Abstract
Colorectal cancer (CRC) is the third most common cancer in males and the second most common in females, worldwide. Currently, 5-fluorouracil (5-FU)-mediated chemotherapy is the adjuvant treatment for patients with CRC following surgical resection. However, a number of patients with CRC develop 5-FU resistance, which is a major challenge for the effective treatment of cancer. Therefore, it is important to investigate the molecular mechanisms underlying chemoresistance and the therapeutic treatments that may improve the treatment of CRC. The present study demonstrated that microRNA (miR)-330 was significantly downregulated in CRC tissues and cell lines. Ectopic miR-330 expression decreased cell proliferation and enhanced cell chemosensitivity to 5-FU via the cell apoptosis pathway in CRC. In addition, thymidylate synthase (TYMS) was revealed to be a direct target gene of miR-330 in CRC. Knockdown of TYMS inhibited CRC cell proliferation, and enhanced cell chemosensitivity to 5-FU by promoting cell apoptosis. In conclusion, the results of the present study indicated that miR-330 targeted TYMS to inhibit the proliferation and enhance the chemosensitivity of CRC cells to 5-FU by promoting cell apoptosis. The present study provided important insight into the molecular mechanism underlying 5-FU-mediated chemoresistance and a novel therapeutic strategy for the enhancement of efficacy in CRC treatment.
Keywords: colorectal cancer, microRNA-330, proliferation, chemosensitivity, 5-fluorouracil, thymidylate synthase
Introduction
Colorectal cancer (CRC) is one of the most common types of malignancy, it is the third most common cancer in males and the second most common in females, worldwide (1). In 2015 it was estimated that there would be 93,090 novel cases of CRC and 49,700 CRC-associated mortalities in the USA (2). The molecular mechanism underlying CRC carcinogenesis and progression involves numerous factors, including genetic instability, hereditary components, increased age, male gender, increased intake of fat, alcohol or red meat, obesity, smoking and a lack of physical exercise (3–5). Currently, the standard therapeutic treatments for CRC are surgical resection, radiotherapy and chemotherapy (6). Despite progress in the development of treatments for CRC, the 5-year overall survival rate has not improved significantly (7–9). The poor prognosis of CRC is associated with an advanced stage of cancer and tumor recurrence and metastasis following surgical resection (10). Patients with advanced stage CRC and metastasis have been treated with 5-fluorouracil (5-FU)-associated chemotherapy, following surgery (11). However, not every case of CRC responds to 5-FU based chemotherapy and the response rate to this treatment is only between 10 and 20% (12). Therefore, overcoming chemoresistance, improving the chemosensitivity of CRC and enhancing the curative effect of chemotherapy, is required for the treatment of CRC.
A number of previous studies have demonstrated that microRNAs (miRNAs) are downregulated in numerous types of human cancer, including CRC (13–15). miRNAs are endogenous, non-protein-coding small RNAs, 18–25 nucleotides in length (16). miRNAs serve important regulatory roles in human, animal, plant and DNA viruses by binding to the 3′untranslated regions (3′UTRs) of target mRNAs, resulting in mRNA degradation or translational repression at the translational or post-transcriptional levels (17,18). miRNAs regulate the expression of ≥20% of human genes, and contribute to a variety of important physiological and pathological processes including, cell cycle, proliferation, migration, invasion, apoptosis, differentiation and development (18–20). A previous study demonstrated that miRNAs are able to function as oncogenes by repressing the expression of tumor target suppressor genes, or tumor suppressors by repressing the expression of target oncogenes (21). Furthermore, miRNAs have previously been identified to serve important functions in the regulation of chemoresistance (22,23). This suggested that miRNAs may be novel potential therapeutic targets for chemoresistance of CRC.
The present study revealed that miRNA (miR)-330 was significantly downregulated in CRC tissues and cell lines. In addition, miR-330 inhibited CRC cell proliferation and enhanced the chemosensitivity of CRC cells to 5-FU by increasing the rate of CRC cell apoptosis induced by 5-FU. Furthermore, thymidylate synthase (TYMS) was identified as a direct target gene of miR-330 in CRC. These results have therapeutic implications and may be exploited for further treatment of CRC.
Materials and methods
Clinical samples
The present study was approved by the Ethics Committee of The Military General Hospital of Beijing PLA (Beijing, China). Written informed consent was obtained from all patients, prior to enrollment in the present study. The patients with CRC had not received chemotherapy or radiotherapy prior to surgery. A total of 59 pairs of CRC tissues and their normal adjacent tissues (NATs) were obtained from patients with CRC, who underwent surgical resection at The Military General Hospital of Beijing PLA. CRC tissues and the NATs were rapidly frozen in liquid nitrogen and stored in at −80°C.
Cell culture and transfection
A total of four human CRC cell lines (HCT116, HT29, SW480 and SW620), the normal human colon epithelium cell line FHC and the HEK293T cell line were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained in Dulbecco's modified Eagle's medium or RPMI-1640 supplemented with 10% fetal bovine serum (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 mg/ml penicillin and 100 mg/ml in a humidified atmosphere of 5% CO2 at 37°C.
