Abstract
INTRODUCTION
In the Drosophila adult midgut, multipotent intestinal stem cells (ISCs) produce two types of daughter cells: nutrient-absorbing enterocytes (ECs) and secretory enteroendocrine (ee) cells. Notch signaling between ISCs and their daughters directs the proper specification of both of these cell types. Previous work suggests that ISCs expressing high levels of the Notch ligand Delta (Dl) strongly activate the Notch signaling pathway in their daughters and result in their differentiation into ECs. By contrast, ISCs that express low levels of Dl direct their daughters to become ee cells. However, in this unidirectional Notch signaling model, the mechanisms regulating differential Dl expression in ISCs are poorly understood.
RATIONALE
During Drosophila pupal midgut development, pupal ISCs only make ee cells. Therefore, we examined how ee cells are made and evaluated the role of Notch signaling function during this developmental time window. On the basis of insights obtained from pupal development, we also asked whether similar mechanisms were used by ISCs in the adult midgut to generate ee cells.
RESULTS
The ee cell fate marker Prospero (Pros) appeared in pupal ISCs at 44 hours after pupal formation (APF). From 44 to 96 hours APF, ISCs first divided asymmetrically, generating one ISC and one ee cell, followed by symmetric division of both ISCs and ee cells, resulting in a pair of ISCs and a pair of ee cells. During ISC asymmetric divisions, Pros was asymmetrically segregated to the basal daughter cell, a process that depended on the function of the Par complex. After ISC asymmetric division, the ee daughter cell expressed the Notch ligand Dl and activated the Notch signaling pathway in ISCs. Loss of Notch signaling in pupal ISCs induced all stem cells to differentiate into ee cells, whereas low-level activation of Notch signaling in pupal ISCs blocked ee cell formation. During ee symmetric divisions, Pros distribution was symmetric; however, cell polarity and Notch signaling remained asymmetric. Loss of Notch signaling between progeny of ee symmetric divisions disrupted expression of peptide hormones in ee cells, indicating a role for Notch signaling in proper ee specification. We also investigated the Notch pathway in adult ISCs and confirmed that postmitotic Notch signaling from ee daughter cells also regulates ISC multipotency.
CONCLUSION
Consistent with previous work, high levels of Dl in ISCs activate high levels of Notch in the daughter cell, promoting EC differentiation. In contrast, after asymmetric localization of Pros, ISCs require a low Notch signal from their immediate ee cell daughters to maintain multipotency. Thus, Notch signaling is both bidirectional and context-dependent. Previous work also has suggested that ISCs remain basal during EC formation and that basal ISCs activate the Notch pathway in daughter cells. Our data show that ISCs are apically located during ee cell formation and that basal ee cells activate the Notch pathway in ISCs. Therefore, Notch signaling is always unidirectional in terms of polarity: Basal daughter cells express the Notch ligand Dl in order to activate the Notch signaling pathway in daughters after asymmetric ISC divisions.
Our work provides further evidence that mechanisms regulating tissue homeostasis are more conserved between the Drosophila and mammalian intestine than previously thought. Inhibition of Notch signaling in the mouse intestine induces crypt base columnar stem cell loss and secretory cell hyperplasia, and ectopic Notch signaling promotes EC differentiation. Loss of Notch signaling in Drosophila ISCs also leads to stem cell loss and premature ee cell formation, whereas high Notch signaling promotes stem cell differentiation into ECs. Because Notch signaling also plays important roles in common lymphoid progenitors making B cells and T cells, and in airway basal cells making secretory cells and ciliated cells, it is tempting to speculate that bidirectional Notch signaling may regulate multipotency in these and other progenitors and stem cells.
Bidirectional Notch signaling and unidirectional polarity: Left: During enteroendocrine cell (ee) formation, the Par complex induces asymmetric segregation of Prospero (Pros), and the Notch signaling ligand Delta (Dl) is expressed in a basal Pros+ ee. Low Notch signaling from a basal ee to an intestinal stem cell (ISC) maintains ISC identity. Right: During enterocyte (EC) production, strong Notch signaling from a basal ISC to an enteroblast (EB) promotes EC differentiation.
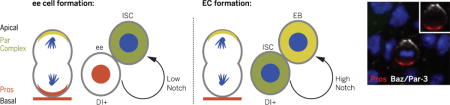
The Drosophila midgut is maintained by multipotent intestinal stem cells (ISCs) that give rise to either absorptive enterocytes (ECs) or hormone-producing enteroendocrine (ee) cells (1, 2). ISCs orchestrate the proper developmental and regenerative ratios of differentiated cell types. However, the mechanisms that control multipotency of ISCs remain an enigma. Previous work (3) suggested that high levels of the contact-dependent Notch signaling pathway ligand Delta (Dl) in ISCs drive high levels of Notch activity in the daughter cell, resulting in EC formation, whereas low levels of Dl in ISCs drive low levels of Notch activity in the daughter cell, resulting in ee cell formation (fig. S1A). How the ISC achieves differential Dl expression remains unknown.
ISC asymmetric divisions specify ee cells
During pupation, ISCs only give rise to ee cells, providing a unique platform to explore Notch signaling in multipotency decision (4, 5). To identify the time during which Notch signaling may be required, we first characterized the generation of pupal ISCs and ee cells. The progenitor driver esg-Gal4 (1), driving UAS-GFP [green fluorescent protein (esg>GFP)], and the ee cell marker Prospero [Pros (1, 2)] were used to quantify wild-type ISC and ee cell numbers between 24 and 96 hours after pupal formation (APF) (Fig. 1A). Punctate Pros staining was first detectable in the anterior midgut at 44 hours APF (Fig. 1, B and B′); by 54 hours APF, Pros was present in the entire intestine (fig. S1B). At 44 hours APF, ISC number was almost 600. From 44 to 54 hours APF, ISC number (GFP+, Pros−) dipped to ~270, followed by an increase to ~1200 (green line, Fig. 1A) by 96 hours APF. From 44 to 96 hours APF, Pros+ cell number increased from 0 to ~1200 (red line, Fig. 1A), resulting in a total of 2400 cells by the end of pupation at 96 hours APF (blue line, Fig. 1A).
