Abstract
Morchella species are well known world-round as popular and prized edible fungi due to their unique culinary flavor. Recently, several species have been successfully cultivated in China. However, their reproductive modes are still unknown, and their basic biology needs to be elucidated. Here, we use the morel genome information to investigate mating systems and life cycles of fourteen black morel species. Mating type-specific primers were developed to screen and genotype ascospores, hymenia and stipes from 223 ascocarps of the 14 species from Asia and Europe. Our data indicated that they are all heterothallic and their life cycles are predominantly haploid, but sterile haploid fruiting also exists. Ascospores in all species are mostly haploid, homokaryotic, and multinuclear, whereas aborted ascospores without any nuclei were also detected. Interestingly, we monitored divergent spatial distribution of both mating types in natural morel populations and cultivated sites, where the fertile tissue of fruiting bodies usually harbored both mating types, whereas sterile tissue of wild morels constantly had one MAT allele, while the sterile tissue of cultivated strains always exhibited both MAT alleles. Furthermore, MAT1-1-1 was detected significantly more commonly than MAT1-2-1 in natural populations, which strongly suggested a competitive advantage for MAT1-1 strains.
Introduction
As iconic famous edible fruiting bodies, true morels, belonging to the genus Morchella, have been highly appreciated, prized and marketed worldwide since antiquity due to their texture and unique scent1. Dramatically increased consumption and the rising market led to over-harvest of wild morel resource over-harvest2. Recently, except M. rufobrunnea 3, several other morel species, such as M. sextelata (unpublished), M. eximia (unpublished) and M. importuna 4 can now be successfully cultivated in China, which greatly alleviates the market pressure. However, artificial cultivation, despite under optimized conditions, faces some big problems, such as unstable production, which has brought serious economic loss to the growers in China and urgently needs to be addressed. Fruiting is a crucial step in the fungal life cycle and occurs by either outcrossing or selfing5. Therefore, determining the reproductive strategies and mating systems of morels has high economical relevance for cultivation and harvesting. However, shedding light on the mechanisms involved in the reproduction of Morchella spp. is a challenging task. Thus far, among morel studies, most of which focused on fungal species richness and distribution of morels, only several were slightly related with the reproductive modes of Morchella species2, 6–8. The weak research progress in this aspect was attributed, on one hand, to the previous confused taxonomy, on the other hand, to the absence of molecular information on MAT genes of morels, because of the low level of conservation among mating type (MAT) genes in ascomycetes and the absence of genomic sequence.
MAT genes are master regulatory loci controlling sexual reproduction and development in fungi5. Heterothallic fungi, namely obligately outcrossing fungi, are self-sterile and usually require the participation of the opposite mating type partner to reproduce. The genes of opposite mating types are located on the same chromosomal locus but are highly divergent and nonhomologous, respectively encoding the alpha-box domain protein (MAT1-1-1) and the high mobility group (HMG) protein (MAT1-2-1)9. Conversely, homothallic fungi are self-fertile and can complete the sexual cycle without a mating partner. Typically a single homothallic strain harbors both MAT genes (linked or unlinked) in the same haploid nucleus9, 10. Over the past several years, the advent of molecular techniques, especially genome sequencing, now paves the way for making a significant leap forward in comprehending the life cycles and reproductive modes of Morchella species.
In this study, we used the genome sequences of morels to reveal the reproductive modes of black morel species. Our objectives were: (1) to infer their phylogeny and genetic diversity of the MAT1-1-1 and MAT1-2-1 genes, (2) to illuminate the reproductive modes of fourteen black morels species and (3) to evaluate both mating types distribution segregation within populations and in ascomatas, and their biparental roles partition. In order to answer these questions, 223 samples belonging to fourteen black morel species were analyzed here, covering wild and cultivated samples, which were assessed by analyzing the presence of both mating types: MAT1-1-1 and MAT1-2-1. All of these results will help to elucidate morel biology and are also of considerable practical impact for optimizing cultivation techniques and increasing production in morels fields.
Results
Searching for mating type genes in the Morel genome
The M. eximia genome sequence has been conducted in Kunming Institute of Botany, CAS recently. Its database was investigated for the presence of orthologs of ascomycetes MAT genes using BLASTN and TBLASTX similarity searches. First, we respectively chose Tuber melanosporum (ADU56595) and T. borchii (AIU38078) to blast Alpha-box and HMG-containing sequences in Morchella genome. Then we revealed one MAT1-1-1 and one MAT1-2-1 which we confirmed by blast to Genbank, resepctively best matching with T. indicum (AHE80940, AHE80941), Stagonosporopsis chrysanthemi (AHY81336) and Penicillium kewense (CBY44653).
Genetic diversity of MAT idiomorphs and phylogenetic analysis
Both MAT1-1-1 and MAT 1-2-1 were successfully amplified and sequenced from the fourteen species. The length of MAT1-1-1 and MAT1-2-1 genes respectively varied from 729 to 736 bp and from 398 to 408 bp among these fourteen species, and respectively contained two exons and one intron, and three exons and two introns. Though the length of both genes among these species varied, the protein length they translated was respectively conserved at 189 amino acids and 89 amino acids.