In order to investigate the function of miR-330 in CRC, CRC cells were transfected with miR-330 mimics or the negative scrambled control (NC), purchased from GenePharma, Inc. (Sunnyvale, CA, USA). Furthermore, TYMS small interfering (si)RNA and NC siRNA were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and transfected into CRC cells. The sequence of the miR-330 mimic was 5′-GCAAAGCACACGGCCUGCAGAGA-3′ and the sequence of the miR-NC mimic was 5′-UUCUCCGAACGUGUCACGUTT-3′. The sequence of the TYMS siRNA was 5′-TACGTCCAAGGTCGGGCAGGAAGA-3′ and the NC siRNA sequence was 5′-AACAGGCACACGTCCCAGCGT-3′. Cell transfection was performed using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc), according to the manufacturer's protocol.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. In order to detect miR-330 expression level, reverse transcription was performed using the TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), followed by RT-qPCR using the TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The temperature protocol for reverse transcription was as follows: 16°C for 30 min; 42°C for 30 min; and 85°C for 5 min. The thermocycling conditions for qPCR were as follows: 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 sec and annealing at 60°C for 1 min; followed by a final elongation step at 72°C for 10 min.
For analysis of TYMS mRNA expression level, reverse transcription was performed using the M-MLV Reverse Transcription system (Promega Corp., Madison, WI, USA), according to the manufacturer's protocol. The temperature protocol for reverse transcription was as follows: 95°C for 2 min; 20 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min; and 72°C for 5 min. Subsequently, SYBR®-Green Master mix was used to determine the mRNA expression level. The thermocycling conditions of qPCR were as follows: 95°C for 10 min; and 40 cycles of 95°C for 15 sec and 60°C for 1 min. U6 small nuclear RNA was used for normalization of miRNA expression and GADPH was used as an internal control for mRNA expression. RT-qPCR was performed using a 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression of miR-330a and TYMS was analyzed by use of the 2−ΔΔCq method (24). This assay was performed in triplicate and repeated three times.
CCK-8 assay
Cell proliferation rates were evaluated using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay. Cells were seeded into 96-well plates at a density of 3,000 cells/well. Subsequently, cells were transfected and incubated in a humidified atmosphere of 5% CO2 at 37°C for 24–96 h. Every 24 h post-transfection, CCK-8 assays was performed. A total of 10 µl CCK8 assay solution was added to each well. Cells were incubated for 2 h at 37°C and the absorbance of each well was determined at 450 nm by an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate.
Chemosensitivity assay
Cells were seeded into 96-well plates at a density of 3,000 cells/well. Subsequently, cells were transfected and incubated for 48 h in a humidified atmosphere of 5% CO2 at 37°C. Cells were then treated with 5-FU (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) at various concentrations (0–32 µM) at room temperature. Following incubation for a further 48 h, a chemosensitivity assay was performed using a CCK8 assay, as aforementioned. The dose-response curve was charted at various concentrations. All experiments were performed in triplicate.
Cell apoptosis assay
Cells were seeded into 6-well plates at a confluence of between 60 and 70%. Cells were transfected and incubated for 24 h in a humidified atmosphere of 5% CO2 at 37°C. Cells were subsequently treated with 5-FU at a concentration of 8 µM. Following a 48-h incubation, the rate of cell apoptosis was determined by Annexin V-fluorescein isothiocyanate (FITC; BD Biosciences, Franklin Lakes, NJ, USA; cat. no. 556419) and propidium iodide (PI; BD Biosciences; cat. no. 556463) staining, according to the manufacturer's protocol. In brief, cells were harvested and washed with ice-cold PBS (Gibco; Thermo Fisher Scientific, Inc.) three times. Cells were resuspended in 100 µl binding buffer, followed by treatment with 2 µl Annexin V-FITC and 5 µl of PI at room temperature. Following a 15 min incubation at room temperature in the dark, 400 µl of binding buffer was added and the mixture was analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) using FCS Express software (version 3.0; De Novo Software, Glendale, CA, USA).
Western blotting
Transfected cells were washed with ice-cold PBS three times and lysed with radioimmunoprecipitaion assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with protease inhibitors (Promega Corp.). Protein concentration was quantified using a bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of protein (20 µg) were separated by 10% SDS-PAGE, followed by transference to polyvinylidene difluoride membranes. The membranes were blocked in TBS-Tween-20 containing 5% non-fat dry milk for 2 h at room temperature, followed by incubation with primary antibodies overnight at 4°C. Subsequently, the membranes were incubated with the goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:3,000 dilution; cat. no. sc-ab6789; Abcam, Cambridge, MA, USA) for 2 h at room temperature. Protein bands were visualized using enhanced chemiluminescence solution (Pierce; Thermo Fisher Scientific, Inc.). The primary antibodies used in the present study were mouse anti-human monoclonal TYMS (cat. no. sc-33679) and mouse anti-human GADPH (cat. no. sc-59540) (both 1:1,000 dilution; both Santa Cruz Biotechnology, Inc., Dallas, TX, USA). GADPH was used as an internal control. Band intensities were imaged using the FluorChem imaging system (alpha innotec GmbH, Kasendorf, Germany) and analyzed using Image Lab version 5.2.1 (Bio-Rad Laboratories, Inc.).