Fig. 1. ee cells are specified by ISCs asymmetric divisions.
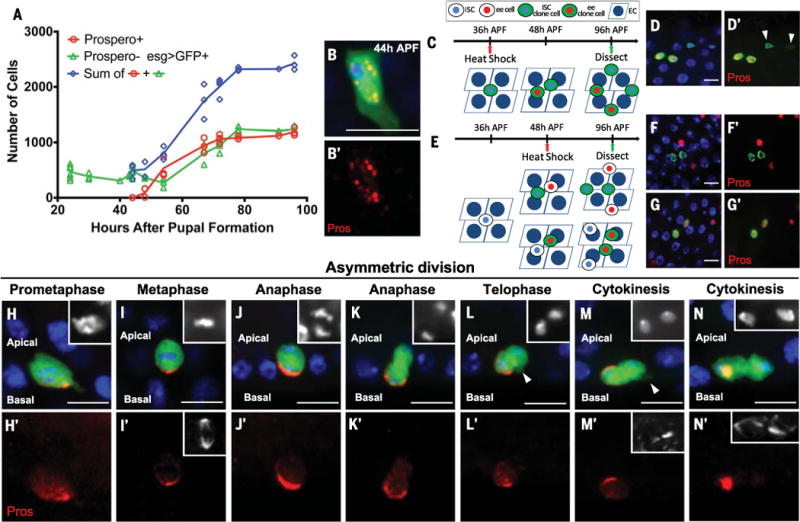
(A) Numbers of ISCs (Prospero- esg>GFP+) and ee cells (Prospero+) during pupal development. Each data point represents one pupal midgut. Curves were generated by connecting adjacent average value positions. (B) Pros staining at 44 hours APF. Here and in all images, blue indicates 4′,6-diamidino-2-phenylindole (DAPI) and red indicates Pros; green indicates esg>GFP except where otherwise specified. (C and E) Schematic representation of wild-type MARCM clones and their outcomes induced at 36 hours APF (C) or 48 hours APF (E) and examined at 96 hours APF. (D) A four-cell clone (green) induced at 36 hours APF contains two Pros+ cells and two ISCs [arrowhead in (D′)]. (F and G) Two-cell clones induced at 48 hours APFcontain either two ISCs (Pros−) (F) or two ee cells (Pros+) (G). (H to N) Pros asymmetrically localizes to the basal side during ISC mitosis. The apical daughter gradually moves toward the basal side during anaphase [(J) and (K)] and telophase [(L) to (N)]. The apical daughter extends a projection toward the basal side during telophase [arrowhead in (L) and (M)]. Insets in (H) to (N), DAPI; insets in (I′), (M′), and (N′), α-tubulin. Scale bars, 10 μm.
To determine how ~600 ISCs amplify by a factor of 4 to give rise to an equal ratio of ISCs and ee cells during this time, we induced wild-type MARCM (mosaic analysis with repressible cell marker) clones (6) at different time points during pupal development and quantified the cellular composition of clones at 96 hours APF (Fig. 1, C and E). Using the MARCM system, homozygous clones become positively labeled with GFP after flippase-catalyzed recombination. When clones were induced at 36 hours APF, 61.2% of them (n = 96) were four-cell clones containing two ISCs and two Pros+ ee cells (Fig. 1, D and D’, and fig. S1C). When clones were induced at 48 hours APF, 67.1% of clones (n = 137) contained two cells (fig. S1D), with 43% containing two ISCs (Fig. 1, F and F′) and 57% containing two ee cells (Fig. 1, G and G′). These data suggest that from 44 to 96 hours APF, ISCs first divide asymmetrically, generating one ISC and one ee cell, followed by symmetric division of both ISCs and ee cells, resulting in a pair of ISCs and a pair of ee cells (Fig. 1C).
To further investigate pupal ISC and ee cell division order and outcomes, we induced wild-type MARCM clones at 30 hours APF and examined the cellular composition of clones at 56 hours APF (fig. S1F). We found that 63.5% of clones (n = 52) were two-cell clones containing one ISC and one Pros+ ee cell (fig. S1, E to H). An additional 13.5% of clones were three-cell clones containing one ISC and two Pros+ ee cells (fig. S1, E, F, I, and J). These data confirmed that the first ISC division was asymmetric and suggested that ee cells symmetrically divide prior to the initiation of ISC symmetric divisions.
After identifying the time period during which ee cells are generated, we next sought to determine the mechanism by which an asymmetric outcome of the initial ISC division is achieved to give rise to an ee daughter. From 44 to 56 hours APF, Pros asymmetrically localized to the basal side of dividing ISCs (Fig. 1, H to N, and fig. S1, L to P). Asymmetric Pros distribution began at pro-metaphase (Fig. 1H) (n = 50), generated a basal crescent pattern during metaphase and anaphase (Fig. 1, I to K) (n = 200), and localized into the basal cell nucleus during cytokinesis (Fig. 1N) (n = 100). During anaphase and telophase, the original apical daughter cell extended a basal protrusion whose presence correlated with its movement toward a basal position (Fig. 1, J to N, and fig. S1K) (n = 50). At the end of the asymmetric division, both daughter cells were basally located (Fig. 1N) (n = 100).
