Abstract
Toll-like receptor 4 (TLR4) is a pattern recognition receptors, a member of the Toll-like receptor family and it serves a role in innate and acquired immunity. It has previously been reported that TLR4 was overexpressed in a variety of tumor tissues and cells, including colorectal cancer, gastric cancer and ovarian cancer. In the tumor microenvironment, the TLR4 signaling pathway may be activated in order to upregulate forkhead box P3 (Foxp3) expression in regulatory T cells (Tregs), and thus enhance the immunosuppressive function of Tregs. Also, inflammatory cytokine release would be increased, which promotes tumor immune system evasion. Additionally, it has previously been reported that TLR4 activation may induce histone methylation changes at multiple sites. However, the effects of the alterations to histone methylation in the process of TLR4-associated tumor immune system evasion are not currently known. Histone methylation serves a critical role in regulating gene expression. Abnormal histone methylation is closely associated with tumor development and progression. In order to investigate the epigenetic mechanisms underlying Foxp3 regulation by TLR4, the human lung adenocarcinoma cell line A549 was used. In the present study, it was revealed that the expression level of H3K9me1/2 histone lysine demethylase 3A (KDM3A) was significantly increased following TLR 4 activation in the lung adenocarcinoma A549 cell line, whereas silencing of KDM3A expression led to significantly reduced Foxp3 expression under TLR4 regulation. This result suggests that KDM3A participates in TLR4 regulation of Foxp3 transcription. Additional analysis revealed that during nuclear transport of Foxp3, KDM3A may directly bind to the Foxp3 promoter and activate its transcription. This results in increased secretion of Foxp3-downstream inhibitory cytokines, including transforming growth factor-β1 (TGF-β1), interleukin 35 (IL-35) and heme oxygenase 1 (HO-1), which have immunosuppressive effects and ultimately facilitate the immune escape of lung cancer cells. From the results, the present study concluded that TLR4 activation promoted the expression of H3K9me1/2 demethylase KDM3A. KDM3A bound directly to the Foxp3 promoter and promoted Foxp3 transcription, thereby inducing the secretion of Foxp3-associated downstream inhibitory cytokines (TGF-β1, IL-35, and HO-1), ultimately facilitating the immune system evasion of lung adenocarcinoma.
Keywords: lysine demethylase 3A, Toll-like receptor 4, forkhead box p3, inhibitory cytokines, lung adenocarcinoma
Introduction
Histone methyltransferase and histone demethylase participate in and maintain different histone methylation states (1). Post-translational histone modifications are involved in various cellular processes through regulation of the chromatin structure and also participate in recognition of histone methylation through various molecules, thereby inducing downstream processes (2). Histone methylation serves a critical role in numerous biological processes such as heterochromatin formation and transcriptional regulation (3,4). Abnormal histone methylation is closely associated with tumor development and progression and is known to serve a role in oncogene activation and tumor suppressor inactivation (5–7). Abnormal histone methylation in tumors often manifests as H3K4 demethylation and H3K27 methylation of tumor suppressor genes, which may lead to the inactivation of these suppressor genes (8). Additionally, H3K27 demethylation and H3K4 methylation of proto-oncogenes can lead to oncogene activation, which ultimately promotes tumor development and progression (9).
Histone lysine demethylase 3A (KDM3A), which contains a Jumonji C-terminal domain (JMJC) -containing enzyme and belongs to the JMJ domain-containing proteins (JMJD) protein family, can specifically catalyze histone H3K9me1/2 demethylation (10). KDM3A has a variety of biological functions and participates in nuclear receptor activation, energy metabolism, spermatogenesis, muscle cell development, hypoxia-induced stress response regulation and stem cell self-renewal (11). Additionally, KDM3A serves an important role in tumor development and progression. It has previously been reported that KDM3A promotes tumor growth and invasion as well as inducing angiogenesis in hypoxia (12,13). However, it remains unclear whether KDM3A performs a role in tumor immune escape, an important factor in tumor development and progression.
It has been reported that Toll-like receptor 4 (TLR4) activation induces histone methylation changes at multiple sites (14). TLR4, as a member of the TLR family, is mainly expressed in immune cells and performs a role in innate and acquired immunity (15). Previous studies have reported that TLR4 is also expressed in a variety of tumor cell lines (16–20). In the tumor microenvironment, the TLR4 signaling pathway can be activated to upregulate forkhead box P3 (Foxp3) expression in regulatory T cells (Tregs) and thereby enhance the immunosuppressive function of Tregs (21). In addition, inflammatory cytokines release is increased to promote tumor immune escape (22). Foxp3 is a member of the forkhead/winged helix transcription factor family and is primarily expressed in cluster of differentiation (CD)4+CD25+ Tregs as a critical factor of cell development and function (23). Tregs mainly perform immunosuppressive functions through the following three pathways: Cell-cell contact inhibition; metabolic disruption; and secretion of the inhibitory cytokines interleukin (IL)-35, IL-10, and transforming growth factor-β (TGF-β) (24). It has previously been revealed that Foxp3 is expressed not only in Tregs, but also in various tumor tissues and cell lines, including pancreatic, breast, prostatic and colon carcinoma (25,26). Additionally, Foxp3 is closely associated with the development, progression and prognosis of certain types of cancer (27). However, the role of Foxp3 differs in various tumor subtypes. To date, few studies have investigated the role of Foxp3 in lung cancer, and the molecular mechanisms involved in the regulation of Foxp3 expression in lung cancer cells have not been elucidated.