The nucleotide sequences alignment of MAT1-1-1 included a total of 744 sites after trimming and contained 191 parsimony-informative sites and three singleton variable sites. Its estimated nucleotide diversity (p) was 0.0488 with 191/744 (25.67%) variable nucleotide sites. The MAT1-2-1 alignment consisted of a total of 411 sites after trimming and included 121 parsimony-informative sites, no singleton variable sites. It had an estimated nucleotide diversity (p) of 0.0574 with 121/411 (29.44%) variable nucleotide sites.
Maximum parsimony phylogenetic analyses were performed with three matrices, viz., MAT1-1-1 sequences, MAT1-2-1 sequences, and the combined two-gene sequences. The phylogenetic trees constructed with each of the three matrices had similar topological structures. Herein, only the phylogenetic tree of their combined matrix is shown (Fig. 1). In this phylogenetic tree, each species could be distinguished and grouped together with high bootstrap support respectively as a monophyletic clade (Fig. 1).
Figure 1.
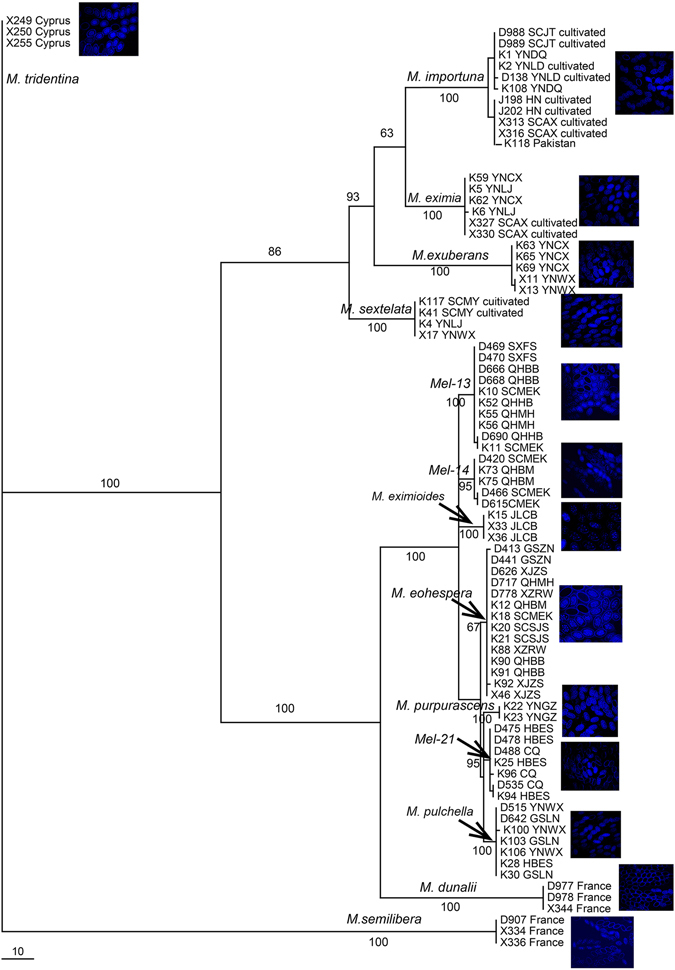
Phylogenetic analyses of 83 collections of fourteen black morel species based on the combined dataset totaling 1155 bp of MAT1-1-1 and MAT1-2-1. Bootstrap values lower than 50% are not shown. The pictures shown beside each species branch are DAPI-staining of ascospores nuclei.
Even mating types ratio of single spore strains in fourteen species
Single ascospores were isolated from the fourteen species, and cultivated as reported in the material and methods section. DNA was successfully extracted from cultures of the single spores, of which the presence and distribution of mating type genes of single spores in each ascomata were assessed using a PCR approach. Each single spore always showed only a single mating type, either the MAT1-1-1 or the MAT1-2-1 gene. No spores harboring both mating types were found. The ratio of MAT1-1-1:MAT1-2-1 of single ascopsores in each species was around 1:1 and consisted with the null hypothesis (no deviation from a theoretical segregation ratio of 1:1, MAT1-1-1:MAT1-2-1) after Fisher’s exact test (Table 1). Therefore, the two mating types occurred equally among ascospores of a single ascomata, and the reproductive modes of these fourteen species were assumed to be heterothallic.
Table 1.