In Drosophila neuroblasts (NBs), the Par complex supplies an apical cue establishing apical-basal (A–B) polarity to ensure asymmetric segregation of Miranda (Mira) into the basal cell (7–9). Mira functions as an adaptor protein that binds Pros and forms a crescent that is basally localized in mitotic cells (10–14). After asymmetric division, Mira is degraded, releasing Pros from the basal cell membrane and allowing Pros to translocate into the nucleus (10, 11, 15–18). By using antibodies to Mira and Bazooka (Baz; the homolog of Par-3 and a core component of the Par complex), we found that Mira colocalized with Pros during ISC asymmetric divisions (Fig. 2, A and B, and fig. S2, A and B). By contrast, Baz localized to an apical crescent, which was mutually exclusive with Pros staining during ISC asymmetric divisions (Fig. 2, C and D, and fig. S2, C to E). To disrupt A–B polarity, we used the temperature-inducible progenitor cell driver esg-Gal4 tub-Gal80ts (esgts) (1) to drive RNA interference (RNAi) against baz, and transferred animals to the permissive temperature (30°C) from 24 to 48 hours APF (see supplementary materials for experimental details of how developmental milestones at 18°C and 30°C were normalized to pupal development at 25°C). Whereas we observed asymmetric localization of Pros in wild-type ISCs, knockdown of Baz resulted in the even distribution of Pros on cell membranes during mitosis (Fig. 2, E and F). In addition to its role in segregating proteins to the basal membrane during NB divisions, Baz controls mitotic spindle orientation (7, 8, 19). We quantified wild-type (esg>GFP) and esgts>baz RNAi ISC metaphase spindle orientation by measuring the angle formed between the basal cell membrane and dividing spindles (Fig. 2, G and H, and fig. S2, F to J). Whereas wild-type ISC metaphase division angles concentrated around 80° (Fig. 2I), esgts>baz RNAi ISC division angles were randomly spread (Fig. 2J). At 64 hours APF, as a consequence of disrupting Pros asymmetric segregation relative to the wild-type control (Fig. 2K), esgts>baz RNAi intestine contained a cluster of four GFP+ cells that were all Pros+ (Fig. 2L). To confirm baz RNAi results, we generated baz4 MARCM clones at 24 hours APF and examined them at 92 hours APF. Every control wild-type MARCM clone had Pros– ISCs (n = 128; Fig. 2M), whereas 45% of baz4 clones contained only Pros+ ee cells (n = 71; Fig. 2N). Therefore, asymmetric distribution of Pros by the Par complex is critical for proper ee cell specification and ISC maintenance.
Fig. 2. Baz is required for asymmetric localization of Prospero during ISC mitosis.
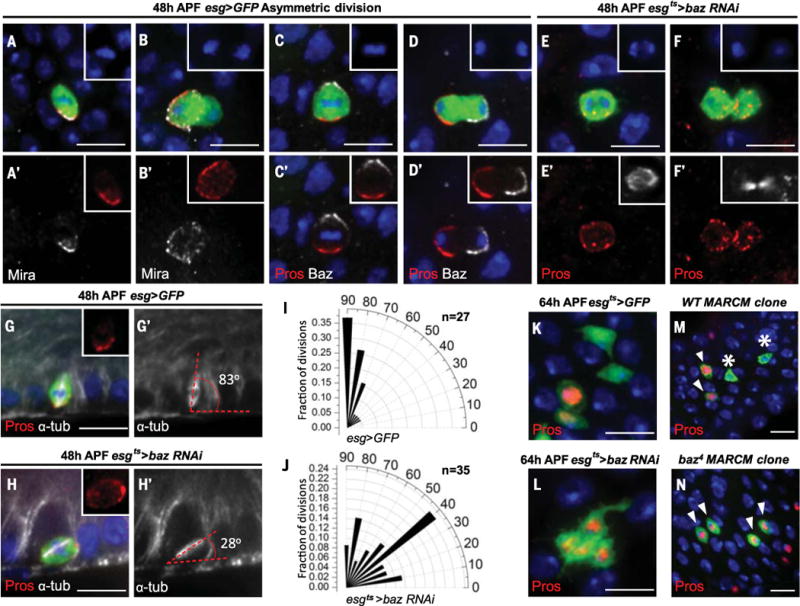
Pupal midguts in (A) to (J) were examined at 48 hours APF. (A and B) Mira (white) colocalizes with Pros (red) during metaphase (A) and telophase (B) of ISC asymmetric divisions. (C and D) Crescent Baz and Pros staining are mutually exclusive in metaphase (C) and telophase (D) ISCs. (E and F) In esgts>baz RNAi animals, Pros evenly distributes on the cell membrane of anaphase (E) and telophase (F) ISCs. Insets in (A) to (F), DAPI; insets in (A′) and (B′), Pros; insets in (C′) and (D′), Pros (red), Baz (white); insets in (E′) and (F′), α-tubulin (white). (G and H) Representative division angles in control (esg>GFP) (G) and esgts>baz RNAi (H) metaphase ISCs. Insets, Pros; white, α-tubulin. (I and J) Radial histogram quantification of division angles in control (esg>GFP) (I) and esgts>baz RNAi (J) metaphase ISCs. (K) After 64 hours of APF control (esgts>GFP), the four GFP+ cells are two Pros+, two Pros−. (L) Segment of esgts>baz RNAi intestine contains four GFP+ cells, all Pros+. Hours in (K) and (L) correspond to developmental times at 25°C. (M and N) Wild-type MARCM clones (M) and baz4 MARCM clones (N), both shown in green, were induced at 24 hours APF and examined at 92 hours APF. Wild-type MARCM clones contain two Pros+ cells (arrowheads) and two Pros− cells (asterisks); all baz4 MARCM clone cells are Pros+ (arrowheads). Scale bars, 10 μm.
ee cells activate Notch in ISCs
To determine the role of Notch signaling during ISC asymmetric divisions, we stained for expression of Dl and the Notch response element LacZ (NRE-lacZ) from 40 to 51 hours APF. At 40 hours APF, Dl was not detectable in ISCs (fig. S3A). However, by 44 hours APF, Dl, but not NRE-lacZ, was present in ISCs (fig. S3, B and C). At 48 to 51 hours APF, after asymmetric division of ISCs, Dl became localized to Pros+ ee daughter cells (Fig. 3A), whereas NRE-lacZ staining was present in pupal ISCs (Fig. 3B and fig. S3D). Together, our data demonstrate that ee cells induce Notch activity in pupal ISCs after asymmetric divisions.
