Abstract
The insulin-like growth factors (IGF) system is involved in tumor proliferation, invasion and metastasis in cancer. The current study investigated the association of IGF-1, IGF-2 and IGF-1 receptor (IGF-1R), IGF binding proteins type 3 (IGFBP-3) mRNA expression levels with clinicopathological characteristics and outcomes of 202 patients with untreated colorectal cancer (CRC). IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels were analyzed in surgical specimens of cancer tissues and adjacent normal mucosa cells using reverse transcription-quantitative polymerase chain reaction. The IGF-1R gene expression level was significantly higher in cancer tissue compared with adjacent normal mucosa. By contrast, IGF-1 gene expression levels were reduced in cancer tissue compared with normal mucosa. IGF-2 and IGFBP-3 gene expression levels did not differ significantly between cancer tissue and adjacent normal mucosa. As for the association of gene expression and clinicopathological characteristics, IGFBP-3 gene expression was significantly associated with lymph node metastasis. High IGFBP-3 gene expression was associated with poor 5-year overall survival compared with patients with low IGFBP-3 expression. Furthermore, IGFBP-3 gene expression was identified as an independent prognostic factor using multivariate analysis. Overexpression of the IGFBP-3 gene is considered an effective independent predictor of outcomes in patients with CRC.
Keywords: colorectal cancer, insulin-like growth factor-binding protein-3, insulin-like growth factor-1, insulin-like growth factor-2, insulin-like growth factor-1 receptor, survival, prognostic factor, biomarker
Introduction
Colorectal cancer (CRC) is one of the most frequently diagnosed types of cancer. Previous studies have suggested that an increased risk of CRC may be associated with dietary factors, blood insulin levels and the bioavailability of insulin-like growth factors (IGFs) (1–3).
The IGF system is a complex network consisting of two ligands (IGF-1 and IGF-2), two cell-surface receptors (IGF-1R and IGF-2R), a family of six high-affinity IGF-binding proteins (IGFBPs-1 to −6) and ≥4 additional low-affinity binding proteins (IGFBP-related proteins). This system is involved in normal cell growth, neoplastic transformation and tumor development. Imbalance of the IGF system has been implicated in the pathogenesis and progression of breast and other malignancies (3,4).
Abnormal expression of IGFs, as well as their receptors and binding proteins, has been identified in several malignancies, including CRC (5). IGF-1 may be able to increase the risk of cancer development (6). IGF-2 is also involved in tumor progression and patient survival and has been suggested to function as an autocrine growth factor in CRC (7). Overexpressed IGF-1R may promote invasion, tumor growth, metastasis and progression (8). Furthermore, ≥6 types of IGFBPs are expressed in most tissues and are present in the circulation in normal patients (9). These IGFBPs bind to IGFs with high affinity and the primary role of IGFBPs is to regulate the availability of IGFs for interactions with IGF-1R (10). From these, IGFBP-3 is the most abundant IGFBP in the circulation under normal circumstances, and has been focused on in numerous studies.
The association of the gene expression of the IGF system in tumors with the prognosis or clinicopathological characteristics of patients with CRC remains to be elucidated. In the present study, mRNA expression levels of the IGF-1, IGF-2, IGF-1R and IGFBP-3 genes were measured in cancer tissue and adjacent normal mucosa obtained from 202 patients with CRC. The focus of the current study was to evaluate the mRNA expression levels of the IGF-1, IGF-2, IGF-1R and IGFBP-3 genes and to determine whether expression levels are associated with clinicopathological characteristics and the clinical outcomes of patients with CRC.
Materials and methods
Patients and surgical specimens
A total of 202 patients with untreated CRC were enrolled into the present study. All patients underwent primary tumor resection at Gastroenterological Center at Yokohama City University Medical Center (Yokohama, Japan) or the Kanagawa Cancer Center (Kanagawa, Japan) between December 2002 and June 2006. Informed consent was obtained from each patient and the Ethical Review Boards at Yokohama City University and Kanagawa Cancer Center approved the present study. None of the patients had received chemotherapy or radiotherapy prior to surgery or had any other malignancies. Tumor staging was evaluated according to the 7th edition of the International Union Against Cancer Tumor-Node-Metastasis classification of malignant tumors (11). The resected tumor and adjacent normal mucosa were obtained from the resected colorectum, embedded in Tissue Tek OCT medium (Sakura Finetek Europe B.V., Felmingweg, Netherlands), frozen in liquid nitrogen and stored at −80°C until used for RNA extraction. Sections of 5-µm thickness were stained with hematoxylin and eosin, and histopathological features were examined using a light microscope (CH30; Olympus Corporation, Tokyo, Japan). Sections that consisted of >80% carcinoma cells were defined as cancer tissue and used for total RNA extraction. The clinicopathological characteristics of the patients with CRC are presented in Table I.
Table I.