Previous studies have demonstrated that the TLR4 expression level is positively correlated with Foxp3 expression level in human non-small cell lung tumor tissues (28,29). Additionally, the TLR4 signaling pathway activation can induce Foxp3 expression in the human lung adenocarcinoma A549 cell line (29). Nonetheless, it remains unknown whether histone methylation changes during TLR4 activation or whether specific molecular mechanisms of TLR4 participate in the regulation of Foxp3 expression in lung cancer cells. The present study demonstrated that TLR4 activation in A549 cells promoted the expression of H3K9me1/2 demethylase KDM3A. KDM3A directly activates Foxp3 transcription through the demethylation of histone H3K9 by binding to its promoter region. This process results in increased secretion of downstream inhibitory cytokines and thereby facilitates the immune escape of lung cancer cells. This is a novel mechanism of tumor immune escape based on the deregulation of the JMJC-domain containing histone demethylase.
Materials and methods
Cell culture
A549 cells were obtained from the Laboratory of Immunology of Jilin University (Changchun, China) maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and cultured at 37°C in 5% CO2.
Reverse transcription quantitative-polymerase chain reaction (RT-qPCR)
Total RNA extraction from A549 cells was performed using TRIzol® reagent (Takara Biotechnology Co., Ltd., Dalian, China). Reverse transcription was performed using the Moloney murine leukemia virus and the oligo deoxy-thymidine primer (Takara Biotechnology Co., Ltd.). The quantity of total RNA was then determined using an Epoch multi-volume spectrophotometer system (BioTek Corporation, Beijing, China). cDNA was synthesized from 1.0 mg total RNA using reverse transcriptase (Takara Biotechnology Co., Ltd.), Moloney murine leukemia virus (Takara Biotechnology Co., Ltd.) and oligonucleotides (dT; Takara Biotechnology Co., Ltd.) in a total volume of 20 ml, according to the manufacturer's protocols. RT-qPCR was performed using an ABI PRISM 7300 sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR Premix Ex Taq™ II (Takara Biotechnology Co., Ltd), following the manufacturer's protocol and using the following conditions: 95°C for 30 sec, 95°C for 5 sec and 60°C for 30 sec, for 40 cycles. The results were analyzed according to the 2−∆∆Cq formula (30). The following primers (Takara Biotechnology Co., Ltd.) were used: Sense, 5′-CTCGAACACCTTTGACAGCTCG-3′ and antisense, 5′-GGGTCATTGATGGCAACAATATC for GAPDH; sense, 5′-TCCCAGAGTTCCTCCACAAC-3′ and antisense, 5′-AGGTTGTGGCGGATGGCGTTCTTC-3′ for Foxp3; sense, 5′-CCAGGCAGAGAATGCTGAGTTC-3′ and antisense, 5′-CCAGGCAGAGAATGCTGAGTTC-3′ for heme oxygenase 1 (HO-1); sense, 5′-TACCTGAACCCGTGTTGCTCTC-3′ and antisense, 5′-GTTGCTGAGGTATCGCCAGGAA-3′ TGFβ1; sense, 5′-TCTCCGAGATGCCTTCAGCAGA-3′ and antisense, 5′-TCAGACAAGGCTTGGCAACCCA-3′ IL-10; sense, 5′-TGCCTTCACCACTCCCAAAACC-3′ and antisense, 5′-CAATCTCTTCAGAAGTGCAAGGG-3′ for IL-12A; sense, 5′-CTGGATCCGTTACAAGCGTCAG-3′ and antisense, 5′-CACTTGGACGTAGTACCTGGCT-3′ for Epstein-Barr virus induced 3 (EBI3; IL-35 is encoded by two separate genes, IL-12α and EBI3); sense, 5′-GCCAACATTGGAGACCACTTCTG-3′ and antisense, 5′-CTCGAACACCTTTGACAGCTCG-3′ for KDM3A; sense, 5′-ACTGCTGACCATTGCTGAACGC-3′ and antisense, 5′-CCTCCTTGAGAGCCTGGATGTT-3′ for KDM3C; sense, 5′-GTGCTTTGTGGTCAGCGGAAGT-3′ and antisense, 5′-TGTGAGACAGCAACCCACGGTG-3′ for KMT2A; sense, 5′-TGTGAGACAGCAACCCACGGTG-3′ and antisense, 5′-TGCCGAATCAGCAGCTCTCGTA-3′ for KMT2D; sense, 5′-CACAGATTGTCAGTGATGCTGAAG-3′ and antisense, 5′-CTGCTGTCCAATGTGAGTCCTAC-3′ for KMT2E; sense 5′-CACCACATAGTCAGTGCTTCCTG-3′ and antisense 5′-AGTCTGACAGCGAGAGTTAGCC-3′ for enhancer of zeste 1 polycomb repressive complex 2 subunit (EZH1); sense, 5′-GACCTCTGTCTTACTTGTGGAGC-3′ and antisense, 5′-CGTCAGATGGTGCCAGCAATAG-3′ for enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2); sense, 5′-CCGATGACTCTTGTGAAGCAGC-3′ and antisense, 5′-GACTTCGTCTGCCAAAGTGGA-3′ for KDM4C. As aforementioned, IL-35 is a dimeric protein composed of IL-12α and EBI3. In order to detect the mRNA level of IL-35, IL-12α and EBI3 were required to be detected at the same time.