Spatial segregation of mating types in ascomatas, MAT ratios of single spores and nucleotide diversity of MAT1-1-1 and MAT1-2-1 in each species.
| Species/N | Site | N | Wild/Cultivated | Year | Spatial segregation types## of MAT/N | MAT1-1-1:MAT1-2-1in maternal tissue* (P value**) | MAT1-1-1:MAT1-2-1of single spores (P value**) | &Pi of MAT1-1-1/Pi of MAT1-2-1 |
|---|---|---|---|---|---|---|---|---|
| M. tridentina/5 | Cyprus | 5 | W | 2015 | II/5 | 1.5:1(P:1) | 24:26 (P:1) | 0/0 |
| M. semilibera/5 | France | 5 | W | 2015 | II/5 | 1.5:1 (P:1) | 25:25 (P:1) | 0/0 |
| M. sextelata/4 | Lijiang# | 1 | W | 2007 | II/1 | 4:1 (P:1) | 25:25 (P:1) | 0/0 |
| Weixi# | 1 | W | 2015 | II/1 | ||||
| Mianyang# | 2 | C | 2015 | I/1, II/1 | ||||
| M. eximia/25 | Lijiang# | 3 | W | 2007 | II/3 | 2.3:1 (P:1) | 26:24 (P:1) | 0.00046/0 |
| Chuxiong# | 6 | W | 2014 | I/1, II/5 | ||||
| Weixi# | 11 | W | 2015 | II/10, III/1 | ||||
| Mianyang# | 5 | C | 2016 | I/3, II/2 | ||||
| M. exuberans/20 | Chuxiong# | 18 | W | 2014 | I/3, II/14 | 1.6:1(P:1) | 25:25 (P:1) | 0/0 |
| Weixi# | 2 | W | 2015 | II/2 | ||||
| M. importuna/39 | Deqin# | 5 | W | 2007 | II/2, III/3 | 1.2:1 (P:1) | 25:25 (P:1) | 0.00189/0.00045 |
| Mianyang# | 1 | W | 2015 | I/1 | ||||
| Pakistan | 1 | W | 2015 | II/1 | ||||
| Chengdu# | 16 | C | 2014 | I/12, II/3, III/1 | ||||
| Nanyang# | 8 | C | 2016 | I/6, II/2 | ||||
| Mianyang# | 8 | C | 2016 | I/5, II/3 | ||||
| Mel-13/20 | Haibei# | 6 | W | 2012 | II/5, III/1 | 1.8:1 (P:1) | 27:23 (P: 0.841) | 0/0.00087 |
| Aba# | 2 | W | 2009 | II/2 | ||||
| Haidong# | 2 | W | 2012 | I/2 | ||||
| Lvliang# | 5 | W | 2009 | II/5 | ||||
| Haibei# | 5 | W | 2012 | II/5 | ||||
| Mel-14/18 | Aba# | 6 | W | 2009 | II/6 | 1.8:1 (P:1) | 25:25 (P:1) | 0.00082/0 |
| Guoluo# | 12 | W | 2012 | I/4, II/8 | ||||
| M. eximioides/20 | Changbai# | 20 | W | 2010 | II/18, III/2 | 5.7:1 (P:0.471) | 24:26 (P:1) | 0/0 |
| M. eohespera/20 | Aba# | 4 | W | 2009 | II/3, III/1 | 2.3:1 (P:1) | 26:24 (P:1) | 0.00039/0 |
| Enshi# | 1 | W | 2010 | III/1 | ||||
| Haibei# | 2 | W | 2012 | II/2 | ||||
| Changdu# | 6 | W | 2012 | II/6 | ||||
| Zhaosu# | 3 | W | 2009 | II/3 | ||||
| Gannan# | 4 | W | 2009 | II/4 | ||||
| M. purpurascens/2 | Deqin# | 2 | W | 2010 | II/2 | 2:0 (P:1) | 24:26 (P:1) | |
| Mel-21/19 | Enshi# | 9 | W | 2010 | I/2, II/7 | 1.3:1 (P:1) | 26:24 (P:1) | 0.00104/0 |
| Chongqing# | 10 | W | 2010 | II/10 | ||||
| M. dunalii/5 | France | 5 | W | 2015 | —&& | —&& | 25:25 (P:1) | 0/0 |
| M. pulchella/21 | Enshi# | 1 | W | 2010 | II/1 | 1.4:1 (P:1) | 25:25 (P:1) | 0/0.00070 |
| Longnan# | 12 | W | 2004 | II/12 | ||||
| Weixi# | 8 | W | 2010 | I/3, II/5 |
#These sites were in China.
##Hymenia and stipes were respectively represented as fertile and sterile tissue. I: both of MAT were found in both hymenia layers and stipes; II: both MAT were detected in hymenia layers, but only one mating type, either MAT1-1-1 or MAT1-2-1, in stipes; III: only one MAT was found in these ascocarps, without ascospores after microscopic observation.
*The sterile tissue was defined as maternal tissue here, due to its possible duty of raising the whole ascomata.
**P value was generated by Fisher’s exact test using SPSS10.0 software.
&Pi: nucleotide diversity analyzed by DnaSP software.
&&Authors only got some part from the hymenia of samples, no stipes, so, the MAT distribution in stipes of M. dunalii could not be analyzed.
Nuclei analysis of ascospores from these fourteen species were performed based on DAPI-staining methods, which indicated most of ascospores in each species are haploid homokaryotic multinuclear, but some aborted ascospores without nuclei were also found in each species (Fig. 1).