Associations between the intratumoral expression levels of IGF-1, IGF-2, IGF-1R and IGFBP-3 genes and the clinicopathological characteristics of patients with colorectal cancer.
| IGF-1 expression | IGF-2 expression | IGF-1R expression | IGFBP-3 expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Low n=101 | High n=101 | P-value | Low n=101 | High n=101 | P-value | Low n=101 | High n=101 | P-value | Low n=101 | High n=101 | P-value |
| Age, years | ||||||||||||
| <60 | 28 | 28 | 1.000 | 27 | 29 | 0.753 | 30 | 26 | 0.530 | 28 | 28 | 1.000 |
| ≥60 | 73 | 73 | 74 | 72 | 71 | 75 | 73 | 73 | ||||
| Gender | ||||||||||||
| Male | 52 | 58 | 0.397 | 51 | 59 | 0.258 | 50 | 60 | 0.158 | 56 | 54 | 0.778 |
| Female | 49 | 43 | 50 | 42 | 51 | 41 | 45 | 47 | ||||
| Tumor location | ||||||||||||
| Colon | 62 | 48 | 0.048 | 62 | 48 | 0.048 | 54 | 56 | 0.778 | 53 | 57 | 0.572 |
| Rectum | 39 | 53 | 39 | 53 | 47 | 45 | 48 | 44 | ||||
| Tumor diameter, cm | ||||||||||||
| ≤5 | 69 | 62 | 0.302 | 73 | 58 | 0.027 | 66 | 65 | 0.883 | 71 | 60 | 0.105 |
| >5 | 32 | 39 | 28 | 43 | 35 | 36 | 30 | 41 | ||||
| Histological type | ||||||||||||
| Well differentiated | 30 | 29 | 0.987 | 30 | 29 | 0.668 | 27 | 32 | 0.605 | 29 | 30 | 0.668 |
| Moderately differentiated | 57 | 58 | 55 | 60 | 58 | 57 | 60 | 55 | ||||
| Poorly differentiated | 14 | 14 | 16 | 12 | 16 | 12 | 12 | 16 | ||||
| Depth of invasion | ||||||||||||
| T1 | 9 | 8 | 0.734 | 9 | 8 | 0.781 | 10 | 7 | 0.232 | 13 | 4 | 0.072 |
| T2 | 19 | 14 | 19 | 14 | 19 | 14 | 18 | 15 | ||||
| T3 | 36 | 42 | 38 | 40 | 32 | 46 | 39 | 39 | ||||
| T4 | 37 | 37 | 35 | 39 | 40 | 34 | 31 | 43 | ||||
| Lymph node metastasis | ||||||||||||
| Absent | 53 | 50 | 0.673 | 52 | 51 | 0.888 | 48 | 55 | 0.325 | 59 | 44 | 0.035 |
| Present | 48 | 51 | 49 | 50 | 53 | 56 | 42 | 57 | ||||
| Lymphatic invasion | ||||||||||||
| Absent | 64 | 68 | 0.554 | 64 | 68 | 0.554 | 62 | 70 | 0.237 | 61 | 71 | 0.139 |
| Present | 37 | 33 | 37 | 33 | 39 | 31 | 40 | 30 | ||||
| Venous invasion | ||||||||||||
| Absent | 41 | 34 | 0.308 | 44 | 31 | 0.058 | 31 | 44 | 0.058 | 43 | 62 | 0.109 |
| Present | 60 | 67 | 57 | 70 | 70 | 57 | 58 | 69 | ||||
| Liver metastasis | ||||||||||||
| Absent | 83 | 77 | 0.298 | 82 | 78 | 0.488 | 80 | 80 | 1.000 | 84 | 76 | 0.165 |
| Present | 18 | 24 | 19 | 23 | 21 | 21 | 17 | 25 | ||||
IGF, insulin.like growth factor; IGF.1R, IGF receptor type 1; IGFBP.3, IGF binding protein type 3.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA in resected CRC and adjacent normal mucosa was isolated with the use of TRIzol® Reagent (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). A total of 10 units of DNase I, RNase-free (Roche Applied Science, Penzberg, Germany) was added and the samples were incubated for 20 min at 37°C. Complementary (c)DNA was synthesized from 0.2 µg of total RNA using the iScript cDNA Synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Reverse transcription was performed in a total volume of 20 µl, which contained 4 µl iScript reaction mix, 1 µl iScript reverse transcriptase, and 15 µl (13.3 ng/µl) total RNA. The complete reaction mix was incubated for 5 min at 25°C, 30 min at 42°C, 5 min at 85°C. Following synthesis, the cDNA was diluted to 0.2 µg/µl with H2O and stored at −20°C until required for experiments.
RT-qPCR was performed with an iQ SYBR-Green Supermix kit (Bio-Rad Laboratories, Inc.). PCR reactions were performed in a total volume of 15 µl, which contained cDNA derived from 75 ng of RNA, 0.27 µM of each primer, 7.5 µl of iQ SYBR-Green Supermix containing dATP, dCTP, dGTP, and dTTP (400 µM each) and iTag DNA polymerase (50 units/ml). The PCR consisted of 10 min at 95°C, followed by 40 cycles of denaturation of the cDNA for 10 sec at 95°C, annealing for 10 sec at an appropriate temperature (Table II) and a primer extension for 20 sec at 72°C, followed by 10 min at 72°C. To distinguish specific from nonspecific products and primer dimmers, melting curve analysis was performed. RT-qPCR experiments were performed in triplicate, with two wells for each gene in each experiment. To evaluate specific mRNA expression in samples, a standard curve was produced for each run, measuring three points of the human control cDNA (Clontech Laboratories, Inc., Mountain View, CA, USA). The concentration of each sample was calculated by relating its crossing point to the standard curve (12).
Table II.