Western blot analysis
Cells were lysed with radioimmunoprecipitation assay lysis buffer, and the protein concentrations in the cell lysates were measured using a BCA protein assay kit (Beyotime Institute of Biotechnology, Jiangsu, China). Equal amounts of cell lysate protein were separated by 10% SDS-PAGE, and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with TBS and Tween 20 (TBST) containing 5% nonfat milk for 2 h at room temperature, and then incubated with anti-KDM3A (dilution, 1:1,000; cat. no. YM0388), anti-Foxp3 (dilution, 1:1,000; cat no. YM0827) or β-actin mouse monoclonal antibody (dilution, 1:4,000; cat. no. YM3039). All antibodies were supplied by Immunoway, Jiangsu, China primary antibodies for 2 h at room temperature. Following washing with TBST three times at room temperature, they were then incubated with horseradish peroxidase (HRP)-labeled goat anti-mouse secondary antibodies (dilution, 1:4,000; cat. no., LK2003) and HPR-labeled goat anti-rabbit secondary antibodies (dilution, 1:4,000; cat. no., K2001); Both were supplied by Immunoway for 45 min at room temperature. Subsequently they were washed with TBST three times at room temperature. Immunoreactive bands were detected using the enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Inc.).
RNA interference
Small interfering RNAs (siRNAs) against KDM3A and Foxp3 plasmids were designed and synthesized from RiboBio (Guangzhou RiboBio Co., Ltd., Guangzhou, China). The A549 cells were resuspended in serum-free DMEM, 3.5×105 cells were seeded in 6-well plate and transfected with small interfering RNA (siRNA)-Foxp3/siRNA-KDM3A or siRNA-negative control (NC); using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), then the medium was changed with DMEM containing 10% FBS 4–6 h later. Cells were harvested at 48 h subsequent to transfection for RT-qPCR or subsequent to 72 h for western blot analysis. A total of 3 different Foxp3/KDM3A-specific siRNAs were screened, and the most efficient one was chosen for experiments. The specific siRNA for Foxp3 was 5′-CAUGGACUACUUCAAGUUCdTdT-3′. The specific siRNA for KDM3A was sense 5′-CCACCUAACCUUGGAGCAAdTdT-3′.
ELISA
A549 cells were seeded at 3.5×105 per well in 6-well plates, incubated overnight at 37°C and transfected with siRNA-Foxp3 as described above. After 36 h, the cells were stimulated with lipopolysaccharide (LPS; Sigma-Aldrich; Merck KGaA; Darmstadt, Germany) for 24 h. The concentrations of TGF-β1, IL-35 and HO-1 in the supernatants were measured with human TGF-β1, human IL-35 and human HO-1 ELISA kits (All from Elabscience, Wuhan, China).
Flow cytometry (FCM)
A549 cells were seeded at 3.5×105 per well in 6-well plates, incubated overnight at 37°C and transfected with siRNA-Foxp3 as described above. A549 cells were collected and counted, and 1×106 cells were suspended in PBS (100 µl total volume). The cells were fixed with 4% paraformaldehyde at 4°C for 1 h, and then treated with 0.1% saponin (Sigma-Aldrich; Merck KGaA) and phycoerythrin (PE) anti-mouse Foxp3 monoclonal antibody (cat. no. M300F8-09A; 5 µl/test; Tianjin Sungene Biotech, Co., Ltd., Tianjin, China) at 4°C for 1 h. The cells were washed twice with PBS and resuspended in 2% paraformaldehyde (Sigma-Aldrich; Merck KGaA). A total of 105 events were assessed using BD Accuri (BD Biosciences, Franklin Lakes, NJ, USA), and the data were analyzed using the BD Accuri C6 Software version 6.0.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Subsequent to blocking with 5% bovine serum albumin, the cells were incubated with rabbit anti-KDM3A (1:1,000; cat. no. YM0388; Immunoway, Jiangsu, China) antibody and mouse anti-Foxp3 antibody (1:500; cat. no. YM0827; Immunoway, Jiangsu, China) overnight at 4°C. Subsequent to washing, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit (dilution, 1:200; cat. no. GR200G-02C; Tianjin Sungene Biotech, Co.) and PE-conjugated goat anti-mouse immunoglobulin G (dilution, 1:150; cat. no. GM200G-09C; Tianjin Sungene Biotech, Co.) were added for 2 h at 37°C in the dark. The nuclei were stained with 1 µg/ml DAPI (Sigma-Aldrich; Merck KGaA) for 3 min. Following washing with PBS 3 times, the cells were observed under an inverted fluorescence microscope (IX71; Olympus Corporation, Tokyo, Japan).
Dual luciferase reporter assay
A549 cells in 96-well plates were transfected with pRL-TK and PGL3-basic or PGL3-Foxp3-promoter luciferase (all supplied by Promega Corporation, Madison, WI, USA), using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers protocol. The pRL-TK (Promega Corporation) plasmid contains the Renilla reniformis luciferase gene under the transcriptional control of constitutively expresses low levels of Renilla luciferase. Transfected cells were lysed using Passive Lysis Buffer (Promega Corporation), and then the luciferase activities in the cell lysates were analyzed using a Dual Luciferase Reporter assay kit (Promega Corporation).