Uneven spatial segregation of mating types in fertile and sterile tissue of wild ascocarps
A total of 186 black morel ascocarps were harvested from twenty-five sites during the spring to autumn collection seasons of 2004–2016 (Table 1). To investigate the MAT distribution difference between fertile and sterile tissues, DNA from both the hymenia and stipes of each ascocarp was analysed, respectively representing fertile and sterile tissues, by PCR using primers specific for the MAT locus. Interestingly, the stipe usually harbors only a single mating type, either the MAT1-1-1 or the MAT1-2-1 gene, but hymenia always displayed both MAT-specific amplicons. Three spatial segregation types of mating types were identified, as shown in detail in Table 1 and Fig. 2. The first one was that both of mating types were found in hymenia and stipes (I); the second and also dominant one was that both mating types were detected in the hymenia, but only one mating type, either MAT1-1-1 or MAT1-2-1, in stipes (II); the last one was that only one mating type was found in the ascocarps, which did not produce ascospores according to microscopic observation (III). By integrating the information from the analysis of ascospores, stipes and the hymenia, we interpreted the dominant component (sterile tissue, namely stipes etc.) of morel ascocarps was haploid, formed by one MAT mycelium, which was defined as ‘maternal’ (M) tissue to support the whole ascomata, and another putative genotype derived from the fertile tissue (hymenia) was defined as ‘paternal’ (P) partner to complete sexual reproduction and life cycle together with the maternal.
Figure 2.
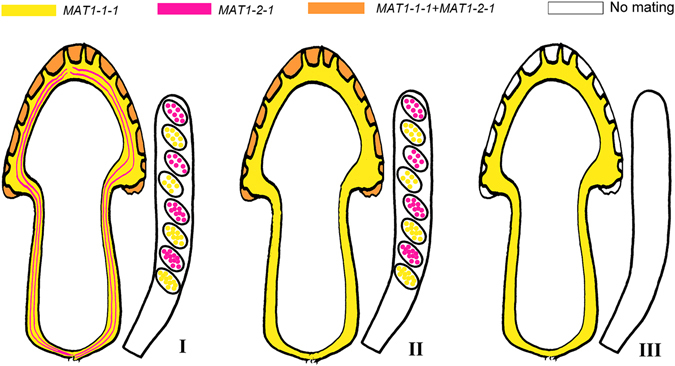
Schematic illustration of mating gene distribution in mature ascomata of morels. Although MAT1-1-1 (depicted here in yellow) was detected much more dominant than MAT1-2-1 (depicted in pink) in the sterile tissues (context of cap and stipe, and ridges on the cap), two mating types occurred equally in fertile tissues (hymenial layers, depicted in orange), and, thus, all 14 studied species must be heterothallic. The hyphae of the dominant MAT1-1-1 served as ‘maternal’ tissue to support the whole ascomata, while MAT1-2-1 served as ‘paternal’ partner to complete sexual reproduction. Three combinations were detected, both MAT loci are equally exhibited (I), MAT1-1-1 acts as the dominant while MAT1-2-1 plays a supporting role (II), and only a single MAT locus, exampled with MAT1-1-1 here, is present and resulted in non-production of ascospores (no mating) (III). Occasionally, MAT1-2-1 played ‘maternal’ while MAT1-2-1 ‘paternal’, which is not illustrated here. Ascospores, when present, are mostly haploid homokaryotic multinuclear.
Unequal MAT idiomorphs distribution in each species
In order to deeply investigate the presence and distribution of mating type genes of ‘the maternal tissue’ within and among the fourteen species, mating type genes detection of ‘the maternal tissue’ were conducted and assessed. Results indicated that, except M. importuna with the ratios of MAT1-1-1:MAT1-2-1 around 1, all the other species were far more than 1, the highest one shown in M. eximioides even almost to 6. Though no rejection in these species, current results strongly indicated that MAT1-1-1 was likely the dominant mating type of maternal tissue for morels ascocarps.
Discrepant distribution of MAT idiomorphs between wild and cultivated populations of three species
The presence and spatiotemporal distribution of opposite mating types was evaluated in different tissues of ascocarps and in different species shown as above. Considering that several species among them have been successfully cultivated, as a control, cultivated ascocarps of M. sextelata, M. eximia and M. importuna were genotyped to investigate the opposite mating types existence and distribution and to be compared with their wild populations.
As expected, the single spores from cultivated ascocarps presented both mating types with approximately equal ratios, similar to the wild populations, indicating the frequencies of the two mating types occurred equally in single spores of wild and cultivated ascocarps in our study.
However, the distributions of MAT idiomorphs were shown to be discrepant between wild and cultivated ascocarps. Our results indicated that though those individuals belonged to the same species, rare type I (3.9%) was found in the wild populations of three species, whereas it was found to be dominant in the cultivated population. The stipe of grown morels always showed the presence of both MAT genes, in contrast, usually only one MAT detected in the stipes of wild ones. In conclusison, an inconsistent distribution of MAT exists between cultivated and wild morels of the same species.