Primers and conditions for the polymerase chain reaction.
| Gene/internal control | Primers | Probes (5′-3′) | Annealing temperature (°C) | Product size (bp) |
|---|---|---|---|---|
| IGF-1 | Forward | GTGGATGAGTGCTGCTTC | 58.0 | 134 |
| Reverse | ACTTCCTTCTGGGTCTTGG | |||
| IGF-2 | Forward | TACCGCCATCTCCCTTCTC | 60.0 | 122 |
| Reverse | TCCCTCTGACTGCTCTGTG | |||
| IGF-1R | Forward | TGCCTTGGTCTCCTTGTC | 58.0 | 154 |
| Reverse | TTTCCCTGCTTTGATGGTC | |||
| IGFBP-3 | Forward | TTTCATCTCTCATCTTTTGTCCTC | 60.0 | 77 |
| Reverse | GCCATTCCTCCTTCCTGTTC | |||
| β-actin | Forward | AGTTGCGTTACACCCTTTCTTGAC | 60.0 | 171 |
| Reverse | GCTCGCTCCAACCGACTGC |
IGF, insulin-like growth factor; IGF-1R, IGF-receptor 1; IGFBP, IGF-binding protein; bp, base pair.
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics 20 (IBM SPSS, Armonk, NY, USA). The gene expression levels in cancer tissue were compared with those in adjacent normal mucosa using the Wilcoxon signed-rank test. The associations between gene expression and potential explanatory variables (including age, gender, tumor location, tumor size, histological type, depth of invasion, lymph node metastasis, lymphatic invasion, venous invasion and liver metastasis) were evaluated using the χ2 test. The gene expression levels in the tumors were compared in the presence or absence of lymph node metastasis. Kaplan-Meier curves for the postoperative survival of patients with CRC were plotted and differences in survival rate between groups were analyzed according to the log-rank test. A Cox proportional-hazards model was used to estimate the hazard ratios of variables for postoperative survival. Univariate and multivariate analyses were conducted using a Cox proportional-hazards model to identify independent prognostic factors for postoperative survival. Variables that had a P-value of <0.05 for at least one endpoint on univariate analysis were subsequently included in multivariate analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of the IGF-1, IGF-2, IGF-1R and IGFBP-3 genes in cancer tissue and adjacent normal mucosa
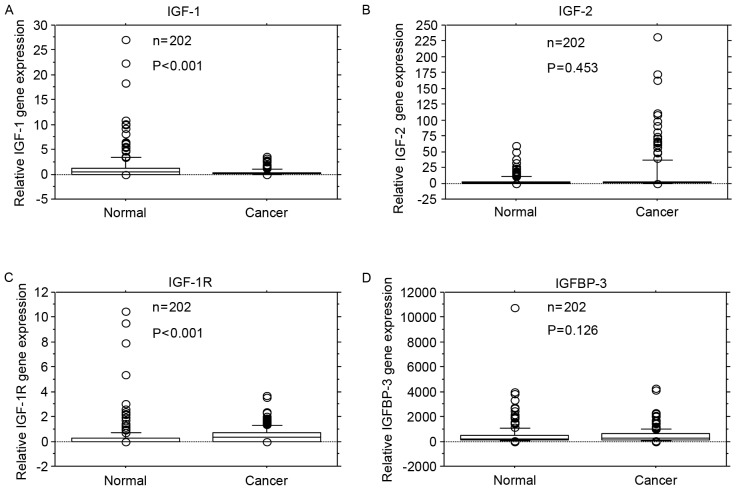
IGF-1 gene expression levels were significantly reduced in cancer tissue compared with adjacent normal mucosa (P<0.001; Fig. 1A). IGF-2 gene expression levels did not differ significantly between cancer tissue and adjacent normal mucosa (P=0.453; Fig. 1B). IGF-1R gene expression levels were significantly higher in cancer tissue compared with adjacent normal mucosa (P<0.001; Fig. 1C). IGFBP-3 gene expression levels did not differ significantly between cancer tissue and adjacent normal mucosa (P=0.126; Fig. 1D).
Figure 1.
Comparison of IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels in colorectal cancer tissue (Cancer) and adjacent normal mucosa (Normal). (A) IGF-1 gene expression levels were lower in cancer tissue compared with adjacent normal mucosa (P<0.001). (B) IGF-2 gene expression levels did not differ significantly between cancer tissue and adjacent normal mucosa (P=0.453). (C) IGF-1R gene expression levels were significantly higher in cancer tissue compared with adjacent normal mucosa (P<0.001). (D) IGFBP-3 gene expression levels did not differ significantly between cancer tissue and adjacent normal mucosa (P=0.126). Box boundaries demonstrate the 25th and 75th percentiles of the observed values. Capped bars indicate the 10th and 90th percentiles and the solid line presents the median average. The P-values were calculated with the Wilcoxon signed-rank test. IGF, insulin-like growth factors; IGF-1R, IGF-1 receptor; IGFBP, IGF binding protein.
Association of IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels to clinicopathological characteristics
Expression levels of the IGF-1, IGF-2, IGF-1R and IGFBP-3 genes were categorized as low or high according to the median values (0.129, 0.362, 0.306 and 292.5, respectively). The associations between gene expression and clinicopathological characteristics were evaluated. Expression levels of the IGFBP-3 gene were significantly associated with lymph node metastasis (P=0.035; Table I). The IGF-1 and IGF-2 gene expression levels were significantly associated with tumor location (P=0.048, P=0.048, respectively).