Dual-luciferase® reporter assay
The Dual-Luciferase Reporter assay (Promega Corporation) was performed in HEK293T cells. PGL3-TYMS-3′UTR wild-type (Wt) and PGL3-TYMS-3′UTR mutant (Mut) were purchased from GenePharma, Inc. Cells were seeded into 24-well plates at a confluence of between 60 and 70%. Following overnight incubation, cells were co-transfected with miR-330 mimics or NC, and PGL3-TYMS-3′UTR Wt or PGL3-TYMS-3′UTR Mut, using Lipofectamine 2000. The firefly and Renilla luciferase activities were determined by Dual-Luciferase reporter assay 48 h following transfection. Luciferase activity was measured at a wavelength of 560 nm using an xMark™ Microplate Absorbance Spectrophotometer (Bio-Rad Laboratories, Inc.). Renilla luciferase activities were used as an internal control. All experiments were performed in triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation, and compared with the Student's t-test or a one-way analysis of the variance using SPSS 16.0 statistical software (SPSS, Inc., Chicago, IL, USA). Two-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
miR-330 expression level in CRC tissues and cell lines
In order to investigate whether miR-330 was downregulated in CRC tissues, the miR-330 expression level was determined by RT-qPCR in CRC tissues and paired NATs. As presented in Fig. 1A, miR-330 was significantly downregulated in CRC tissues compared with paired NATs (P<0.05). Furthermore, the expression level of miR-330 was evaluated in CRC and FHC cell lines. Consistent with the result that the miR-330 expression level was decreased in CRC tissues, miR-330 was downregulated in the HCT116, HT29, SW480 and SW620 cell lines, compared with the FHC cell line (all P<0.05; Fig. 1B). These findings suggested that miR-330 may act as a tumor suppressor in CRC.
Figure 1.
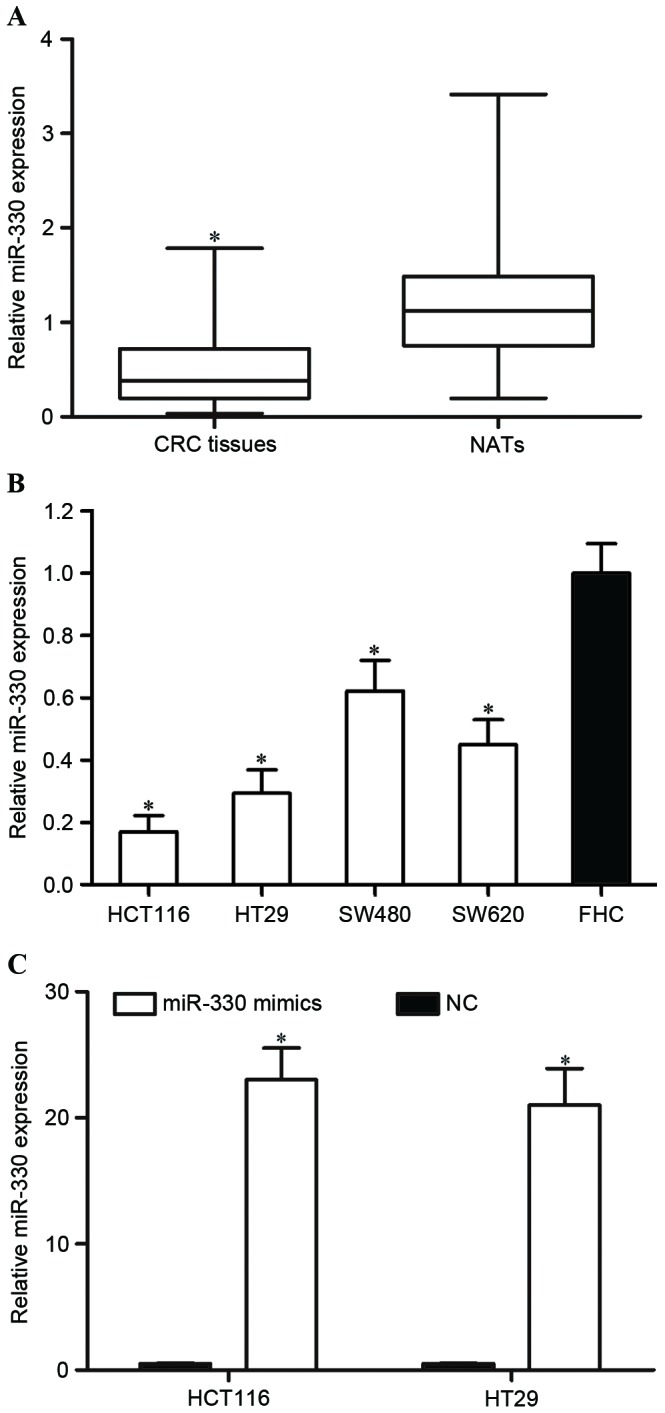
The expression level of miR-330 in CRC tissues. (A) miR-330 was downregulated in CRC tissues compared with matched NATs. (B) The expression level of miR-330 was decreased in CRC cell lines compared with the FHC cell line. (C) miR-330 was significantly upregulated in HCT119 and HT29 cells following transfection with miR-330 mimics. *P<0.05 compared with their respective controls. miR-330, microRNA-330; CRC, colorectal cancer; NATs, normal adjacent tissues; NC, negative control.