Fig. 3. ee cells induce Notch activity in ISCs to prevent ISCs from differentiating into ee cells during pupal development.
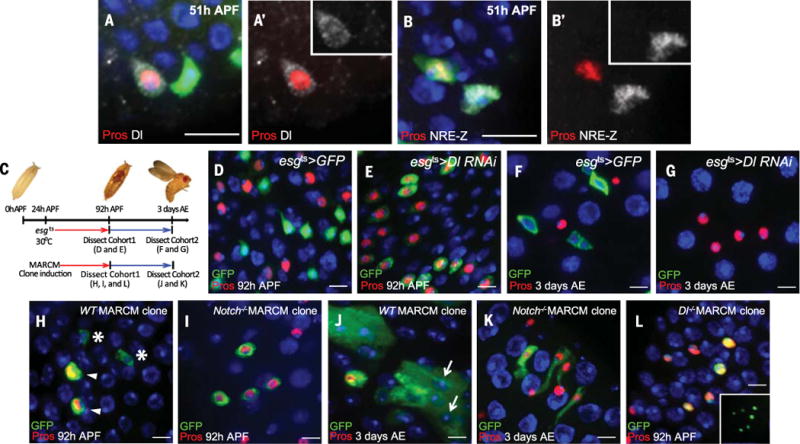
Hours correspond to developmental times at 25°C (see supplementary materials for experiment details). (A) Dl [inset in (A′)] is present in a Pros+ cell at 51 hours APF. (B) The Notch signaling reporter NRE-lacZ [inset in (B′)] is present in a pupal ISC (Pros−) at 51 hours APF. (C) Schematic of the genetic manipulations carried out during pupal development. (D and E) Cohort 1: Midguts were examined at 92 hours APF. In control animals (D), strong esgts>GFP+ cells are Pros−; in esgts>Dl RNAi animals (E), all GFP+ cells are Pros+. (F and G) Cohort 2: Midguts were examined at 3 days AE. In control animals (F), progenitor cells (ISCs and enteroblasts) are esgts>GFP+. (G) In esgts>Dl RNAi animals, GFP+ cells are absent and all diploid cells are Pros+ ee cells. (H and I) Cohort 1: MARCM clones (green) were examined at 92 hours APF. Wild-type clone (H) contains two Pros+ cells (arrowheads) and two Pros− cells (asterisks); all Notch mutant clone cells (I) are Pros+. (J and K) Cohort 2: MARCM clones were examined at 3 days AE. Wild-type clone (J) contains multiple ECs (arrows); Notch mutant clone (K) contains four Pros+ cells (sagittal view). (L) Dl mutant MARCM clones were examined at 92 hours APF. All mutant cells are Pros+. Inset, clonal marker. Scale bars, 10 μm.
To knock down Notch signaling during pupation, we used esgts to drive UAS-Dl RNAi and transferred animals to the permissive temperature (30°C) from 24 to 92 hours APF (Fig. 3C). Relative to control intestine (Fig. 3D), knockdown of Notch signaling in pupal ISCs led to all esgts>Dl RNAi; GFP+ cells expressing Pros at the end of pupation (Fig. 3E and fig. S3E), which suggests that knockdown of Notch signaling during pupal development results in loss of pupal ISCs. We next followed the fate of pupal GFP+ cells into adulthood (Fig. 3C). In control esgts intestine, all intestinal progenitors were GFP+ (Fig. 3F and fig. S3G) at 3 days after eclosion (AE). By contrast, esgts>Dl RNAi intestine contained either no GFP+ cells (Fig. 3G) or only rare ones (fig. S3F) at 3 days AE. Similar results were obtained using UAS-Notch RNAi and UAS-NotchDN (a Notch receptor that can bind the ligand Dl but lacks the transcription regulation domain Cdc10) (20) (fig. S3, H and I), which suggests that knockdown of Notch signaling during pupal development results in loss of ISCs.
In addition, to identify the precise time window in which Notch is required for maintaining stem cell identity, we cultured esgts> Dl RNAi and esgts>Notch RNAi flies at 18°C for various times corresponding to 42, 53, 64, 76, and 87 hours APF at 25°C. Pupae were then transferred to 30°C to permit esg-Gal4 expression. Intestines were examined before eclosion, as assessed by the appearance of black wings and mature bristles (21) (fig. S4A). At or before 53 hours APF, knockdown of Notch signaling in ISCs resulted in ISC loss (fig. S4, B and C). After 64 hours APF, knockdown of Notch signaling had no effect on ISC maintenance (fig. S4D). Therefore, Notch signaling is required to maintain ISC fate for only a brief period after asymmetric division.
To corroborate the RNAi knockdown results, we generated Notch55e11 null MARCM clones at 24 hours APF and examined them at 92 hours APF (Fig. 3C). In wild-type MARCM clones, about 50% of the cells were Pros+ ee cells (356 ee cells in 744 clone cells, clone number n = 128) (Fig. 3H). However in Notch55e11 clones, 100% of cells were ee cells (479 clone cells, n = 79) (Fig. 3I). ISCs in wild-type clones had generated ECs by 3 days AE (Fig. 3J). By contrast, Notch55e11 clones contained only Pros+ cells (Fig. 3K). Furthermore, at 10 days AE, Notch55e11 clones were no longer present, presumably replaced by normal tissue turnover (2). Similar results were obtained with a null mutant allele of Delta (DlRevF10) (3) (607 ee cells in 622 clone cells, n = 73) (Fig. 3L). Together, our results demonstrate that Notch signaling is required for preventing pupal ISC differentiation into ee cells, thereby maintaining ISC identity.