Association of IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels and lymph node metastasis
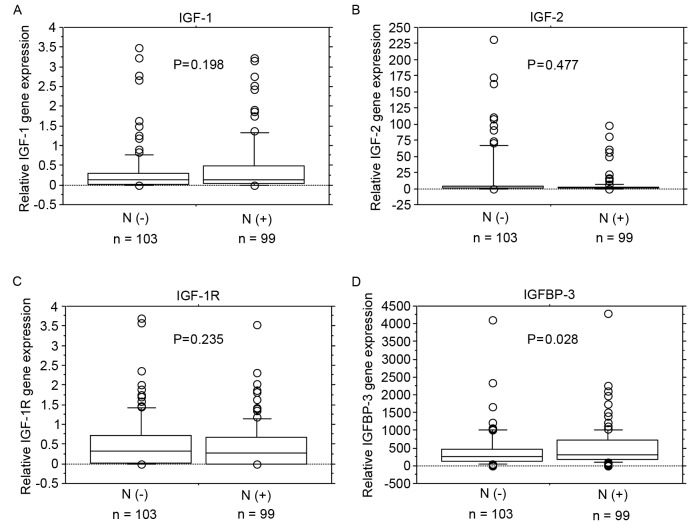
No significant associations were identified between IGF-1, IGF-2 and IGF-1R mRNA expression levels and lymph node metastasis (Fig. 2A-C). IGFBP-3 mRNA expression levels were significantly increased in patients with lymph node metastasis (P=0.028; Fig. 2D).
Figure 2.
mRNA expression levels of IGF-1, IGF-2, IGF-1R and IGFBP-3 in lymph node metastasis. No significant associations were identified between (A) IGF-1, (B) IGF-2 or (C) IGF-1R mRNA expression levels and lymph node metastasis. (D) IGFBP-3 mRNA expression levels were significantly higher in patients with lymph node metastasis (P=0.028). Box boundaries demonstrate the 25th and 75th percentiles of the observed values. Capped bars indicate the 10th and 90th percentiles and the solid line presents the median average. The P-values were calculated with the Mann-Whitney U test. IGF, insulin-like growth factors; IGF-1R, IGF-1 receptor; IGFBP, IGF binding protein; N(−), negative for lymph node metastasis; N(+), positive for lymph node metastasis.
Association between IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels and postoperative survival rate
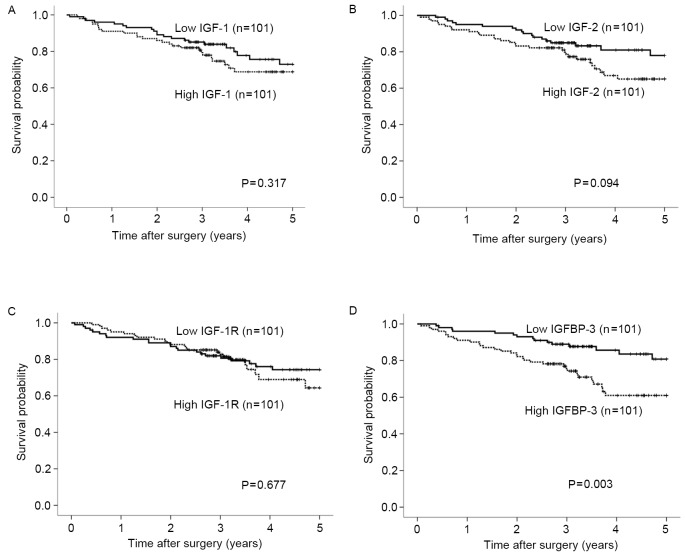
The expression levels of IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA were categorized as low or high according to the respective median values. Post-operative survival did not differ significantly according to expression levels of the IGF-1, IGF-2 or IGF-1R genes (Fig. 3A-C). By contrast, postoperative survival was significantly poorer in patients with high expression levels of the IGFBP-3 gene compared with those with low expression levels (Fig. 3D; P=0.003). Univariate analysis revealed that the depth of invasion, lymph node metastasis, lymphatic invasion, liver metastasis, tumor diameter and IGFBP-3 expression were associated with clinical outcomes (P<0.01). On multivariate Cox proportional-hazards regression analysis, lymph node metastasis, liver metastasis and IGFBP-3 gene expression were independently inversely correlated with patient outcomes (P=0.011, P<0.001, P=0.026, respectively; Table III).
Figure 3.
Association of IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels with postoperative survival rate. Postoperative survival did not differ significantly according to expression levels of the (A) IGF-1, (B) IGF-2 or (C) IGF-1R genes. (D) By contrast, postoperative survival rate was significantly poorer in patients with high expression levels of the IGFBP-3 gene compared with those with low expression levels (P=0.003). Expression levels of IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA were categorized as low or high according to the respective median values. The postoperative survival rate was analyzed using the Kaplan-Meier method and differences in survival rates were assessed with the use of the log-rank test. Results have a median follow-up of 3.24 years. IGF, insulin-like growth factors; IGF-1R, IGF-1 receptor; IGFBP, IGF binding protein.
Table III.