HCT116 and HT29 expression levels were lower compared with SW480 and SW620 expression levels. Therefore, the present study transfected miR-330 mimics or NC into HCT116 and HT29 cells in order to explore the functions of miR-330 in CRC cells. RT-qPCR was performed to determine the miR-330 expression level following transfection. As presented in Fig. 1C, miR-330 was significantly upregulated in miR-330 mimic transfected HCT116 and HT29 cells, compared with in NC cells (both P<0.05).
miR-330 inhibited cell proliferation in HCT116 and HT29 cells
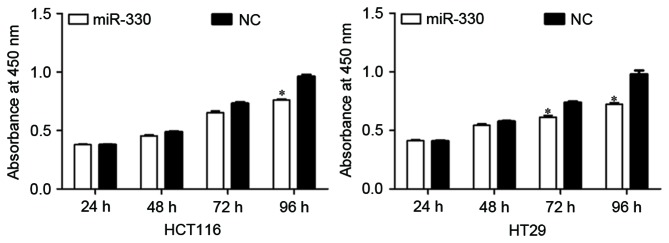
The effect of miR-330 expression on CRC cell proliferation was evaluated by performing a CCK-8 assay. As presented in Fig. 2, the upregulation of miR-330 resulted in significant growth inhibition of HCT116 and HT29 cells (both P<0.05) at 96 h compared with the NC. The results suggested that miR-330 may be a negative regulator of cell proliferation in CRC cells.
Figure 2.
CCK8 assays revealed that the upregulation of miR-330 resulted in the growth inhibition of HCT116 and HT29 cells. *P<0.05 compared with the NC. CCK-8, Cell Counting Kit-8; miR-330, microRNA-330; NC, negative control.
miR-330 enhanced cell chemosensitivity to 5-FU in HCT116 and HT29 cells
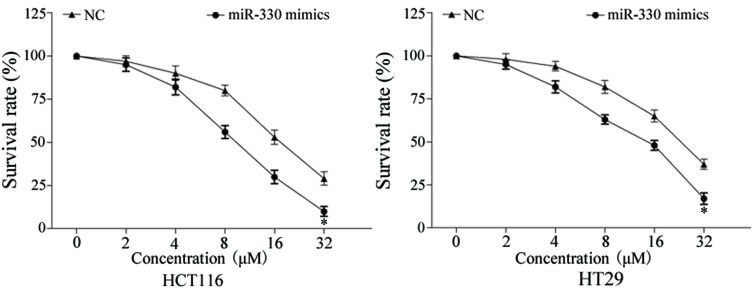
In order to evaluate the effect of miR-330 on CRC cell chemosensitivity to 5-FU, a chemosensitivity assay was performed. As presented in Fig. 3, enforced miR-330 expression decreased the survival rate of HCT116 and HT29 cells, compared with NC-transfected HCT116 and HT29 cells (P<0.05). These results indicated that miR-330 increased cell chemosensitivity of CRC cells to 5-FU.
Figure 3.
Chemosensitivity assays demonstrated that enforced miR-330 expression decreased the survival rate of HCT116 and HT29 cells compared with NC-transfected HCT116 and HT29 cells. *P<0.05 compared with the NC. miR-330, microRNA-330; NC, negative control.
miR-330 increased the rate of cell apoptosis induced by 5-FU in HCT116 and HT29 cells
Cell-cycle arrest at the G1 phase and/or apoptosis of cancer cells is enhanced by 5-FU (25,26). Therefore, the rate of cell apoptosis induced by 5-FU was evaluated. As presented in Fig. 4, the overexpression of miR-330 significantly increased the rate of apoptosis of HCT116 and HT29 cells induced by 5-FU (P<0.05) compared with the NC. This supports the observation that miR-330 increases HCT116 and HT29 cell chemosensitivity to 5-FU. These results suggested that miR-330 increased cell chemosensitivity of HCT116 and HT29 cells to 5-FU via the cell apoptosis pathway.
Figure 4.
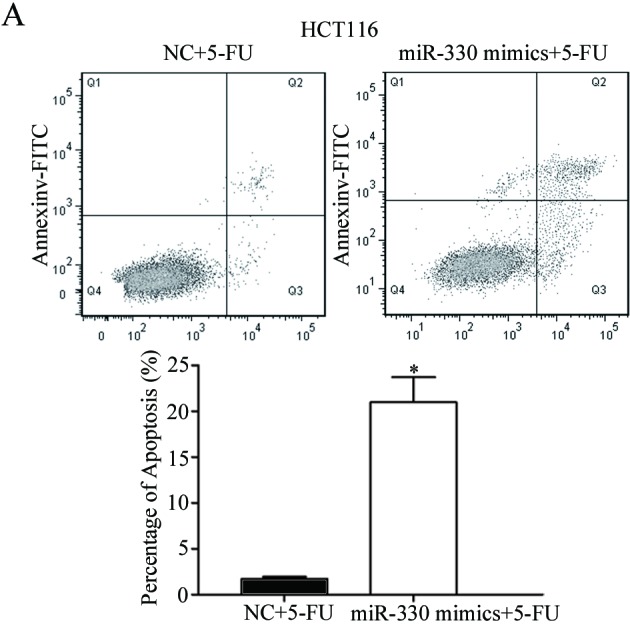
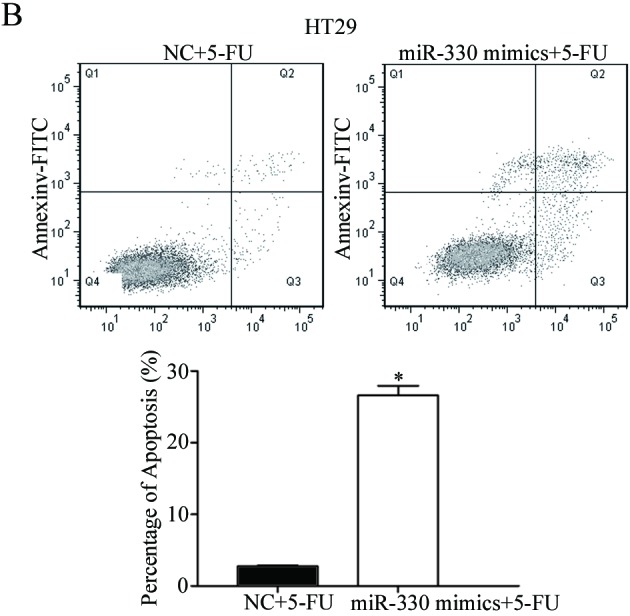
Overexpression of miR-330 significantly increased the rate of apoptosis of (A) HCT116 and (B) HT29 cells induced by 5-FU. *P<0.05 compared with the NC. miR-330, microRNA-330; 5-FU, 5-fluorouracil; FITC, fluorescein isothiocyanate; NC, negative control.