Discussion
Sequencing of the M. eximia genome paved the way and promoted the progress to characterize the MAT locus in some true morel species in this study. Further progress on understanding the life cycle and mating behavior of true morels is necessitated by the recent successful cultivation of several morel species. To date, their mating biology and conditions required for fruiting remains unknown, even for these successfully cultivated species. Thus, understanding the mating biology of them is critical both to conservation of natural populations as well as to development of artificial cultivation systems.
Attainment of mating types and assessment of their genetic diversity and phylogeny
Notably, both mating type genes were first identified in fourteen black morel species after multiple attempts, including M. tridentina, M. semilibera, M. sextelata, M. eximia, M. exuberans, M. importuna, Mel-13, Mel-14, M. eximioides, M. eohespera, M. purpurascens, Mel-21, M. dunalii and M. pulchella, though they showed different length in both MAT1-1-1 and MAT1-2-1 nucleotide sequences. Analyses of MAT sequence showed high levels of nucleotide diversity (pi) in both MAT1-1-1 (0.0488) and MAT1-2-1 (0.0574) within these species, but low levels of nucleotide diversity (pi) within intraspecies as shown in Table 1. This indicated that MAT genes evolve rapidly among morel species but highly conserved within species. Strong purifying selection against deleterious mutations were suggested in MAT genes11, which probably resulted in low intraspecific polymorphism observed in MAT genes of morels. MAT1-2-1 was found to be more variable than MAT1-1-1 in the fourteen species. Differences in function, selective pressures, and expression levels of the two mating type genes may account for this difference12–14, and our analyses indicated different sexual competence between both mating types in morels, but functional and transcriptional assays for the MAT1-1-1 and MAT1-2-1 genes of morel species are needed in the future to assess the potential contribution of these factors.
Phylogenetic analysis of the MAT1-1-1 and MAT1-2-1 combined sequences matrix supported each of the fourteen species as a monophyletic group (Fig. 1) corresponding to the accepted species relationships within Morchella genus15, 16. Both MAT genes are recommended as good candidate markers for phylogenetic inferences in Morchella genus including those closely related species, as reported in other fungi17–20.
Heterothallic reproductive modes of fourteen morel species
Fungal mating type systems play a pivotal role in controlling survival and sexuality. A better understanding of the reproductive modes that control fruiting in these morel species is therefore of both practical cultivation and economical relevance.
Here, we provide evidence to support the hypothesis that these fourteen species are heterothallic. Firstly, the findings that single spores from fourteen species harbor single opposite mating types with almost equal ratios strongly supports a heterothallic life style for each. Second, the presence of both mating type genes within the fertile tissue hymenia, paralleled with the dominant presence of only a single mating-type gene in the corresponding sterile tissue (type II), provides evidence of the heterothallic nature of these species and its prevalently haploid life cycle typical with other Ascomycota in which a maternal mycelium is fertilized by a paternal.
However, very unusually, haploid fruiting can occur without an opposite mating-type partner, given the existence of rare type III ascocarps (Table 1), where only one MAT gene was detected and no asci and ascospores in their mature hymenia were found after microscopic observation. It’s the first reported in Morchella genus to fruit without an opposite mating-type partner, which was also found in Cordyceps militaris, Botrytis cinerea and Sordaria brevicollis, where their haploid fruiting bodies were sterile and their progeny were usually undeveloped (no perithecia and ascospores)14, 21–23. They show a mixed mating system; i.e., individuals have the ability to outcross and haploid fruiting, though rare for the latter.
Our results supported that the fertile tissue (hymenia) of most morels carried both mating types, but, this contrasts with the expectations based on previous results where no heterozygosity was identified in many studies even when evaluated with codominant markers2, 6, 7, 15, 16, 24. Considering that these ascocarps were derived from outcrossing, as demonstrated by the presence of both mating types, till now, only difference at MAT locus was identified between their biparental genotypes. Usually, heterozygosity was expected in this case unless the fertilizing parent is the same as the maternal parent (homothallism or selfing) or the maternal and the fertilizing parents are closely related (inbred). Therefore, our current results suggest that these morels may have resulted from biparental inbreeding. Mating between closely-related parents has led to their progeny that may not display marker segregation except at the mating type locus25, 26. Our previous population genetic study is also consistent with this finding, which supported that inbreeding was prevalent in both Mel-13 and M. eohespera species2.
For the first time in Morchella, karyological staining in parallel with MAT screening among and within spores was conducted and have allow us to determine their haploid multinuclear homokaryotic level (Fig. 1). These nuclei in each spore are all copies of a single meiotic product and are not representatives of different meiotic products. But, it is still unknown what reasons lead to the production of aborted ascospores in each species or why fruiting bodies with no viable ascospores are produced.