Univariate and multivariate analyses using the Cox proportional hazard model of variables associated with the postoperative survival of patients with colorectal cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
| Gender | ||||||
| Male vs. female | 1.330 | 0.742–2.742 | 0.338 | – | – | – |
| Age, years | ||||||
| ≥60 vs. <60 | 1.367 | 0.695–2.695 | 0.365 | – | – | – |
| Depth of invasion | ||||||
| T3/4 vs. T1/2 | 17.687 | 2.439–128.439 | 0.004 | 5.650 | 0.746–42.746 | 0.094 |
| Lymph node metastasis | ||||||
| Present vs. absent | 6.383 | 2.979–13.979 | <0.001 | 3.038 | 1.292–7.292 | 0.011 |
| Tumor location | ||||||
| Rectum vs. colon | 1.513 | 0.851–2.851 | 0.158 | – | – | – |
| Lymphatic invasion | ||||||
| Present vs. absent | 3.307 | 1.849–5.849 | <0.001 | 1.604 | 0.834–3.834 | 0.157 |
| Venous invasion | ||||||
| Present vs. absent | 1.601 | 0.827–3.827 | 0.163 | – | – | – |
| Liver metastasis | ||||||
| Present vs. absent | 7.258 | 4.033–13.033 | <0.001 | 4.695 | 2.511–8.511 | <0.001 |
| Histological type | ||||||
| Moderate and poor vs. well | 2.045 | 0.955–4.955 | 0.066 | – | – | – |
| Tumor diameter, cm | ||||||
| ≤5 vs. >5 | 2.191 | 1.236–3.236 | 0.007 | 1.225 | 0.677–2.677 | 0.502 |
| IGF-1 expression level | ||||||
| High vs. low | 1.340 | 0.754–2.754 | 0.319 | – | – | – |
| IGF-2 expression level | ||||||
| High vs. low | 1.645 | 0.914–2.914 | 0.097 | – | – | – |
| IGF-1R expression level | ||||||
| High vs. low | 1.130 | 0.635–2.635 | 0.677 | – | – | – |
| IGFBP-3 expression level | ||||||
| High vs. low | 2.439 | 1.320–4.320 | 0.004 | 2.033 | 1.087–3.087 | 0.026 |
IGF-1, insulin-like growth factor type 1; IGF-1R, IGF receptor type 1; IGFBP-3, IGF binding protein type 3; CI, confidence interval.
Discussion
The IGF system serves an important role in the pathogenesis of dysplasia and neoplasia (8,13). The present study investigated tissue expression levels of the IGF-1, IGF-2, IGF-1R and IGFBP-3 genes, the clinicopathological characteristics of 202 patients with untreated CRC, and the associations of these expression levels with postoperative survival.
A number of previous studies have compared the mRNA expression levels of the IGF-1, IGF-2, IGF-1R and IGFBP-3 genes in CRC tissue and adjacent normal mucosa. Freier et al (14) reported that there was no identifiable IGF-1 mRNA in healthy or malignant human colonic tissue. Nosho et al (15) reported significantly increased mRNA expression levels of the IGF-2 gene in colorectal tumor tissue compared with those in adjacent normal mucosa. The mRNA expression levels of the IGF-1R gene were higher in adenocarcinoma tissue of the colon compared with adjacent normal mucosa (14). Another previous study demonstrated that mRNA expression of the IGF-IR gene was detected in ~40% of CRC tissue specimens but was undetectable in adjacent normal mucosa (15). Keku et al (16) reported that IGFBP-3 gene expression was significantly lower in colorectal adenoma tissue compared with normal mucosa. In the current study, the mRNA expression level of the IGF-1R gene was higher in CRC tissue compared with adjacent normal mucosa. However, the mRNA expression levels of the IGF-1 gene were reduced in cancer tissue compared with adjacent normal mucosa. The mRNA expression levels of the IGF-2 and IGFBP-3 genes did not differ significantly between cancer tissue and adjacent normal mucosa.
The association between IGF-1, IGF-2, IGF-1R and IGFBP-3 mRNA expression levels and clinicopathological characteristics were examined in patients with CRC. Peters et al (17) reported that IGF-1 gene expression was not associated with any clinicopathological characteristic in CRC, whilst Shiratsuchi et al (18) reported that IGF-1 gene expression in CRC was associated with tumor size, depth of tumor invasion, lymphatic invasion and venous invasion in CRC. In another previous study, IGF-2 gene expression was correlated with age and tumor size, whilst IGF-1R gene expression did not associate with any clinicopathological characteristic in patients with early CRC (15). Although IGF-1R expression correlated with tumor size and depth of invasion in CRC (18), it did not associate any of the clinicopathological characteristics in patients with prostate cancer (19). Increased postoperative tumor growth and the presence of liver metastasis were associated with significantly elevated IGF-1R gene expression in gastrinoma (20). In the present study, IGF-1 gene expression was significantly associated with tumor location. IGF-2 gene expression was significantly associated with tumor location and tumor size; whilst IGF-1R gene expression was not associated with any clinicopathological characteristic in patients with CRC. In a previous study, IGFBP-3 gene expression was significantly associated with age and positively correlated with tumor stage in CRC (21). Higher mRNA levels of the IGFBP-3 gene were associated with reduced levels of apoptosis (16). Positive associations of IGFBP-3 expression with tumor size and lymph node metastasis have been identified in oral squamous cancer (22). An increased expression level of IGFBP-3 has been associated with lymph node metastasis in pancreatic endocrine neoplasms (23). The IGFBP-3 gene was overexpressed in advanced pancreatic cancer and the intratumoral levels of the IGFBP-3 gene was associated with tumor size (24). The results of the current study are in accordance with the literature that IGFBP-3 gene overexpression is associated with lymph node metastasis.