TYMS was a direct target gene of miR-330 in vitro
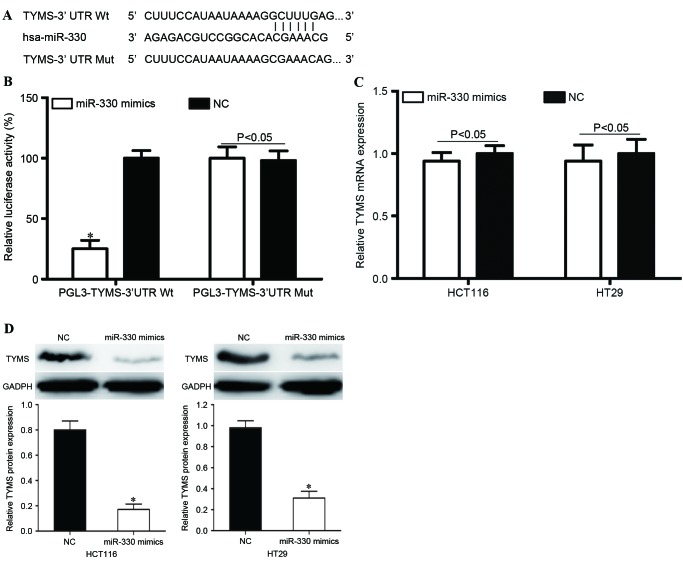
In order to investigate the molecular mechanisms underlying the functions of miR-330 in CRC cells, bioinformatic predication algorithms (TargetScan; www.targetscan.org) were used to determine the direct target gene of miR-330. Amongst the hypothetical target mRNAs identified for miR-330, TYMS was selected for further investigation (Fig. 5A). In addition, a Dual-Luciferase reporter assay was performed to determine whether TYMS was a direct target mRNA of miR-330 in vitro. As presented in Fig. 5B, miR-330 significantly inhibited PGL3-TYMS-3′UTR Wt luciferase activity in HEK293T cells (P<0.05) compared with the NC, whereas PGL3-TYMS-3′UTR Mut demonstrated no significant response to miR-330 (P>0.05).
Figure 5.
TYMS is a direct target gene of miR-330 in vitro. (A) The miR-330 binding site in the 3′UTR of TYMS and the TYMS 3′UTR mutant sequence. (B) miR-330 inhibited PGL3-TYMS-3′UTR Wt luciferase activity in HEK293T cells (P<0.05), whereas PGL3-TYMS-3′UTR Mut revealed no response to miR-330. (C) TYMS mRNA expression levels were not significantly altered following transfection with miR-330 mimics. (D) The TYMS protein was downregulated in miR-330 mimic-transfected HCT116 and HT29 cells compared with the NC groups. *P<0.05 compared with the NC. TYMS, thymidylate synthase; miR-330, microRNA-330; 3′UTR, 3′untraslated region; Wt, wild-type; Mut, mutant; NC, negative control.
In order to further investigate the potential effects of miR-330 in the regulation of TYMS, RT-qPCR and western blot analysis were performed to evaluate TYMS mRNA and protein levels following transfection with miR-330 mimics or NC. As presented in Fig. 5C, TYMS mRNA expression levels were not significantly altered following transfection with miR-330 mimics (P>0.05). However, TYMS protein expression was significantly downregulated in miR-330 mimic-transfected HCT116 and HT29 cells, compared with NC groups (both P<0.05; Fig. 5D). This indicated that miR-330 regulated TYMS expression at the post-transcriptional level. Therefore, TYMS was a direct target gene of miR-330 in CRC cells.