EMC divisions are symmetric and asymmetric
Consistent with our cell counting and wild-type clonal analysis, from 56 to 78 hours APF, Pros+ ee cell division was symmetric in terms of Pros distribution (Fig. 4, A and B). We refer to ee cells capable of dividing as enteroendocrine mother cells (EMCs). We quantified metaphase division angles of dividing EMCs and found that they centered around 50° (Fig. 4C and fig. S5, A and B). We next determined the pattern of Mira and Baz localization during EMC division. Whereas Mira was not detectable (Fig. 4D), Baz was still present in an apical crescent (Fig. 4E and fig. S5, C and D). Our data demonstrate that EMC divisions are symmetric for Pros distribution but asymmetric for cell polarity.
Fig. 4. EMC divisions are symmetric for Prospero distribution but asymmetric for cell polarity and Notch signaling.
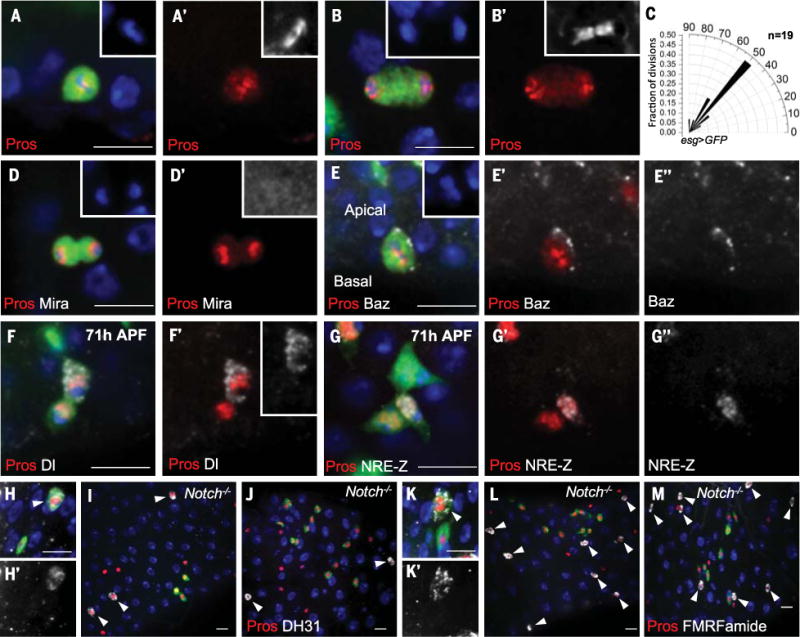
(A and B) Pros symmetrically localizes to both daughters during EMC metaphase (A) and telophase (B). Insets in (A) and (B), DAPI; insets in (A′) and (B′), α-tubulin. (C) Radial histogram quantification of division angles in metaphase of esg>GFP EMC mitosis. (D) Mira staining is absent during EMC division. Inset in (D), DAPI; inset in (D′), Mira (enhanced levels). (E) Baz staining (E″) localizes as a crescent on the apical cell membrane during EMC metaphase. Inset in (E), DAPI. (F) After EMC division (71 hours APF), strong Dl staining [inset in (F′)] is present in one of the Pros+ pair of cells. (G) After EMC division (71 hours APF), one of the two Pros+ cells is NRE-lacZ+ (G´´). (H to M) Wild-type [(H), (H´), (K), and (K´)] and Notch mutant [(I), (J), (L), and (M)] MARCM clones were induced at 24 hours APF, and midguts were dissected at 92 hours APF. In (H) and (H′), the peptide hormone DH31 (arrowhead) is present in a wild-type MARCM ee clone cell (green). Red, Pros; white, DH31. In (I) and (J), Notch mutant MARCM clones (green) are Pros+ but fail to stain for DH31. Arrowheads denote all DH31+ cells (white). [(K) and (K′)] The neuropeptide motif FMRFamide is present in a wild-type MARCM ee clone cell (green). Red, Pros; white, FMRFamide. [(L) and (M)] Notch mutant MARCM clones (green) are Pros+ but fail to stain for FMRFamide. Arrowheads denote all FMRFamide+ cells (white). Scale bars, 10 μm.
Because Notch signaling is asymmetric during specification of EMCs, we next asked whether Notch signaling is also asymmetric during EMC divisions. At 71 hours APF, after EMC division, one daughter in each pair of Pros+ cells was Dl+ (Fig. 4F and fig. S5E) and one daughter was NRE-lacZ+ (Fig. 4G and fig. S5, F to H), demonstrating that Notch signaling is asymmetric. To determine whether asymmetric Notch signaling is functionally relevant, we induced Notch55e11 clones at 24 hours APF and found that mutant cells at 92 hours APF did not express the hormone DH31 or the neuropeptide motif FMRFamide (Fig. 4, H to M). Consistent with this result, culturing esgts>Dl RNAi or esgts>Notch RNAi pupae at 30°C before 64 hours APF blocked DH31 expression in ee cells (fig. S4, A to D). Hence, Notch signaling is required by EMC daughters to properly specify ee cell fate.