Finally, the association of IGF-1, IGF-2, IGF-1R and IGFBP-3 gene expression levels and the outcomes of patients with CRC were assessed. In previous studies, IGF-1 expression was not observed to be significantly associated with overall survival rate (17,25). IGF-2 expression has been associated with poorer clinical outcomes in patients with CRC (7,17,26). IGF-1R expression in primary CRC may promote an increased risk of recurrence (27), but was not associated with patients' 5-year survival rate (28). Increased tissue expression levels of the IGFBP-3 gene have been associated with the rapid growth of breast cancer and poor patient outcomes (29,30). Previous immunohistochemical studies of breast cancer demonstrated that IGFBP-3 expression was associated with shorter overall survival (31). High expression levels of the IGFBP-3 gene were associated with unfavorable prognostic characteristics in breast cancer (32). Santosh et al (33) reported that IGFBP-3 overexpression in tumor tissues was an independent predictor of reduced survival rate in patients with newly diagnosed glioblastoma. By contrast, Aishima et al (34) identified that high expression levels of IGFBP-3 were independently associated with an improved survival rate in patients with hepatocellular carcinoma. In patients with squamous cell carcinoma of the tongue, IGFBP-3 expression was associated with favorable outcomes (35). In the present study, the postoperative survival rate was significantly poorer in patients with high expression levels of the IGFBP-3 gene compared with those with low expression levels of the IGFBP-3 gene.
The molecular mechanisms underlying the association between IGFBP-3 expression and poor outcomes in cancer remain to be fully elucidated. Schmid et al (36) reported that IGFBP-3 was overexpressed in the endothelial cells of mouse breast tumor vessels. Granata et al (37) reported that IGFBP-3 dose-dependently promoted neovessels in subcutaneous implants in vivo and suggested that IGFBP-3 promotes angiogenesis and positively regulates the expression of proangiogenic molecules, including vascular endothelial growth factor (VEGF) (37). Yu et al (38) reported a positive correlation between the high expression of epidermal growth factor receptor (EGFR) and the high expression of IGFBP-3 in breast cancer tissue, and Butt et al (10) identified that a blockade of EGFR kinase activity restored the inhibitory effects of IGFBP-3 in vitro. Additionally, Martin et al (39) demonstrated that IGFBP-3 enhanced EGF signaling and its proliferative effects via increased EGFR phosphorylation and the activation of MAP kinase signaling pathways in breast cancer cells in vitro. An associated previous study also reported that IGFBP-3 promotes the proliferation of breast cancer cells through increasing EGFR signaling mediated by SphK1 (40). Therefore, IGFBP-3 has been suggested to promote angiogenesis by inducing VEGF, thereby inducing EGFR signaling mediated by SphK1 and activation of MAP kinase signaling pathways. These effects are considered to promote proliferation of cancer cells, potentially leading to poor survival outcomes. Molecular investigations are required to additionally investigate the role of IGFBP-3 as a prognostic factor and to elucidate the pleiotropic functions of this protein.
In conclusion, high expression of the IGFBP-3 gene was significantly associated with lymph node metastasis and poor outcomes. The results of the present study suggest that overexpression of the IGFBP-3 gene is an important independent predictor of outcomes following surgery in patients with CRC.
References
- 1.Ma J, Pollak MN, Giovannucci E, Chan JM, Tao Y, Hennekens CH, Stampfer MJ. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J Natl Cancer Inst. 1999;91:620–625. doi: 10.1093/jnci/91.23.2052. [DOI] [PubMed] [Google Scholar]
- 2.Shikata K, Ninomiya T, Kiyohara Y. Diabetes mellitus and cancer risk: Review of the epidemiological evidence. Cancer Sci. 2013;104:9–14. doi: 10.1111/cas.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton PE, Banerjee I, Murray PG, Renehan AG. Growth hormone, the insulin-like growth factor axis, insulin and cancer risk. Nat Rev Endocrinol. 2011;7:11–24. doi: 10.1038/nrendo.2010.171. [DOI] [PubMed] [Google Scholar]
- 4.Renehan AG, Zwahlen M, Minder C, O'Dwyer ST, Shalet SM, Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: Systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 5.Durai R, Yang W, Gupta S, Seifalian AM, Winslet MC. The role of the insulin-like growth factor system in colorectal cancer: Review of current knowledge. Int J Colorectal Dis. 2005;20:203–220. doi: 10.1007/s00384-004-0675-4. [DOI] [PubMed] [Google Scholar]