TYMS served a role in miR-330-mediated functions in HCT116 and HT29 cells
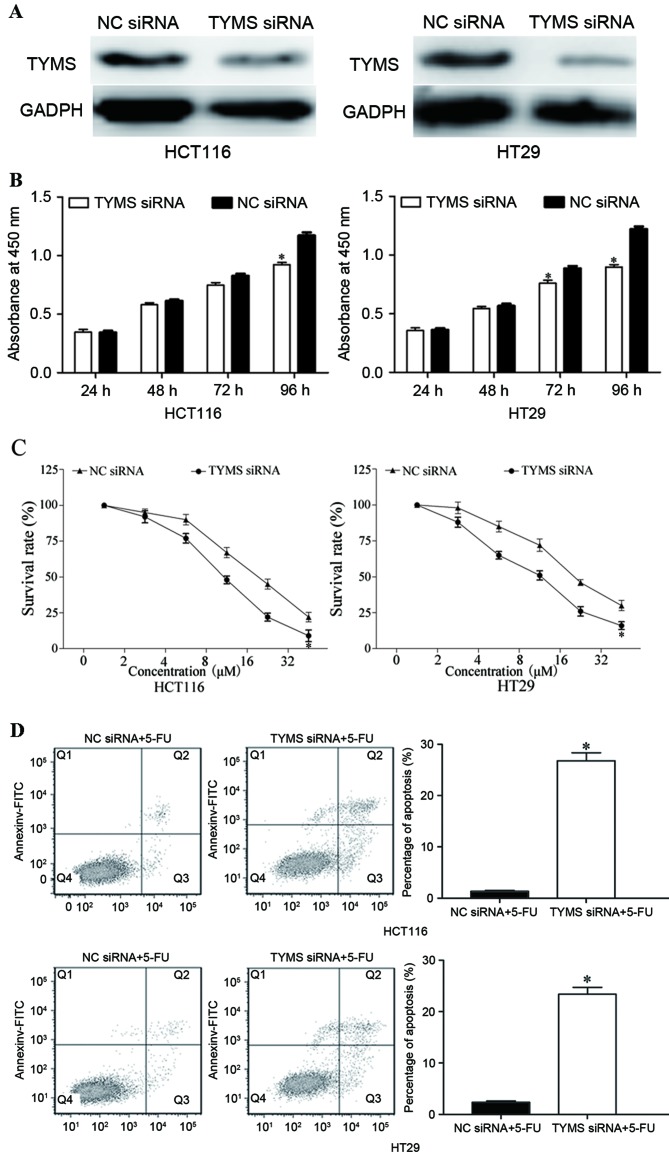
In order to investigate the function of TYMS in CRC, HCT116 and HT29 cells were transfected with TYMS siRNA or NC siRNA. As presented in Fig. 6A, TYMS siRNA decreased the expression level of TYMS in HCT116 and HT29 cells (P<0.05). Following transfection with TYMS siRNA, CCK-8, chemosensitivity and cell apoptosis assays were performed. As presented in Fig. 6B, relative cell growth was significantly suppressed in TYMS siRNA transfected HCT116 and HT29 cells, compared with NC siRNA cells (both P<0.05). In addition, chemosensitivity assays demonstrated that TYMS siRNA significantly improved chemosensitivity of HCT116 and HT29 cells to 5-FU (both P<0.05; Fig. 6C). Furthermore, TYMS siRNA significantly increased the rate of HCT116 and HT29 cell apoptosis induced by 5-FU (both P<0.05; Fig. 6D). These results revealed that the effects of TYMS siRNA were similar to those induced by miR-330 in CRC cells, suggesting that TYMS was a functional target of miR-330 in CRC cells.
Figure 6.
Effects of TYMS expression in HCT116 and HT29 cells. (A) TYMS siRNA decreased the expression level of TYMS in HCT116 and HT29 cells. (B) Cell growth was suppressed in TYMS siRNA transfected HCT116 and HT29 cells compared with the NC siRNA groups. (C) Chemosensitivity assays demonstrated that TYMS siRNA enhanced the chemosensitivity of HCT116 and HT29 cells to 5-FU. (D) Inhibition of TYMS increased apoptosis of HCT116 and HT29 cells induced by 5-FU. *P<0.05 compared with the NC. TYMS, thymidylate synthase; siRNA, short interfering RNA; NC, negative control; 5-FU, 5-fluorouracil.
Discussion
CRC is the third leading cause of cancer-associated mortality worldwide, and the 5-year survival rate is mostly dependent on the stage of cancer, resulting in a survival rate of between 10 and 95% (27,28). In China, the incidence rate of CRC is relatively lower; however, an increased prevalence in recent years has been observed (29). For patients with CRC following surgical resection, 5-FU based chemotherapy is the adjuvant treatment (30). In addition, chemotherapy is the first-line of treatment for a number of patients with metastatic CRC (31). However, numerous CRC cases develop 5-FU resistance, which is a major challenge to the effective treatment of cancer (32). Therefore, it is essential to explore the molecular mechanism underlying chemoresistance in CRC and investigate novel therapeutic strategies for patients with CRC.
miR-330 has been identified to be downregulated in prostate cancer, and inhibited cell migration and invasion through the downregulation of SP1 (33). However, miR-330 was also revealed to be upregulated in human esophageal cancer cells (34), non-small cell lung cancer cells (35) and glioblastoma cells (36). In previous functional studies, miR-330 was identified to be an oncogene. In esophageal cancer, miR-330 targeted programmed cell death protein 4 in order to enhance cell proliferation, migration and invasion, and to decrease the rate of cisplatin-induced apoptosis (34). Liu et al (35) demonstrated that miR-330 increased non-small cell lung cancer cell proliferation by directly targeting early growth response protein 2 (35). In glioblastoma tissues, the upregulation of miR-330 promoted cell growth, migration and invasion, and suppressed cell apoptosis by regulating SH3 domain containing GRB2 like 2 (36). These conflicting results revealed that the expression and functions of miR-330 were tissue-type dependent. The present study demonstrated that miR-330 was downregulated in CRC tissues and cell lines. In addition, enforced miR-330 expression inhibited CRC cell proliferation and enhanced the chemosensitivity of CRC cells to 5-FU by the cell apoptosis pathway. These findings may facilitate the development of a novel strategy for 5-FU-based chemotherapy.