Two thresholds of Notch signaling
Expression of an activated form of Notch in adult stem cells promotes their differentiation into ECs (1, 3). Consistent with a previous report (5), we found that expression of an activated form of the Notch receptor (Notchintra1790; see Fig. 5J) by esgts at 30°C was sufficient to promote differentiation of all pupal ISCs into Pdm1+ ECs (Fig. 5, G and H, and fig. S6, H and I) (22). Given that pupal ISCs are NRE-lacZ+ after asymmetric divisions, we wondered why they did not differentiate into ECs. We noticed after ISC asymmetric division that NRE-lacZ staining in ISCs decreased or became absent as pupal development proceeded (fig. S5, F to H), which suggests that (i) endogenous Notch signaling in pupal ISCs is weaker than 30°C misexpression of Notchintra1790, and (ii) weak Notch signaling is sufficient to block ee cell differentiation in pupal ISCs but not to induce EC differentiation. Because the level of Notchintra1790 expression by esgts depends on temperature (23), we created a Notch signaling gradient in pupal ISCs by culturing esgts>Notchintra1790 flies at 30°, 27°, 25°, and 18°C (fig. S6, A to G) and examined their intestines before eclosion. Whereas esgts>GFP control intestine contained both ISCs and ee cells, expression of Notchintra1790 at 30° and 27°C completely blocked ee cell formation (Fig. 5, A, B, C, and F). Pros+ cells were also drastically decreased in esgts>Notchintra1790 intestine reared at 25°C (Fig. 5, D and F), whereas esgts>Notchintra1790 had no effect on ee cell number in animals raised at 18°C (Fig. 5, E and F). Although GFP+ ISCs in esgts>Notchintra1790 intestine reared at 27°C had enough Notch signaling to block ee cell formation, ISCs did not differentiate into Pdm1+ ECs (Fig. 5I). In addition, we ectopically expressed two additional isoforms of the Notch intra cellular domain (Fig. 5J) at 30°C. In descending order of transcriptional activation activity, the three Notchintra isoforms are Notchintra1790 > Notch1792–2156 > Notch1895–2116 (24). Whereas Notch1792–2156 driven by esgts induced all ISCs to differentiate into Pdm1+ ECs (Fig. 5K), esgts>Notch1895–2116 blocked ee cell formation (Fig. 5L) but failed to drive ISC differentiation into ECs (Fig. 5M). These experiments reveal two thresholds of Notch signaling: High Notch signaling promotes EC differentiation, whereas low Notch signaling in ISCs maintains ISCs by preventing their differentiation into ee cells.
Fig. 5. High Notch signaling promotes EC differentiation, whereas low Notch signaling inhibits ee differentiation.

(A to E) Flies were cultured at 18°C until 48 hours APF. Pupae were then moved to various temperatures, and their intestines were analyzed immediately before eclosion. (A) Pros+ cells are present in esgts>GFP control intestine. [(B) to (E)] Pros+ cells are absent in esgts>Notchintra1790 midguts from animals reared at 30°C (B) and 27°C (C). Rare Pros+ cells are present at 25°C (D). Pros+ cells are readily apparent at 18°C (E). (F) Quantification of Pros+ cell numbers in esgts>Notchintra1790 intestine for each of the tested temperatures. ***P < 0.0001. (G) esgts>GFP cells are negative for the EC maker Pdm1. (H) esgts>Notchintra1790 ISCs differentiate into Pdm1+ cells (arrowheads) at 30°C. (I) esgts>Notchintra1790 ISCs (GFP+) are Pdm1− at 27°C. (J) Diagram of different Notch protein isoforms used. (K) esgts>Notchintra1792–2156 ISCs differentiate into Pdm1+ cells (arrowheads) at 30°C. (L) esgts>Notchintra1895–2116 guts lack Pros+ cells at 30°C. (M) esgts>Notchintra1895–2116 ISCs (GFP+) do not differentiate into ECs (Pdm1+) at 30°C. Scale bars, 10 μm.
Adult ee formation is conserved
To determine whether a similar mechanism is used by adult ISCs to generate ee cells, we counted all Pros+ cells that were also positive for the mitotic marker PH3 in esg>GFP adult intestine. Similar to previous reports (25–27), 7.9% (44/558) of PH3+ cells were Pros+. In 27.3% of these cells (12/44), Pros was asymmetrically localized to the basal side of dividing ISCs (Fig. 6, A and A′, and fig. S7, A to C, G, and H). In the remaining 72.7% (32/44), Pros was symmetrically localized to both daughters (fig. S8), similar to what we observed during pupation (Fig. 4, A and B). We next generated two-cell wild-type MARCM clones containing one ISC and one Pros+ ee cell and examined the pattern of Dl and NRE-lacZ expression. As was the case during pupal development, Dl was present in the ee cell but not the ISC (Fig. 6, B and B′). Furthermore, the ISC, but not the ee cell, was NRE-lacZ+ (Fig. 6, C and C′).
Fig. 6. Postmitotic Notch signaling from ee daughters regulates ISC identity.