- 6.Rinaldi S, Cleveland R, Norat T, Biessy C, Rohrmann S, Linseisen J, Boeing H, Pischon T, Panico S, Agnoli C, et al. Serum levels of IGF-I, IGFBP-3 and colorectal cancer risk: Results from the EPIC cohort, plus a meta-analysis of prospective studies. Int J Cancer. 2010;126:1702–1715. doi: 10.1002/ijc.24927. [DOI] [PubMed] [Google Scholar]
- 7.Kawamoto K, Onodera H, Kondo S, Kan S, Ikeuchi D, Maetani S, Imamura M. Expression of insulin-like growth factor-2 can predict the prognosis of human colorectal cancer patients: Correlation with tumor progression, proliferative activity and survival. Oncology. 1998;55:242–248. doi: 10.1159/000011858. [DOI] [PubMed] [Google Scholar]
- 8.Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, Church D, Hassan AB. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol. 2005;205:145–153. doi: 10.1002/path.1712. [DOI] [PubMed] [Google Scholar]
- 9.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 10.Butt AJ, Martin JL, Dickson KA, McDougall F, Firth SM, Baxter RC. Insulin-like growth factor binding protein-3 expression is associated with growth stimulation of T47D human breast cancer cells: The role of altered epidermal growth factor signaling. J Clin Endocrinol Metab. 2004;89:1950–1956. doi: 10.1210/jc.2003-030914. [DOI] [PubMed] [Google Scholar]
- 11.Sobin LH, Wittekind C. TNM: Classification of Malignant Tumours. 6th. John Wiley & Sons; Hoboken, NJ: 2002. [Google Scholar]
- 12.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollak M. Insulin-like growth factor physiology and cancer risk. Eur J Cancer. 2000;36:1224–1228. doi: 10.1016/S0959-8049(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 14.Freier S, Weiss O, Eran M, Flyvbjerg A, Dahan R, Nephesh I, Safra T, Shiloni E, Raz I. Expression of the insulin-like growth factors and their receptors in adenocarcinoma of the colon. Gut. 1999;44:704–708. doi: 10.1136/gut.44.5.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosho K, Yamamoto H, Taniguchi H, Adachi Y, Yoshida Y, Arimura Y, Endo T, Hinoda Y, Imai K. Interplay of insulin-like growth factor-II, insulin-like growth factor-I, insulin-like growth factor-I receptor, COX-2, and matrix metalloproteinase-7, play key roles in the early stage of colorectal carcinogenesis. Clin Cancer Res. 2004;10:7950–7957. doi: 10.1158/1078-0432.CCR-04-0875. [DOI] [PubMed] [Google Scholar]
- 16.Keku TO, Sandler RS, Simmons JG, Galanko J, Woosley JT, Proffitt M, Omofoye O, McDoom M, Lund PK. Local IGFBP-3 mRNA expression, apoptosis and risk of colorectal adenomas. BMC Cancer. 2008;8:143. doi: 10.1186/1471-2407-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters G, Gongoll S, Langner C, Mengel M, Piso P, Klempnauer J, Rüschoff J, Kreipe H, von Wasielewski R. IGF-1R, IGF-1 and IGF-2 expression as potential prognostic and predictive markers in colorectal-cancer. Virchows Arch. 2003;443:139–145. doi: 10.1007/s00428-003-0856-5. [DOI] [PubMed] [Google Scholar]
- 18.Shiratsuchi I, Akagi Y, Kawahara A, Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M, Shirouzu K. Expression of IGF-1 and IGF-1R and their relation to clinicopathological factors in colorectal cancer. Anticancer Res. 2011;31:2541–2545. [PubMed] [Google Scholar]
- 19.Mita K, Nakahara M, Usui T. Expression of the insulin-like growth factor system and cancer progression in hormone-treated prostate cancer patients. Int J Urol. 2000;7:321–329. doi: 10.1046/j.1442-2042.2000.00200.x. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa M, Raffeld M, Mateo C, Sakamoto A, Moody TW, Ito T, Venzon DJ, Serrano J, Jensen RT. Increased expression of insulin-like growth factor I and/or its receptor in gastrinomas is associated with low curability, increased growth, and development of metastases. Clin Cancer Res. 2005;11:3233–3242. doi: 10.1158/1078-0432.CCR-04-1915. [DOI] [PubMed] [Google Scholar]
- 21.Georges RB, Adwan H, Hamdi H, Hielscher T, Linnemann U, Berger MR. The insulin-like growth factor binding proteins 3 and 7 are associated with colorectal cancer and liver metastasis. Cancer Biol Ther. 2011;12:69–79. doi: 10.4161/cbt.12.1.15719. [DOI] [PubMed] [Google Scholar]
- 22.Zhong LP, Yang X, Zhang L, Wei KJ, Pan HY, Zhou XJ, Li J, Chen WT, Zhang ZY. Overexpression of insulin-like growth factor binding protein 3 in oral squamous cell carcinoma. Oncol Rep. 2008;20:1441–1447. [PubMed] [Google Scholar]
- 23.Hansel DE, Rahman A, House M, Ashfaq R, Berg K, Yeo CJ, Maitra A. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res. 2004;10:6152–6158. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- 24.Xue A, Scarlett CJ, Jackson CJ, Allen BJ, Smith RC. Prognostic significance of growth factors and the urokinase-type plasminogen activator system in pancreatic ductal adenocarcinoma. Pancreas. 2008;36:160–167. doi: 10.1097/MPA.0b013e31815750f0. [DOI] [PubMed] [Google Scholar]