In order to explore the molecular mechanisms of miR-330 underlying the cellular response to 5-FU, the present study identified that TYMS was a direct target gene of miR-330 in CRC. TargetScan predicated that TYMS contained a miR-330 seed match at position 192–197 of the TYMS-3′UTR. miR-330 decreased the luciferase activity of TYMS-3′UTR. However, co-transfection of miR-330 mimics and TYMS-3′UTR Mut did not induce a decrease in luciferase activity. Subsequently, miR-330 inhibited TYMS expression at the post-transcriptional level. Finally, functional studies revealed that TYMS siRNA enhanced cell chemosensitivity of CRC cells to 5-FU and mediated the suppressive role of miR-330 in CRC cell growth, and enhanced cell apoptosis due to exposure to 5-FU. Identification of miR-330 target genes is essential for understanding its function in chemoresistance.
In CRC chemotherapy, 5-FU is the most commonly administered chemotherapeutic agent alone or combined with other chemotherapeutic agents (37). The structure of 5-FU is similar to pyrimidine, a nucleoside required for DNA replication (38). Therefore, 5-FU primarily affects nucleoside metabolism and may be incorporated into RNA and DNA molecules. Consequently, 5-FU enhances cell-cycle arrest at the G1 phase and/or apoptosis in cancer cells (25,26). TYMS, a cytosolic enzyme that alters the methylation of deoxyuridine monophosphate to deoxythymidine monophosphate, is an important therapeutic target of 5-FU (39). Previous studies verified that patients with CRC and low TYMS expression have an improved response to 5-FU and 5-year overall survival rate (40,41). In comparison, the upregulation of TYMS in patients with CRC induces 5-FU chemoresistance (42,43). The present study demonstrated that the downregulation of TYMS significantly inhibited CRC cell proliferation and enhanced cell chemosensitivity of CRC cells to 5-FU by inducing apoptosis under 5-FU exposure. Therefore, TYMS may serve as a predictive biomarker of the cellular response to 5-FU and a therapeutic target for 5-FU-based chemotherapy. The present study provides evidence for the use of 5-FU in combination with miR-330 in the chemotherapeutic treatment of CRC.
In conclusion, the present study revealed that miR-330 was downregulated in CRC tissues and cell lines. Increased expression of miR-330 suppressed CRC cell proliferation. In addition, the present study demonstrated that miR-330 enhanced the chemosensitivity of CRC cells to 5-FU by the cell apoptosis pathway. Furthermore, TYMS was identified as a direct target gene of miR-330 in CRC. These results have therapeutic implications and may be exploited for further treatment of CRC.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Andrews L. Dietary flavonoids for the prevention of colorectal cancer. Clin J Oncol Nurs. 2013;17:671–672. doi: 10.1188/13.CJON.671-672. [DOI] [PubMed] [Google Scholar]
- 4.Altobelli E, Lattanzi A, Paduano R, Varassi G, di Orio F. Colorectal cancer prevention in Europe: Burden of disease and status of screening programs. Prev Med. 2014;62:132–141. doi: 10.1016/j.ypmed.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Sugarbaker PH. Colorectal cancer: Prevention and management of metastatic disease. Biomed Res Int. 2014;2014:782890. doi: 10.1155/2014/782890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Z, Zhou N, Han Q, Zhao L, Bai C, Chen Y, Zhou J, Zhao RC. MicroRNA-197 influences 5-fluorouracil resistance via thymidylate synthase in colorectal cancer. Clin Transl Oncol. 2015;17:876–883. doi: 10.1007/s12094-015-1318-7. [DOI] [PubMed] [Google Scholar]
- 8.Brouquet A, Nordlinger B. Metastatic colorectal cancer outcome and fatty liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:266–267. doi: 10.1038/nrgastro.2013.53. [DOI] [PubMed] [Google Scholar]
- 9.Kuipers EJ, Rösch T, Bretthauer M. Colorectal cancer screening-optimizing current strategies and new directions. Nat Rev Clin Oncol. 2013;10:130–142. doi: 10.1038/nrclinonc.2013.12. [DOI] [PubMed] [Google Scholar]
- 10.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 12.De Angelis PM, Svendsrud DH, Kravik KL, Stokke T. Cellular response to 5-fluorouracil (5-FU) in 5-FU-resistant colon cancer cell lines during treatment and recovery. Mol Cancer. 2006;5:20. doi: 10.1186/1476-4598-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng K, Liu W, Liu Y, Jiang C, Qian Q. MicroRNA-133a suppresses colorectal cancer cell invasion by targeting Fascin1. Oncol Lett. 2015;9:869–874. doi: 10.3892/ol.2014.2753. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Yuan W, Sui C, Liu Q, Tang W, An H, Ma J. Up-regulation of microRNA-145 associates with lymph node metastasis in colorectal cancer. PLoS One. 2014;9:e102017. doi: 10.1371/journal.pone.0102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Wang Z, Chen M, Peng L, Wang X, Ma Q, Ma F, Jiang B. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol Cancer. 2012;11:23. doi: 10.1186/1476-4598-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 17.O'Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: Key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104–1110. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Lu F, Zhang J, Ji M, Li P, Du Y, Wang H, Zang S, Ma D, Sun X, Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J Oncol. 2014;45:383–392. doi: 10.3892/ijo.2014.2390. [DOI] [PubMed] [Google Scholar]