Considering the haploid level occupying the predominant life cycle of morels, we suggested that heterokaryons are only transiently formed within a limited phase for the production of ascospores. The absence of heterokaryons during the dominant period of the morel life cycle implies that these species may contain a vegetative incompatibility system, as found in many other ascomycetes27–29. Genetic nonself recognition systems, probably work at the pre-fusion level, preventing the formation of anastomosis between strains to maintain the genetic integrity of each strain29–31 and protect resources within hyphae from exploitation by non-kin individuals during vegetative growth27. Moreover, the MAT locus was reported to be one of the loci governing vegetative incompatibility in Neurospora crassa, Sordaria brevicollis, and Ascobolus stercorarius 32, however, whether they play the same role and act on the vegetative incompatibility in Morchella spp. remains unknown. Also, the factors which mandate and underpin the transition from the vegetative to reproductive stages in morels still awaits to be uncovered. Additionally, an anamorphic phase of morels was recently described33 and also found in M. sextelata, M. eximia, M. importuna and several other cultivated species at their cultivation sites in China, however, what role asexual conidia played and whether act as fertilizing agents in morels are open questions.
Orchestration of maternal and parental tissue with MAT segregation in morel ascocarps
In some ascomycetes, genetic material is usually typified with maternal and parental partitioning, and the dominant partner of the ascocarp is suggested to be the maternal one feeding the fruiting body26, 34, 35. Sexual identity, i.e., which mating type acts as male or female, is not defined. In order to determine whether both mating types play equivalent sexual roles in morel ascocarps, the stipes representing sterile part and hymenia representing fertile part were respectively detected for MAT diversity in this study.
Our results indicated that the hymenia of wild morels always displayed both MAT genes, whereas in the corresponding stipes a single mating type was always preferentially or exclusively amplified (Type II, Table 1). This provides definitive evidence for the following conclusions: 1) that most morels developed as a consequence of mating between strains of opposite sexual partners; 2) the stipes of morels, likely including other sterile tissue, resulted from homokaryotic/haploid mycelia and could be formed by mycelial strains harboring either MAT1-1-1 or MAT1-2-1, behaving as ‘maternal’ partners in mating events; 3) both kinds of morel MAT mycelial strains are capable of producing the ‘maternal’-driven apothecial structures. We still did not find sexual structures (gametangia) in morels, but the current results indicated that a morel strain with either mating type might be hermaphroditic and have the capability of differentiating into ‘maternal’ and ‘paternal’ sexual structures.
However, for cultivated morels, based on the investigation in three cultivated species, not only their hymenia, but also their corresponding stipes always displayed both MAT genes (Type I, Table 1); this was greatly different from wild morels. Evidence of segregation of mating types among haploid single spores rejects the possibility of secondary homothallism and supports heterothallism. Given that our results above supported that competitions existed between both MAT mycelium in natural sites, we presumed the current special phenomenon might be related with adequate nutrition and optimal environmental conditions at cultivated sites, however the exact reasons remain unclear.
Additionally, we also found some ascocarps from natural sites, though this type was rare (Type III, Table 1), exhibited only one MAT gene and could not complete their life cycle, due to lacking of ascospores. This type, as we presumed above, was resulted from one unique mating behavior, sterile haploid fruiting.
Spatial competition and unbalanced distribution of mating types in natural sites
Many studies carried out on the frequency distribution of mating types in natural field sites and revealed that, for some fungi, they are always reported to be patchy and unbalanced, such as C. militaris 23, Cryptococcus neoformans 36, Mycosphaerella graminicola 13, 37, Phytophthora infestans 38, 39, and T. melanosporum 26, 29, 35. In the current study, we monitored the distribution of morel strains with different mating types respectively from fourteen species in different morels grounds. Given that mother tissue discussed above usually were haploid and included one MAT, so we genotyped MAT of mother tissue (stipes) within different populations of different species to investigate the distribution and competition between both MAT mycelia.
The distribution analysis of strains with different mating types provides an intriguing scenario of morels. Firstly, based on the analysis of the total samples from each of the fourteen species, except M. importuna where MAT1-1-1 was almost equal to MAT1-2-1 which might be associated with many cultivated morels included, MAT1-1-1 was detected significantly more commonly than MAT1-2-1 in all the other species, especially in M. eximioides (almost 6:1), indicating a competitive advantage for MAT1-1-1 strains, as raised in many studies5, 26, 29, 37. In some populations of these species, such as, Lvliang population in Mel-13, Changdu and Gannan populations in M. eohespera, unexpectedly extreme unbalance was observed where none of MAT1-2-1 detected. Generally and apparently, MAT types frequency was skewed and unbalanced with dominantly more frequent MAT1-1-1 than MAT1-2-1. Moreover, the diverse distribution types of mating types from wild and cultivated populations in this study indicated that a large-size population of strains is necessary.
Studies have shown, in some fungi, divergences in ecological, physiological, pathogenic or other functional importance are indicated between opposite mating types5, 13, 40, which likely affect the spatial distribution frequency of mating types in natural populations. Additionally, our results show clearly that MAT segregates equally in ascospores which means mating types should be in equal frequency in the population of ascopsores. However, MAT1-1-1 is more common in the stipes than MAT1-2-1, indicating that MAT1-1-1 is more competent to play a female sexual role. This might suggest mating types in Morchella genus have become more like “sexes” than the typical fungal mating types.
