of tissue properties on excised samples of melanoma and skin without the need for histological staining,” IEEE Trans. Ultrason., Ferroelect., Freq. Control, vol. 60, no. 2, pp. 320–331, Feb. 2013. [DOI] [PubMed] [Google Scholar]
- [34].Zhuo S. and Sim T., “Defocus map estimation from a single image,” Pattern Recognit., vol. 44, no. 9, pp. 1852–1858, 2011. [Google Scholar]
- [35].(2015). Cancer Research UK. [Online]. Available: http://www.cancerresearchuk.org/about-cancer/type/melanoma/treatment/stagesof-melanoma#bres
- [36].Dana-Farber Cancer Institute. (2015). [Online]. Available: http://www.dana-farber.org/Adult-Care/Treatment-and-Support/Melanoma.aspx
- [37].Argenziano G. and Soyer H., Interactive Atlas of Dermoscopy. Milan, Italy: EDRA-Medical New Media, 2000. [Google Scholar]
- [38].Pereyra M., Dobigeon N., Batatia H., and Tourneret J., “Segmentation of skin lesions in 2-D and 3-D ultrasound images using a spatially coherent generalized Rayleigh mixture model,” IEEE Trans. Med. Imag., vol. 31, no. 8, pp. 1509–1520, Aug. 2012. [DOI] [PubMed] [Google Scholar]
- [39].McLachlan G. and Krishnan T., The EM Algorithm and Extensions. New York, NY, USA: Wiley, 1997. [Google Scholar]
- [40].Nascimento J. and Marques J. S., “Adaptive snakes using the em algorithm,” IEEE Trans. Image Process., vol. 14, no. 11, pp. 1678–1686, Nov. 2005. [DOI] [PubMed] [Google Scholar]
- [41].Marghoob A., Braun R., and Kopf A., Interactive CD-ROM of Dermoscopy. London, U.K.: Informa Healthcare; 2007. [Google Scholar]
- [42].Mendonça T., Ferreira P. M., Marques J. S., Marcal A. R. S., and Rozeira J., “PH2-A dermoscopic image database for research and benchmarking,” in Proc. IEEE Eng. Med. Biol. Soc. 35th Annu. Int. Conf. (EMBC), Jul. 2013, pp. 5437–5440. [DOI] [PubMed] [Google Scholar]
- [43].Perona P. and Malik J., “Scale-space and edge detection using anisotropic diffusion,” in Proc. IEEE Trans. Pattern Anal. Mach. Intell., vol. 12, no. 7, pp. 629–639, Jul. 1990. [Google Scholar]
- [44].Levin A., Lischinski D., and Weiss Y., “A closed-form solution to natural image matting,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 30, no. 2, pp. 228–242, Feb. 2008. [DOI] [PubMed] [Google Scholar]
- [45].Ercal F., Chawla A., Stoecker W. V., Lee H.-C., and Moss R. H., “Neural network diagnosis of malignant melanoma from color images,” IEEE Trans. Biomed. Eng., vol. 41, no. 9, pp. 837–845, Sep. 1994. [DOI] [PubMed] [Google Scholar]
- [46].Haralick R. M., Shanmugam K., and Dinstein I. H., “Textural features for image classification,” IEEE Trans. Syst., Man, Cybern., Syst., vol. 3, no. 6, pp. 610–621, Nov. 1973. [Google Scholar]
- [47].Smaoui N. and Bessassi S., “A developed system for melanoma diagnosis,” Int. J. Comput. Vis. Signal Process., vol. 3, no. 1, pp. 10–17, 2013. [Google Scholar]
- [48].Hu M. K., “Visual pattern recognition by moment invariants,” IRE Trans. Inf. Theory, vol. 8, no. 2, pp. 179–187, Feb. 1962. [Google Scholar]
- [49].Flusser J. and Suk T., “Pattern recognition by affine moment invariants,” Pattern Recognit., vol. 26, no. 1, pp. 167–174, 1993. [Google Scholar]
- [50].Cortes C. W. and Vapnik V., “Support-vector networks,” Mach. Learn., vol. 20, no. 3, pp. 273–297, 1995. [Google Scholar]
- [51].Christianini N. and Shawe-Taylor J., An Introduction to Support Vector Machines. Cambridge, U.K.: Cambridge Univ. Press, 2000. [Google Scholar]
- [52].Viola P. and Jones M. J., “Robust real-time face detection,” Int. J. Comput. Vis., vol. 57, no. 2, pp. 137–154, May 2004. [Google Scholar]
- [53].Leung T. and Malik J., “Representing and recognizing the visual appearance of materials using three-dimensional textons,” Int. J. Comput. Vis., vol. 43, no. 1, pp. 29–44, Jun. 2001. [Google Scholar]
- [54].Fergus R., Perona P., and Zisserman A., “Object class recognition by unsupervised scale-invariant learning,” in Proc. IEEE Int. Conf. Comput. Vis. Pattern Recog., vol. 2 Jun. 2003, pp. II-264–II-271. [Google Scholar]
- [55].Hsu C.-W., Chang C.-C., and Lin C.-J. (2010). A Practical Guide to Support Vector Classification. [Online]. Available: https://www.csie.ntu.edu.tw/ cjlin/papers/guide/guide.pdf [Google Scholar]
- [56].Chang C.-C. and Lin C.-J., “LIBSVM: A library for support vector machines,” ACM Trans. Intell. Syst. Technol., vol. 2, no. 3, pp. 27-1–27-27, 2011. [Online]. Available: http://www.csie.ntu.edu.tw/~cjlin/libsvm [Google Scholar]
- [57].Abuzaghleh O., Faezipour M., and Barkana B. D., “A comparison of feature sets for an automated skin lesion analysis system for melanoma early detection and prevention,” in IEEE Long Island Syst., Appl. Technol. Conf. (LISAT), May 2015, pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Barata C., Celebi M. E., and Marques J. S., “Melanoma detection algorithm based on feature fusion,” in Proc. 37th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. (EMBC), Aug. 2015, pp. 2653–2656. [DOI] [PubMed] [Google Scholar]
- [59].Rastgoo M., et al. , “Classification of melanoma lesions using sparse coded features and random forests,” Proc. SPIE, vol. 9785, p. 97850C, Mar. 2016. [Google Scholar]
- [60].Silva V. P. M., Ikino J. K., Sens M. M., Nunes D. H., and Di Giunta G., “Dermoscopic features of thin melanomas: a comparative study of melanoma in situ and invasive melanomas smaller than or equal to 1mm,” Anais Brasileiros Dermatol., vol. 88, no. 5, pp. 712–717, Oct. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kittler H. and Menzies S. W., “Follow-up of melanocytic skin lesions with digital dermoscopy,” in An Atlas of Dermoscopy, 2nd ed. Jul. 2012, pp. 354–361.
- [62].Gutman D., et al. (May 2016). “Skin lesion analysis toward melanoma detection:Achallenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC)” [Online]. Available: https://arxiv.org/abs/1605.01397 [Google Scholar]
- [63].The International Skin Imaging Collaboration: Melanoma Project. ISIC Archive. [Online]. Available: https://isic-archive.com/