- 23.Nagano H, Tomimaru Y, Eguchi H, Hama N, Wada H, Kawamoto K, Kobayashi S, Mori M, Doki Y. MicroRNA-29a induces resistance to gemcitabine through the Wnt/β-catenin signaling pathway in pancreatic cancer cells. Int J Oncol. 2013;43:1066–1072. doi: 10.3892/ijo.2013.2037. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Thomas DM, Zalcberg JR. 5-fluorouracil: A pharmacological paradigm in the use of cytotoxics. Clin Exp Pharmacol Physiol. 1998;25:887–895. doi: 10.1111/j.1440-1681.1998.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 26.Noordhuis P, Holwerda U, Van der Wilt CL, Van Groeningen CJ, Smid K, Meijer S, Pinedo HM, Peters GJ. 5-Fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Ann Oncol. 2004;15:1025–1032. doi: 10.1093/annonc/mdh264. [DOI] [PubMed] [Google Scholar]
- 27.Coget J, Borrini F, Susman S, Sabourin JC. Colorectal carcinomas in 2013: The search for powerful prognostic markers is still on the go! Cancer Biomark. 2014;14:145–150. doi: 10.3233/CBM-130378. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Gao F, Zhang XP. miR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol Rep. 2015;33:607–614. doi: 10.3892/or.2014.3646. [DOI] [PubMed] [Google Scholar]
- 29.Xu AG, Yu ZJ, Jiang B, Wang XY, Zhong XH, Liu JH, Lou QY, Gan AH. Colorectal cancer in Guangdong province of China: A demographic and anatomic survey. World J Gastroenterol. 2010;16:960–965. doi: 10.3748/wjg.v16.i8.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ades S. Adjuvant chemotherapy for colon cancer in the elderly: Moving from evidence to practice. Oncology (Williston Park) 2009;23:162–167. [PubMed] [Google Scholar]
- 31.McEwan DG, Brunton VG, Baillie GS, Leslie NR, Houslay MD, Frame MC. Chemoresistant KM12C colon cancer cells are addicted to low cyclic AMP levels in a phosphodiesterase 4-regulated compartment via effects on phosphoinositide 3-kinase. Cancer Res. 2007;67:5248–5257. doi: 10.1158/0008-5472.CAN-07-0097. [DOI] [PubMed] [Google Scholar]
- 32.He Y, Wang J, Wang J, Yung VY, Hsu E, Li A, Kang Q, Ma J, Han Q, Jin P, et al. MicroRNA-135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am J Cancer Res. 2015;5:1382–1395. [PMC free article] [PubMed] [Google Scholar]
- 33.Mao Y, Chen H, Lin Y, Xu X, Hu Z, Zhu Y, Wu J, Xu X, Zheng X, Xie L. microRNA-330 inhibits cell motility by downregulating Sp1 in prostate cancer cells. Oncol Rep. 2013;30:327–333. doi: 10.3892/or.2013.2452. [DOI] [PubMed] [Google Scholar]
- 34.Meng H, Wang K, Chen X, Guan X, Hu L, Xiong G, Li J, Bai Y. MicroRNA-330-3p functions as an oncogene in human esophageal cancer by targeting programmed cell death 4. Am J Cancer Res. 2015;5:1062–1075. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Shi H, Liu B, Li J, Liu Y, Yu B. miR-330-3p controls cell proliferation by targeting early growth response 2 in non-small-cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 2015;47:431–440. doi: 10.1093/abbs/gmv032. [DOI] [PubMed] [Google Scholar]
- 36.Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y. MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One. 2012;7:e46010. doi: 10.1371/journal.pone.0046010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, Schnitzer RJ, Pleven E, Scheiner J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–666. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Du F, Zhao Q, Jin J, Ma X, Li H. Acquisition of 5-fluorouracil resistance induces epithelial-mesenchymal transitions through the Hedgehog signaling pathway in HCT-8 colon cancer cells. Oncol Lett. 2015;9:2675–2679. doi: 10.3892/ol.2015.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehman NL. Future potential of thymidylate synthase inhibitors in cancer therapy. Expert Opin Investig Drugs. 2002;11:1775–1787. doi: 10.1517/13543784.11.12.1775. [DOI] [PubMed] [Google Scholar]
- 40.Popat S, Matakidou A, Houlston RS. Thymidylate synthase expression and prognosis in colorectal cancer: A systematic review and meta-analysis. J Clin Oncol. 2004;22:529–536. doi: 10.1200/jco.2004.22.14_suppl.9576. [DOI] [PubMed] [Google Scholar]
- 41.Lenz HJ, Hayashi K, Salonga D, Danenberg KD, Danenberg PV, Metzger R, Banerjee D, Bertino JR, Groshen S, Leichman LP, Leichman CG. p53 point mutations and thymidylate synthase messenger RNA levels in disseminated colorectal cancer: An analysis of response and survival. Clin Cancer Res. 1998;4:1243–1250. [PubMed] [Google Scholar]
- 42.Copur S, Aiba K, Drake JC, Allegra CJ, Chu E. Thymidylate synthase gene amplification in human colon cancer cell lines resistant to 5-fluorouracil. Biochem Pharmacol. 1995;49:1419–1426. doi: 10.1016/0006-2952(95)00067-A. [DOI] [PubMed] [Google Scholar]
- 43.Cho YB, Chung HJ, Lee WY, Choi SH, Kim HC, Yun SH, Chun HK. Relationship between TYMS and ERCC1 mRNA expression and in vitro chemosensitivity in colorectal cancer. Anticancer Res. 2011;31:3843–3849. [PubMed] [Google Scholar]