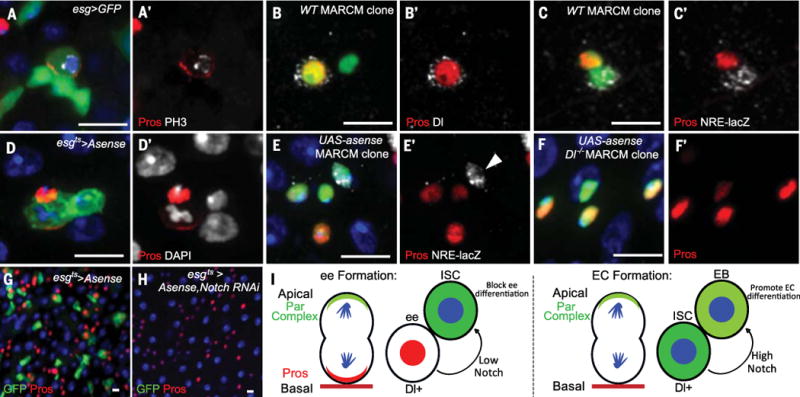
(A) Pros localizes to a crescent in an adult dividing esg>GFP+ cell. (B) A two-cell wild-type MARCM clone contains an ee cell (nuclear Pros+) positive for Dl. (C) A two-cell MARCM clone contains one ee (nuclear Pros+) and an ISC positive for NRE-lacZ. (D) Asymmetric localization of Pros in an esg>Asense cell undergoing mitosis. (E) A MARCM clone driving Asense overexpression contains three ee cells (Pros+) and a NRE-lacZ+ ISC [arrowhead in (E′)]. Green, clone marker. (F) A Dl mutant MARCM clone driving Asense mis-expression. All clone cells are Pros+. Green, clone marker. (G) Asense over-expression by esgts for 8 days in the adult midgut. ISCs are GFP+ Pros– cells. (H) Overexpression of Asense and Notch RNAi by esgts for 8 days during the adult period. esgts>GFP+ cells are absent; all remaining diploid cells are Pros+. (I) Model of how bidirectional Notch regulates ISC multipotency. Left: Notch signaling from a basal ee cell to an apical ISC blocks ee differentiation and maintains ISC identity. Right: Notch signaling from a basal ISC to an apical enteroblast (EB) promotes EC differentiation. Scale bars, 10 μm.
In the Drosophila adult midgut, the proneural gene asense is necessary and sufficient for ee differentiation (27–29). Occasionally, adult ISCs express Asense and make ee cells (27–29). To force all adult ISCs to express Pros and generate ee cells, we ectopically expressed Asense in adult ISCs. Pros asymmetrically localized in every dividing ISC within 36 hours after the onset of over-expression (Fig. 6, D and D′, and fig. S7, D to F). After asymmetric divisions, Dl was present in ee cells (fig. S9A) and ISCs were positive for NRE-lacZ (fig. S9B).
To gain further evidence that Notch signaling functions the same way in adult and pupal ISCs during the ee cell–making process, we overexpressed UAS-asense and an RNAi against Notch together in adult ISCs by using esgts. After 8 days, most ISCs were lost (Fig. 6, G and H, and fig. S9, C and D). To demonstrate that lost ISCs differentiated into ee cells, we made UAS-asense MARCM clones and UAS-asense; Dl RevF10 MARCM clones in adults. UAS-asense clones always contained one Pros– NRE-lacZ′+ cell, which corresponded to the ISC (Fig. 6, E and E′, and fig. S9, E to G). In UAS-asense; DlRevF10 clones, all cells were Pros+ (Fig. 6, F and F′, and fig. S9, H and I). Moreover, relative to 5 days after clone induction (ACI), UAS-asense; DlRevF10 clone number in the posterior midgut was markedly decreased by 10 days ACI (fig. S9, J to L), indicating that ISCs were lost after differentiating into ee cells. Overall, our results demonstrate Notch signaling is required in adult ISCs to maintain stem cell identity during ee cell production.
Discussion
Our findings provide insight into the molecular mechanisms regulating ISC multipotency. Consistent with previous work, high levels of Dl in ISCs activate high levels of Notch in the daughter cell, promoting EC differentiation (3, 27, 30). By contrast, after asymmetric localization of Pros, ISCs require a low Notch signal from their immediate ee cell daughters to remain multipotent. Thus, Notch signaling is both bidirectional and context-dependent (Fig. 6I and fig. S10). Previous work also has suggested that ISCs remain basal during EC formation and that basal ISCs activate the Notch pathway in apical daughter cells (3, 30). However, our data show that ISCs are apically located during ee cell formation and that basal ee cells activate the Notch pathway in apical ISCs. Therefore, Notch signaling is always unidirectional in terms of polarity: Basal daughter cells express Dl, the Notch ligand, in order to activate the Notch signaling pathway in apical daughters during asymmetric ISC divisions.
How might asymmetric Notch signaling between ISCs and ee cells be established and maintained? A recent adult ISC study described the asymmetric segregation of Sara endosomes into the enteroblast, where Notch signaling is activated by Dl and Notch receptor in Sara endosomes (31). Therefore, asymmetric segregation of Sara may also play a role in Notch activation similar to that played by ISCs during ee cell production. In addition, other intracellular trafficking processes, such as asymmetric activation of Dl recycling endosome Rab11 (32) and Arp3 actin polymerization–dependent Dl transportation (33), may also be involved. After mitotic divisions, the basal cell always expresses higher levels of Dl. Although it is unknown why this is the case, the high levels of Dl could inhibit the Notch receptor in the basal cell through a process known as cis inhibition, thereby biasing the direction of Notch signaling toward the apical cell.
After activation of Notch signaling, ISCs toggle from making ee cells to making ECs (fig. S10). Yet although the ISC transiently experiences Notch signaling, it will continue to make only ECs for many divisions afterward. Why might that be? One intriguing possibility is that ISCs would retain an epigenetic memory of Notch signaling, which could act to continuously inhibit Asense expression (27, 29) and repress ee cell formation. That memory might then be reversed by expression of an ee cell–promoting signal, currently unidentified, and/or diluted after a set number of ISC divisions. In addition, loss of the Slit-Robo signaling pathway has recently been shown to result in a modest increase in ee cell production by ISCs (25, 34), raising the possibility that Slit-Robo signaling may prolong the effect of the Notch signaling pathway once ee cells and ISCs are no longer in contact.