Statistical analysis
Student's t-test for independent samples was used to compare data between the experimental groups. All data were analyzed by statistical software SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) and are presented as the mean ± standard error. P<0.05 was considered to indicate a statistically significant difference.
Results
TLR4 activation promotes Foxp3 expression and inhibitory cytokines secretion in A549 cells
To observe the effect of TLR4 activation on Foxp3 expression and the relevant inhibitory cytokines secretion in A549 cells, the present study first determined Foxp3 expression changes following LPS-induced activation of the TLR4 signaling pathway in A549 cells (Fig. 1A and B). Subsequently, the present study examined TLR4 activation-induced expression changes of the inhibitory cytokines, TGF-β1, IL-35, IL-10 and HO-1 that promote tumor immune escape. The mRNA level changes were detected by RT-qPCR, and the protein expression level changes were detected by ELISA. The results demonstrated that except for IL-10 expression (not detected, data no shown), the inhibitory cytokines were increased at the mRNA or protein level following LPS activation of the TLR4 signaling pathway in A549 cells (P<0.05; Fig. 1C and D). These results suggest that following TLR4 activation in A549 cells, these cytokines are secreted to inhibit the functions of effector T cells and dendritic cells (DCs) and facilitate immune escape of lung cancer cells.
Figure 1.
LPS induces the increased expression of Foxp3 and production of inhibitory cytokines in A549 cells. (A) A549 cells were stimulated with LPS (5 µg/ml) for 24 h, mRNA expression of Foxp3 was detected by RT-qPCR. (B) Protein level of Foxp3 was detected using flow cytometry. (C) mRNA expression of inhibitory cytokines including TGF-β1, IL-35 and HO-1 were detected by RT-qPCR. IL-35 is a dimeric protein composed of IL-12α and EBI3. In order to detect the mRNA level of IL-35, IL-12α and EBI3 were required to be detected at the same time. (D) Protein amounts of these inhibitory cytokines in the supernatants were detected using ELISA. Data are represented as the mean ± standard deviation of 3 independent experiments. *P<0.05 compared with the control group. Foxp3, forkhead box P3; LPS, lipopolysaccharide; RT-qPCR, reverse transcription quantitative-polymerase chain reaction; TGF-β1, transforming growth factor-β1; IL-35, interleukin 35; HO-1, heme oxygenase 1.
TLR4 activation regulates inhibitory cytokines secretion through Foxp3 in A549 cells
To additionally determine whether TLR4 performs an immunosuppressive function by promoting inhibitory cytokines secretion during the regulation of Foxp3 expression in A549 cells, the present study downregulated Foxp3 expression using siRNA (Fig. 2A and B). TLR4 expression on A549 was activated with LPS (5 µg/ml) 48 h subsequent to transfection with siRNA-Foxp3 or control siRNA. The results demonstrated that TGF-β1, IL-35, and HO-1 expression levels were significantly reduced at both the mRNA and protein levels following Foxp3 silencing in A549 cells (P<0.05; Fig. 2C and D). This observation suggests that Foxp3 performs a role in tumor immune escape by secreting inhibitory cytokines in A549 cells, and this process is regulated by TLR4.
Figure 2.
Decreased inhibitory cytokines production in Foxp3-silenced A549 cells in response to LPS stimulation. (A) A549 cells were transfected with siRNA-NC or siRNA-Foxp3 and mRNA expression of Foxp3 was detected by RT-qPCR. (B) Protein amount of Foxp3 was detected using western blot analysis. IL-35 is a dimeric protein composed of IL-12α and EBI3. In order to detect the mRNA level of IL-35, IL-12α and EBI3 were required to detected at the same time. (C) A549 cells were harvested and transfected with siRNA-NC or siRNA-Foxp3, 48 h subsequent to interference, A549 were stimulated with LPS (5 µg/ml) for another 24 h. The mRNA expression of the inhibitory cytokines TGF-β1, IL-35 and HO-1 were detected by RT-qPCR at the indicated times. (D) TGF-β1, IL-35 and HO-1 in the supernatants were detected using ELISA. Data in C and D are represented as the mean ± standard deviation of 3 independent experiments. *P<0.05 compared with the control group. Foxp3, forkhead box P3; LPS, lipopolysaccharide; siRNA, small interfering RNA; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TGF-β1, transforming growth factor-β1; IL-35, interleukin 35; HO-1, heme oxygenase 1.
Expression of histone demethylase KDM3A is increased following TLR4 activation
To observe the histone methylation changes of TLR4 during regulation of Foxp3 expression, the present study detected the expression levels of associated histone methyltransferases and demethylases (KDM3A, KDM5C, KMT2A, KMT2D, KMT2E, KDM4C, EZH1 and EZH2) during TLR4 activation in A549 cells (14). The expression changes of these KMTs or KDMs in A549 cells were analyzed by RT-qPCR following LPS stimulation for 24 h. The results revealed that KDM3A was increased significantly following TLR4 activation (P<0.05; Fig. 3). This finding suggests that the H3K9me1/2 demethylase KDM3A is associated with TLR4 activation in A549 cells. KDM3A likely participates in TLR4-mediated transcriptional regulation of Foxp3 and thus performs a role in the Foxp3-induced promotion of tumor immune escape.