In China, morels are successfully cultivated in many sites, but the biggest problem growers face is the unstable production. No prior detection and screening of mating types during the strains breeding processs might be one of the reasons. We speculated the predominance of one MAT may be important as a reason for degeneration of strains due to strains with a particular mating type competing to survive. The spatial segregation of mating type could decrease the probability of sexually compatible partners meeting, lower the odds of mating and eventually lead to declining production.
Finally, here, we use molecular markers and current results to reconsider the potential morels life cycle (Fig. 3). Heterothallism is shown here to be the dominant reproductive mode in all of these fourteen species, with low proportional sterile haploid fruiting. However, the morphology of the fertilization process in the morel life cycle still remains elusive.
Figure 3.
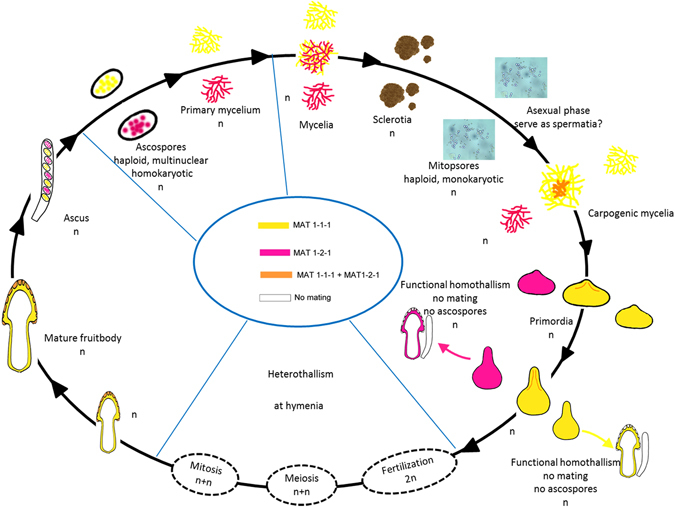
Proposed life cycle of the genus Morchella. Here, we illustrated two kinds of life cycles identified in morels, viz. haploid fruiting and heterothallism. Considering two types (I and II, shown in Fig. 2) were found in their heterthallism life cycle, we took type II with MAT1-1-1 serving as maternal tissue for example in this figure.
Conclusions and perspectives
To the best of our knowledge, this is the first study to determine mating types of morels and to assess their distribution in nature and in cultivation. The selection of morel strains, without clearly characterized genetic background, are prone to producing fruiting bodies susceptible to degeneration, ultimately resulting in wasted human resources and time. To avoid such problems in production efforts, the prior screening and determination of mating types with the aid of molecular markers is the important part of morel cultivation. The methods and results presented in this study could effectively facilitate selection, domestication, and management of strains to decrease the possibility of degeneration and also provide a theoretical reference for the artificial cultivation of morels and even the industrial-scale production in the future.
Material and Methods
Obtainment of mating type genes
Based on the genome sequence of Morchella eximia, MAT1-1-1 and MAT1-2-1 were identified using BLASTn and BLASTx against the NCBI nucleotide and protein database by sequence similarity searches (http://www.ncbi.nlm.nih.gov/BLAST/).
Sampling
168 wild samples collected in 25 populations of 10 provinces in China from 2004 to 2016; 10 wild samples from France; 5 wild samples from Cyprus; and an additional from Pakistan were used here to represent as wide geographical differences as possible. Based on the multi-gene phylogenetic species recognition results2, 16, the above 186 specimens belong to 14 black morel species, namely M. tridentina, M. semilibera, M. sextelata, M. eximia, M. exuberans, M. importuna, Mel-13, Mel-14, M. eximioides, M. eohespera, M. purpurascens, Mel-21, M. dunalii and M. pulchella. Additionally, 39 cultivated samples from five growth sites of M. sextelata, M. eximia and M. importuna were included here in order to detect if the ratio of mating types is consistent with a theoretical ratio of 1:1 (MAT1-1-1:MAT1-2-1) between wild and cultivated populations. Detailed species and geographical information of these 223 samples are presented in Table 1.
Single spore isolates and cultural conditions
Fifty single ascospores were randomly isolated from each species. Ascospores were washed, suspended in sterilized water, and 200 μL of a solution adjusted to a concentration of 200–300 ascospores mL−1 was spread on potato dextrose agar (PDA) and incubated at 23–25 °C for 1–2 d. Single germinated ascospores were picked using a dissecting needle under a dissecting microscope (Zeiss 455094) and transferred to a new PDA Petri dish, which was incubated at 23–25 °C for 1 week.
DNA extraction
DNA extraction from single spore cultures
A culture plug bearing actively growing mycelia was transferred onto freshly-prepared PDA. When the growth of the fungus reached the edge of the plate, mycelia were scraped from the surface with a sterilized inoculating shovel and transferred into a 1.5 ml microcentrifuge tube. Samples were immediately frozen in liquid nitrogen and stored at −80 °C until used.