Drosophila NBs are derived from ectoderm, whereas ISCs originate from endodermal tissue (35). Despite their distinct germ layer origins, pupal ISC asymmetric divisions share many similarities with embryo and larval type I NBs. Both ISCs and NBs express Dl relative to their neighbors before asymmetric divisions (36). During mitosis, both ISCs and NBs use Baz/Par-3 to define apical-basal polarity, and they both segregate Mira and Pros to the basal daughter cell to direct cell type specification (37). After asymmetric division, ganglion mother cells (GMCs) and EMCs are Notch signaling pathway–negative, whereas NBs and ISCs are Notch signaling pathway– positive (38, 39). Moreover, both GMCs and EMCs divide once more using asymmetric Notch signaling to establish different cell fates between their daughters (19).
Pupal ISCs are, however, different from NBs in a number of ways. Throughout mitosis, the NB remains in an apical position, whereas the location of the ISC depends on the phase of the cell cycle and the type of progenitor produced. NBs give rise to two daughters of unequal size (19)—a larger NB and a smaller GMC—whereas the two daughters generated by ISC divisions are similar in cell size. Furthermore, NBs give rise to only one type of progenitor, the GMC. ISCs, on the other hand, are capable of producing two types of progenitors: an enteroblast or an EMC (fig. S10).
Our data provide evidence that mechanisms regulating tissue homeostasis are more conserved between the Drosophila and mammalian intestine than previously thought. Inhibition of Notch signaling in the mouse intestine induces crypt base columnar stem cell loss and secretory cell hyperplasia, and ectopic Notch signaling promotes EC differentiation (40–43). We have shown that loss of Notch signaling in Drosophila ISCs also leads to stem cell loss and premature ee cell formation, whereas high Notch signaling promotes stem cell differentiation into ECs. Because Notch signaling also plays important roles in common lymphoid progenitors making B cells and T cells (44) and in airway basal cells making secretory cells and ciliated cells (45), it is tempting to speculate that bidirectional Notch signaling may regulate multipotency in these and other progenitors and stem cells.
Supplementary Material
Acknowledgments
We thank S. Feng, R. Mann, S. Kidd, T. Lieber, M. Zecca, G. Struhl, H. Bellen, G. Veenstra, C. Doe, Y. Jan, T. Nystul, and M. Muskavitch for their generous gifts of fly stocks and reagents; I. Driver for production of the Pdm1 antibody; and E. Lucchetta, N. Rafel, C. Montagne, S. Kidd, and T. Lieber for helpful discussions. Supported by NIH grant R01 DK082456-05 (B.O.).
Footnotes
REFERENCES AND NOTES
- 1.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 2.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 3.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 4.Micchelli CA, Sudmeier L, Perrimon N, Tang S, Beehler-Evans R. Identification of adult midgut precursors in Drosophila. Gene Expr Patterns. 2011;11:12–21. doi: 10.1016/j.gep.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Takashima S, et al. Development of the Drosophila enteroendocrine lineage and its specification by the Notch signaling pathway. Dev Biol. 2011;353:161–172. doi: 10.1016/j.ydbio.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/S0166-22360001791-4. [DOI] [PubMed] [Google Scholar]
- 7.Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- 8.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 9.Izumi Y, Ohta N, Itoh-Furuya A, Fuse N, Matsuzaki F. Differential functions of G protein and Baz-aPKC signaling pathways in Drosophila neuroblast asymmetric division. J Cell Biol. 2004;164:729–738. doi: 10.1083/jcb.200309162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 11.Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/S0092-86740080505-X. [DOI] [PubMed] [Google Scholar]
- 12.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 13.Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 14.Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- 15.Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol Cell Neurosci. 1998;12:325–339. doi: 10.1006/mcne.1998.0724. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 17.Schuldt AJ, et al. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen CP, et al. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Zecchini V, Brennan K, Martinez-Arias A. An activity of Notch regulates JNK signalling and affects dorsal closure in Drosophila. Curr Biol. 1999;9:460. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- 22.Lee WC, Beebe K, Sudmeier L, Micchelli CA. Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development. 2009;136:2255–2264. doi: 10.1242/dev.035196. [DOI] [PubMed] [Google Scholar]
- 23.Zeidler MP, et al. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat Biotechnol. 2004;22:871–876. doi: 10.1038/nbt979. [DOI] [PubMed] [Google Scholar]
- 24.Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biteau B, Jasper H. Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 2014;7:1867–1875. doi: 10.1016/j.celrep.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zielke N, et al. Fly-FUCCI: A versatile tool for studying cell proliferation in complex tissues. Cell Rep. 2014;7:588–598. doi: 10.1016/j.celrep.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Zeng X, Hou SX. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142:644–653. doi: 10.1242/dev.113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng X, Lin X, Hou SX. The Osa-containing SWI/SNF chromatin-remodeling complex regulates stem cell commitment in the adult Drosophila intestine. Development. 2013;140:3532–3540. doi: 10.1242/dev.096891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulas S, Conder R, Knoblich JA. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montagne C, Gonzalez-Gaitan M. Sara endosomes and the asymmetric division of intestinal stem cells. Development. 2014;141:2014–2023. doi: 10.1242/dev.104240. [DOI] [PubMed] [Google Scholar]
- 32.Emery G, et al. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng X, et al. Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Rep. 2015;10:1226–1238. doi: 10.1016/j.celrep.2015.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagoshi H. Functional specification in the Drosophila endoderm. Dev Growth Differ. 2005;47:383–392. doi: 10.1111/j.1440-169X.2005.00811.x. [DOI] [PubMed] [Google Scholar]
- 36.Artavanis-Tsakonas S, Delidakis C, Fehon RG. The Notch locus and the cell biology of neuroblast segregation. Annu Rev Cell Biol. 1991;7:427–452. doi: 10.1146/annurev.cb.07.110191.002235. [DOI] [PubMed] [Google Scholar]
- 37.Homem CC, Knoblich JA. Drosophila neuroblasts: A model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 40.Fre S, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 41.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci USA. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 43.VanDussen KL, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 45.Mori M, et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development. 2015;142:258–267. doi: 10.1242/dev.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