- 25.Sheen-Chen SM, Chou FF, Hsu W, Huang CC, Eng HL, Tang RP. Lack of prognostic value of insulin-like growth factor-1 in patients with breast cancer: Analysis with tissue microarray. Anticancer Res. 2007;27:3541–3544. [PubMed] [Google Scholar]
- 26.Xu Z, Liu F, Qi X, Li J. Relationship between insulin-like growth factor II and prognosis of colorectal cancer. Zhonghua Wai Ke Za Zhi. 1999;37:718–720. 743. (In Chinese) [PubMed] [Google Scholar]
- 27.Nakamura M, Miyamoto S, Maeda H, Zhang SC, Sangai T, Ishii G, Hasebe T, Endoh Y, Saito N, Asaka M, Ochiai A. Low levels of insulin-like growth factor type 1 receptor expression at cancer cell membrane predict liver metastasis in Dukes' C human colorectal cancers. Clin Cancer Res. 2004;10:8434–8441. doi: 10.1158/1078-0432.CCR-04-0430. [DOI] [PubMed] [Google Scholar]
- 28.Shiono S, Ishii G, Nagai K, Murata Y, Tsuta K, Nitadori J, Kodama T, Ochiai A. Immunohistochemical prognostic factors in resected colorectal lung metastases using tissue microarray analysis. Eur J Surg Oncol. 2006;32:308–309. doi: 10.1016/j.ejso.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Vestey SB, Perks CM, Sen C, Calder CJ, Holly JM, Winters ZE. Immunohistochemical expression of insulin-like growth factor binding protein-3 in invasive breast cancers and ductal carcinoma in situ: Implications for clinicopathology and patient outcome. Breast Cancer Res. 2005;7:R119–R129. doi: 10.1186/bcr963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheen-Chen SM, Zhang H, Huang CC, Tang RP. Insulin-like growth factor-binding protein-3 in breast cancer: Analysis with tissue microarray. Anticancer Res. 2009;29:1131–1135. [PubMed] [Google Scholar]
- 31.Kim JH, Cho YH, Park YL, Sohn JH, Kim HS. Prognostic significance of insulin growth factor-I receptor and insulin growth factor binding protein-3 expression in primary breast cancer. Oncol Rep. 2010;23:989–995. doi: 10.3892/or_00000724. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Levesque MA, Khosravi MJ, Papanastasiou-Diamandi A, Clark GM, Diamandis EP. Insulin-like growth factor-binding protein-3 and breast cancer survival. Int J Cancer. 1998;79:624–628. doi: 10.1002/(SICI)1097-0215(19981218)79:6<624::AID-IJC12>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Santosh V, Arivazhagan A, Sreekanthreddy P, Srinivasan H, Thota B, Srividya MR, Vrinda M, Sridevi S, Shailaja BC, Samuel C, et al. Grade-specific expression of insulin-like growth factor-binding proteins-2, −3, and −5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol Biomarkers Prev. 2010;19:1399–1408. doi: 10.1158/1055-9965.EPI-09-1213. [DOI] [PubMed] [Google Scholar]
- 34.Aishima S, Basaki Y, Oda Y, Kuroda Y, Nishihara Y, Taguchi K, Taketomi A, Maehara Y, Hosoi F, Maruyama Y, et al. High expression of insulin-like growth factor binding protein-3 is correlated with lower portal invasion and better prognosis in human hepatocellular carcinoma. Cancer Sci. 2006;97:1182–1190. doi: 10.1111/j.1349-7006.2006.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadimitrakopoulou VA, Brown EN, Liu DD, El-Naggar AK, Lee Jack J, Hong WK, Lee HY. The prognostic role of loss of insulin-like growth factor-binding protein-3 expression in head and neck carcinogenesis. Cancer Lett. 2006;239:136–143. doi: 10.1016/j.canlet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Schmid MC, Bisoffi M, Wetterwald A, Gautschi E, Thalmann GN, Mitola S, Bussolino F, Cecchini MG. Insulin-like growth factor binding protein-3 is overexpressed in endothelial cells of mouse breast tumor vessels. Int J Cancer. 2003;103:577–586. doi: 10.1002/ijc.10874. [DOI] [PubMed] [Google Scholar]
- 37.Granata R, Trovato L, Lupia E, Sala G, Settanni F, Camussi G, Ghidoni R, Ghigo E. Insulin-like growth factor binding protein-3 induces angiogenesis through IGF-I- and SphK1-dependent mechanisms. J Thromb Haemost. 2007;5:835–845. doi: 10.1111/j.1538-7836.2007.02431.x. [DOI] [PubMed] [Google Scholar]
- 38.Yu H, Levesque MA, Khosravi MJ, Papanastasiou-Diamandi A, Clark GM, Diamandis EP. Associations between insulin-like growth factors and their binding proteins and other prognostic indicators in breast cancer. Br J Cancer. 1996;74:1242–1247. doi: 10.1038/bjc.1996.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin JL, Weenink SM, Baxter RC. Insulin-like growth factor-binding protein-3 potentiates epidermal growth factor action in MCF-10A mammary epithelial cells. Involvement of p44/42 and p38 mitogen-activated protein kinases. J Biol Chem. 2003;278:2969–2976. doi: 10.1074/jbc.M210739200. [DOI] [PubMed] [Google Scholar]
- 40.Martin JL, de Silva HC, Lin MZ, Scott CD, Baxter RC. Inhibition of insulin-like growth factor-binding protein-3 signaling through sphingosine kinase-1 sensitizes triple-negative breast cancer cells to EGF receptor blockade. Mol Cancer Ther. 2014;13:316–328. doi: 10.1158/1535-7163.MCT-13-0367. [DOI] [PubMed] [Google Scholar]