Figure 3.
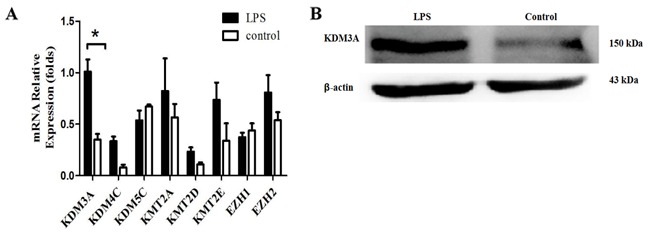
Upregulation of KDM3A expression in A549 cells by LPS stimulation. (A) A549 cells were stimulated or not with LPS (5 µg/ml) for 24 h. RT-PCR analysis of KDMs and KMTs mRNA amounts at indicated time. Data are represented as the mean ± standard deviation of 3 independent experiments. *P<0.05 compared with the control group. (B) Protein amount of KDM3A in A549 cells subsequent to LPS stimulation for 24 h was detected using western blot analysis. β-actin is loading control. Data are shown as one typical result from 3 independent experiments with similar results. KDM3A, lysine demethylase 3A; LPS, lipopolysaccharide; RT-PCR, reverse transcription-polymerase chain reaction; KDMs, lysine methyltransferases; KMTs, demethylases.
KDM3A facilitates TLR4 activation-induced Foxp3 expression and inhibitory cytokines secretion in A549 cells
To examine the role of KDM3A in the TLR4-mediated regulation of Foxp3, the present study designed a specific siRNA for the human KDM3A gene (siRNA-KDM3A) and transfected it into A549 cells. The silencing efficiency was determined by RT-qPCR and western blot assays 48 h following transfection. The results showed that the silencing effect of KDM3A was evident in A549 cells transfected with siRNA-KDM3A (Fig. 4A and B), which can be used to study cellular signaling in the absence of KDM3A.
Figure 4.
Decreased inhibitory cytokines production in KDM3A-silenced A549 cells in response to LPS stimulation. (A) A549 cells were transfected with siRNA-NC or siRNA-KDM3A. The mRNA expression of KDM3A was detected by RT-qPCR. The protein amount of KDM3A was detected using western blot analysis. (B) A549 cells were harvested and transfected with control siRNA or siRNA-KDM3A. Subsequently, 48 h following interference, A549 were stimulated with LPS (5 µg/ml) for another 24 h. The mRNA expression of Foxp3 and inhibitory cytokines TGF-β1, EBI3, IL-12α and HO-1 were detected by RT-qPCR at the indicated times. IL-35 is a dimeric protein composed of IL-12α and EBI3. In order to detect the mRNA level of IL-35, IL-12α and EBI3 were required to detected at the same time. (C) Protein level of Foxp3 was detected by flow cytometry. (D) TGF-β1, IL-35 and HO-1 expression levels in the supernatants were detected using ELISA. Data in C and D are represented as the mean ± standard deviation of 3 independent experiments. *P<0.05 compared with the control group. KDM3A, lysine demethylase 3A; siRNA, small interfering RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; LPS, lipopolysaccharide; IL-12α, interleukin 12α; TGF-β1, transforming growth factor-β1; HO-1, heme oxygenase 1.
siRNA-KDM3A and siRNA-NC were transfected into A549 cells
Subsequent to 24 h of LPS stimulation, the mRNA level changes of Foxp3 and its downstream cytokines, TGF-β1, IL-35 and HO-1, were examined by RT-qPCR. The expression level changes of associated proteins were assayed by FCM and ELISA. The results showed that Foxp3 expression was significantly reduced in A549 cells following TLR4 activation when KDM3A was silenced. Meanwhile, TGF-β1, HO-1 and IL-35 secretion was significantly reduced (P<0.05; Fig. 4C and D). These findings indicate that KDM3A performs a positive regulatory role during TLR4 regulation of Foxp3 and inhibitory cytokines secretion in A549 cells.
KDM3A regulates Foxp3 by activating its promoter activity in A549 cells
To additionally study the role of KDM3A in the TLR4-mediated regulation of Foxp3 transcription, A549 cells were stained by rabbit anti-KDM3A and mouse anti-Foxp3 primary antibody, and FITC-conjugated goat anti-rabbit and PE-conjugated goat anti-mouse secondary antibody. Certain overlying yellow fluorescence illustrated that KDM3A and Foxp3 were co-localized in A549 cells (Fig. 5A). The present study then generated a construct in which luciferase expression was driven by regulatory regions of Foxp3 (Fig. 5B). The results demonstrated that KDM3A silencing significantly suppressed luciferase activity driven by the Foxp3 promoter (Fig. 5C). This finding elucidates that KDM3A regulates Foxp3 transcription by binding its promoter directly.
Figure 5.
KDM3A facilitates Foxp3 expression in A549 cells. (A) A549 cells were detected by immunofluorescence staining with the rabbit anti-KDM3A and mouse anti-Foxp3 primary antibodies and fluorescein isothiocyanate-conjugated goat anti-rabbit (green) and phycoerythrin-conjugated goat anti-mouse (red) secondary antibodies. The nuclei were stained with DAPI (blue), and the cells were observed by inverted fluorescence microscopy. The merged image indicates the overlap of KDM3A (green), Foxp3 (red) and DAPI (blue), and the yellow in the merged image indicates overlap of green and red labels. (B) The construct containing luciferase expression was driven by promoters of Foxp3. (C) A549 cells were co-transfected with siRNA-NC or siRNA-KDM3A with luciferase reporter plasmids containing either PGL3-basic or PGL3-Foxp3-promoter. Luciferase was measured. The fold changes of relative luciferase activity in siRNA-KDM3A with indicated plasmids transfected cells were normalized to siRNA-NC with corresponding indicated plasmids transfected cells, respectively. Data are represented as the mean ± standard deviation of 3 independent experiments. *P<0.05 compared with the control group. KDM3A, lysine demethylase 3A; Foxp3, forkhead box P3; siRNA, small interfering RNA; NC, negative control; UTR, untranslated region; siRNA, small interfering RNA; NC, negative control.
Together, the findings of the present study have demonstrated that TLR4 activation can promote the expression of the H3K9me1/2 demethylase KDM3A in A549 cells
KDM3A binds directly to the Foxp3 promoter and promotes Foxp3 transcription, thereby increasing the secretion of Foxp3-related downstream inhibitory cytokines such as TGF-β1, IL-35, and HO-1. These cytokines can suppress effector T cells and DCs, ultimately facilitating the immune escape of lung cancer cells.
Discussion
TLR4 is a member of the type I transmembrane glycoprotein receptor family and serves as a pattern recognition receptor. TLR4 was originally identified in immune cells, and it performs a vital role in the primary and secondary immune responses. LPS as an exogenous ligand of TLR4, once recognized and specifically bound by TLR4, can activate the TLR4 signaling pathway (31). High expression levels of TLR4 have recently been implicated in colorectal, stomach and ovarian tumor tissues and cell lines (32–34). In the tumor microenvironment, TLR4 activation promotes tumor immune escape by releasing increased amounts of inflammatory cytokines and is closely associated with the prognosis of tumors (35,36). Once activated, the TLR4 pathway can also induce activation and enhance the immunosuppressive function of Tregs (37). Foxp3 is the key driver of Tregs differentiation and immunosuppressive function (38). One of the major immunosuppressive pathways of Tregs is the secretion of inhibitory cytokines (e.g. IL-35, IL-10, TGF-β1 and HO-1) and the suppression of the function of effector T cells and DCs.
The present study demonstrated that TLR4 activation in A549 cells can promote Foxp3 expression and IL-35, TGF-β1 and HO-1 secretion (IL-10 expression was not detected; data not shown). The secretion of these inhibitory cytokines was significantly reduced subsequent to silencing Foxp3 expression. The results indicate that, similar to its expression in Tregs, the Foxp3 expressed in lung adenocarcinoma cells serves a role in promoting tumor immune escape, and this process is regulated by TLR4. Additionally, histone methylation performs a critical role in the activation of the TLR4 signaling pathway. As previously reported, JMJD3 can participate in LPS-induced transcription of various genes (39,40). The H3K4me3 level of transcriptional genes induced by TLR4 activation is relatively high, and MLL performs a key role in this process. EZH1 catalyzes the TLR4 signaling pathway to negatively regulate the H3K427 methylation level at the transcription start site (TSS) of the Tollip gene. This mechanism results in a lower transcription level of the Tollip gene and thereby promotes TLR4 pathway activation and downstream cytokines secretion (14).
Histone H3 methylation at sites K4, K9, K27, K36, K79 and H4K20 changes the looseness of chromatin and the accessibility of gene loci, and thus regulating gene transcription activity. Demethylation of histone H3K9 is a sign of transcriptional activation. Lysine demethylase 1A that contains JMJC is capable of specifically removing methyl groups on the lysine residue of histones. KDM3A, as the first member of the JMJD family to be discovered, mainly demethylates mono- and dimethyl-H3K9, resulting in H3K9 demethylation and transcriptional activation (41). KDM3A regulates the expression of various genes and extensively participates in physiopathological processes by catalyzing histone H3K9 demethylation.
Furthermore, KDM3A serves an important role in tumor development and progression. For example, KDM3A is expressed in non-small cell lung and bladder cancer tissues and cell lines. KDM3A can directly activate transcription of homeobox A1 (HOXA1) through the demethylation of histone H3K9 by binding to its promoter region. In addition, KDM3A regulates the expression of HOXA1-dependent cyclin D1, activates G1/S phase transformation of tumor cells and promotes tumor growth (18). KDM3A is also expressed in hepatoma cell lines to promote cellular proliferation and invasion capabilities. KDM3A facilitates the inter-epithelial-mesenchymal transition of tumor cells, which is closely related to the metastasis of the hepatoma (19). In Ewing's sarcoma (EWS), overexpressed KDM3A inhibits the expression of microRNA (miR) −122, an EWS/friend leukemia integration 1 transcription factor-repressed miR, and thereby promotes cancer development and progression (42). Furthermore, hypoxia-inducible factor 1α (HIF-1α) is an upstream regulatory gene of KDM3A; when the tumor volume exceeds 1–2 mm3, the hypoxic microenvironment in the tumor center stimulates HIF-1α activation, induces pro-angiogenesis factors (e.g. vascular endothelial growth factor and platelet-derived growth factor), and maintains continuous tumor growth (43). The KDM3A promoter region, HRE, binds to HIF-1α to upregulate KDM3A expression and thereby changes the methylation level of histone H3K9 in downstream target genes, regulates the expression of hypoxia-related genes and promotes tumor cell growth. The present study revealed that the expression of the H3K9me1/2 demethylase KDM3A was significantly increased following LPS-induced TLR activation in A549 cells Therefore, it may be inferred that KDM3A is associated with TLR4 activation in A549 cells and likely participates in TLR4 regulation transcriptional of Foxp3. In subsequent assays, the present study revealed that TLR4 activation-induced Foxp3 expression was reduced by specific interference with KDM3A expression, which affected TGF-β1, IL-35 and HO-1 secretion. The dual luciferase reporter assay demonstrated that KDM3A directly binds to the Foxp3 promoter to activate its transcription, leading to increased inhibitory cytokines secretion under Foxp3 regulation. However, the KDM3A-binding site of Foxp3 requires further confirmation.
Based on the aforementioned findings, the present study has demonstrated that during TLR4 activation-induced nuclear transport of Foxp3, the H3K9me1/2 demethylase KDM3A directly binds to the Foxp3 promoter and thereby stimulates Foxp3 transcription through the demethylation of histone H3K9 in lung carcinoma cells. Additionally, KDM3A facilitates the secretion of TGF-β1, IL-35 and HO-1 that is regulated by Foxp3. These cytokines suppress the function of effector T cells and DCs and ultimately promote immune escape of lung carcinoma cells. The findings of the present study reveal an important role of epigenetic histone methylation in regulating the immune escape of lung carcinoma cells. The present data also suggest that KDM3A can provide new clues for the treatment of lung cancer and serve as a useful therapeutic target in patients with lung adenocarcinoma.
References
- 1.Alam H, Gu B, Lee MG. Histone methylation modifiers in cellular signaling pathways. Cell Mol Life Sci. 2015;72:4577–4592. doi: 10.1007/s00018-015-2023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herzog M, Josseaux E, Dedeurwaerder S, Calonne E, Volkmar M, Fuks F. The histone demethylase Kdm3a is essential to progression through differentiation. Nucleic Acids Research. 2012;40:7219–7232. doi: 10.1093/nar/gks399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rea S, Eisenhaber F, OCarroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chormatin structure by site-specific histone H3 methyltransferase. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 4.Gol IMG, Bestor TH. Histone modification and replacement in chromatin activation. Genes Dev. 2002;16:1739–1742. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- 5.Suikki HE, Kujala PM, Tammela TL, van Weerden WM, Vessella RL, Visakorpi T. Genetic alterations and changes in expression of histone demethylases in prostate cancer. Prostate. 2010;70:889–898. doi: 10.1002/pros.21123. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Ledaki I, Turley H, Gatter KC, Montero JC, Li JL, Harris AL. Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann N Y Acad Sci. 2009;1177:185–197. doi: 10.1111/j.1749-6632.2009.05027.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part I: Covalent histone modification. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 9.Nichol N, Dupéré-Richer D, Ezponda T, Licht JD, Miller WH., Jr H3K27 methylation: A focal point of epigenetic deregulation in cancer. Adv Cancer Res. 2016;131:59–95. doi: 10.1016/bs.acr.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a Jmjc-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Zhang Q, Ding Y, Li X, Zhao D, Zhao K, Guo Z, Cao X. Histone lysine methyltransferase Ezh1 promotes TLR-triggered inflammatory cytokines production by suppressing tollip. J Immunol. 2015;194:2838–2846. doi: 10.4049/jimmunol.1402087. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–141. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 13.Li TT, Ogiono S, Qian ZR. Toll-like receptor signaling in colorectal cancer: Carcinogenesis to cancer therapy. Word J Gastroenterol. 2014;20:17699–17708. doi: 10.3748/wjg.v20.i47.17699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Zhao Y, Qian J, Sun L, Lu Y, Li H, Li Y, Yang J, Cai Z, Yi Q. Toll-like receptor-4 signaling in mantle cell lymphoma: Effects on tumor growth and immune evasion. Cancer. 2013;119:782–791. doi: 10.1002/cncr.27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmausser B, Andrulis M, Endrich S, Mȕller-Hermelink HK, Eck M. Toll-like receptors TLR4, TLR5 and TLR9 on gastric carcinoma cells: An implication for interaction with Helicobacter pylori. Int J Med Microbiol. 2005;295:179–185. doi: 10.1016/j.ijmm.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, McFarland-Mancini MM, Funk HM, Husseinzadeh N, Mounajjed T, Drew AF. Toll-like receptor expression in normal ovary and ovarian tumors. Cancer Immunol Immunother. 2009;58:1375–2385. doi: 10.1007/s00262-008-0650-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, Bando Y, Kudo E, Shimada M, Sano T. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer. 2010;102:908–915. doi: 10.1038/sj.bjc.6605558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milkova L, Voelcker V, Forstreuter I, Sack U, Andereqq U, Simon JC, Maier-Simon C. The NF-kappaB signaling pathway is involved in the LPS/IL-2-induced upregulation of Foxp3 expression in human CD4+CD25high regulatory T cells. Exp Dermatol. 2010;19:29–37. doi: 10.1111/j.1600-0625.2009.00953.x. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Samanta A, Song X, Furuuchi K, Iacono KT, Kennedy S, Katsumata M, Saouaf SJ, Greene MI. FOXP3 ensembles in T-cell regulation. Immunol Rev. 2006;212:99–131. doi: 10.1111/j.0105-2896.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt EG, Williams CB. Generation and function of induced regulatory T cells. Front Immunol. 2013;4:152. doi: 10.3389/fimmu.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karanikas V, Speletas M, Zamanakou M, Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI, Germenis AE. Foxp3 expression in human cancer cells. J Transl Med. 2008;6:19. doi: 10.1186/1479-5876-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He W, Liu Q, Wang L, Chen W, Li N, Cao X. TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol Immunol. 2007;44:2850–2859. doi: 10.1016/j.molimm.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Hinz S, Pagerols-Raluy L, Oberg HH, Ammerpohl O, Grȕssel S, Sipos B, Grȕtzmann R, Pilarsky C, Unqefroren H, Saeqer HD, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res. 2007;67:8344–8350. doi: 10.1158/0008-5472.CAN-06-3304. [DOI] [PubMed] [Google Scholar]
- 24.Merlo A, Casalini P, Carcangiu ML, Malventano C, Triulzi T, Mènard S, Tagliabue E, Balsari A. FOXP3 expression and overall survival in breast cancer. J Clin Oncol. 2009;27:1746–1752. doi: 10.1200/JCO.2008.17.9036. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, McNally B, Lin L, Zhou P, Zuo T, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16:336–346. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Triulzi T, Tagliabue E, Balsari A, Casalini P. FOXP3 expression in tumor cells and implications for cancer progression. J Cell Physiol. 2013;228:30–35. doi: 10.1002/jcp.24125. [DOI] [PubMed] [Google Scholar]
- 27.Fu HY, Li C, Yang W, Gai XD, Jia T, Lei YM, Li Y. FOXP3 and TLR4 protein expression are correlated in non-small cell lung cancer: Implications for tumor progression and escape. Acta Histochem. 2013;115:151–157. doi: 10.1016/j.acthis.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Jia T, Fu H, Sun J, Zhang Y, Yang W, Li Y. Foxp3 expression in A549 cells is regulated by Toll-like receptor 4 through nuclear factor-κB. Mol Med Rep. 2012;6:167–172. doi: 10.3892/mmr.2012.877. [DOI] [PubMed] [Google Scholar]
- 29.Κawai T, Akira S. The role of pattern recognition receptors in innate immunity: Update on Toll like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 30.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 31.Kim KH, Jo MS, Suh DS, Yoon MS, Shin DH, Lee JH, Choi KU. Expression and signification of the TLR4/MyD88 signaling pathway in ovarian epithelial cancers. Word J Surg Oncol. 2012;10:193. doi: 10.1186/1477-7819-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang B, Zhao J, Li H, He KL, Chen Y, Chen SH, Mayer L, Unkeless JC, Xiong H. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005;65:5009–5014. doi: 10.1158/0008-5472.CAN-05-0784. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Zhao Y, Qian J, Sun L, Lu Y, Li H, Li Y, Yanq J, Cai Z, Yi Q. Toll-like receptor-4 signaling in mantle cell lymphoma: Effects on tumor growth and immune evasion. Cancer. 2013;119:782–791. doi: 10.1002/cncr.27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goto Y, Arigami T, Kitago M, Nquyen SL, Narita N, Ferrone S, Morton DL, Irie RF, Hoon DS. Activation of Toll-like receptors 2, 3 and 4 on human melanoma cells induces inflammatory factors. Mol Cancer Ther. 2008;7:3642–3653. doi: 10.1158/1535-7163.MCT-08-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 36.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19:903–912. doi: 10.1016/S1074-7613(03)00332-7. [DOI] [PubMed] [Google Scholar]
- 37.Welte T, Koch F, Schuler G, Lechner J, Doppler W, Heufler C. Granulocyte-macrophage colony-stimulating factor induces a unique set of STAT factors in murine dendritic cells. Eur J Immunol. 1997;27:2737–2740. doi: 10.1002/eji.1830271038. [DOI] [PubMed] [Google Scholar]
- 38.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of Jmjc domain-containing protein. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 39.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z, Zhou S, Liao L, Chen X, Meisrrich M, Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J Biol Chem. 2010;285:2758–2770. doi: 10.1074/jbc.M109.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrish JK, Sechler M, Winn RA, Jedicka P. The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in ewing sarcoma. Oncogene. 2015;34:257–262. doi: 10.1038/onc.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, Mori M. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: In vivo identification from hypoxic tumor cells. Clin Cancer Res. 2010;16:4636–4646. doi: 10.1158/1078-0432.CCR-10-0407. [DOI] [PubMed] [Google Scholar]
- 43.Mahajan K, Lawrence HR, Lawrence NJ, Mahajan NP. ACK1 tyrosine kinase interacts with histone demethylase KDM3A to regulate the mammary tumor oncogene HOXA1. J Biol Chem. 2014;289:28179–28191. doi: 10.1074/jbc.M114.584425. [DOI] [PMC free article] [PubMed] [Google Scholar]