DNA extraction of fruiting bodies
To detect whether there are differences in mating type genes between fertile tissue and sterile tissue, the hymenia and stipe of each ascomata were separated for DNA extraction to respectively represent fertile and sterile tissue and also used for comparison of different individuals within and among species. But, for M. dunalii, we just got some pieces of tissue from the hymenia of samples, so, no stipes DNA could be extracted and no analysis on MAT distribution of stipes in this species could be conducted.
Samples (mycelia/hymenia/stipe) were ground to a fine powder in a 1.5 ml microcentrifuge tube using a Kontes pellet pestle (Kaimu, China). Once pulverized, the samples were suspended in 700 μl of CTAB extraction buffer (100 mM Tris–Cl pH 8.4, 1.4 M NaCl, 25 mM EDTA, 2% CTAB), and incubated for 1.5–2.0 h at 65 °C, during which time they were gently inverted 3–5 times. After the samples were cooled to room temperature, 700 μl of chloroform-isoamyl alcohol (24:1) was added to each tube. The mixture was vortexed briefly, centrifuged at 12,000 g for 10 min, and then 500 μl of the upper phase was carefully transferred to a new 1.5 ml microcentrifuge tube. After a second chloroform-isoamyl alcohol (24:1) extraction was performed, the supernatant was transferred to a new 1.5 ml microcentrifuge tube and an equal volume of 100% isopropanol at 20 °C was added to each tube. The tube contents were mixed briefly by inversion to obtain a homogeneous solution and then they were stored overnight at 20 °C to precipitate total genomic DNA. After the tubes were warmed to room temperature they were centrifuged at 12,000 g for 10 min and the supernatant was discarded. The DNA pellet was washed consecutively with 70% and 100% ethanol, air-dried and then resuspended in 100 μl of sterile double distilled H2O. All genomic DNA samples were stored frozen at 20 °C until ready for use.
MAT PCR amplification
The mating type-specific primers for MAT1-1-1 and MAT1-2-1 genes were designed according to morel genome sequencing. The primers’ sequences are reported in Table S1 and used for the following PCR and DNA sequencing. Each PCR reaction contained 1 μl of 20 ng/μl genomic DNA, 2.5 μl of 10 × PCR reaction buffer, 0.5 μl dNTP mix (10 mmol), 2 μl each of primers (5 μmol), 1.5 μl bovine serum albumin (20 mg/ml) and 1.5 U of Taq DNA polymerase (Biomed, China). The final volume was adjusted to 25 μl with sterile distilled H2O. PCRs were conducted in an Applied Biosystems 2720 thermocycler (ABI, Foster City, CA), using the following cycling parameters: 94 °C for 3 min, 35 cycles of 94 °C for 1 min, 50 °C for 30 s, 72 °C for 1 min, followed by a final extension of 10 min at 72 °C. Amplicons were electrophoresed in 1.2% agarose in 1 × TAE, stained with GoldView™ (Guangzhou Geneshun Biotech Ltd., Guangdong, China), and then photographed over a ultraviolet transilluminator. PCR products were purified using a Bioteke DNA Purification Kit (Bioteke Corporation, Beijing, China), sequenced with ABI BigDye ver 3.1 (Sangon Co., Ltd., Shanghai, China), and then run on an ABI 3730 DNA Analyzer. The raw DNA sequences of MAT1-1-1 and MAT1-2-1 genes amplified were aligned with SeqMan (DNAStar Package, Madison, WI). Sequences generated in the present study have been deposited in GenBank under accession numbers KY508074-KY508239.
Mating type detection and screening for single ascospores, hymenia and stipes
According to the current results, the sequence length of MAT1-1-1 and MAT1-2-1 are respectively 729–736 bp and 398–408 bp, so mating type gene detection of the 700 single ascospores, 223 hymenia and 218 stipe cultures could be performed by observing amplicon length over a ultraviolet transilluminator after electrophoresis for first screening. This method greatly reduced the sequencing cost and accelerated mating type detection. Then, some samples in each species (totaled eighty-three samples) were chosen for sequence analysis of their MAT1-1-1 and MAT1-2-1 genes for genetic diversity and phylogenetic analysis.
Sequence diversity of MAT1-1-1 and MAT1-2-1 and phylogenetic analysis
MAT1-1-1 and MAT1-2-1 datasets were respectively aligned automatically with MAFFT41, manually adjusted using BioEdit ver. 7.0.9 42 (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and then manually trimmed to eliminate uneven ends. Nucleotide diversity and variable sites were estimated for each gene using DnaSP43. Maximum parsimony (MP) bootstrap analyses were conducted for the two-locus, MAT1-1-1 and MAT1-2-1 matrices.
MP analyses were performed in PAUP* V 4.0b10 44 based on a heuristic search of 1,000 replicates with random stepwise addition using tree-bisection-reconnection branch swapping and starting with trees obtained by the stepwise addition of sequences. All of the characteristics were equally weighted. Parsimony bootstrap analyses with 1,000 replicates were used to assess clade support.
Ratio assessment of mating types and Statistical analysis
The presence ratios of both mating types were assessed respectively for intra and inter-species single spore isolates, and among hymenia and stipes of intra and inter-species. The mating type ratios within and among species, and between wild populations and cultivated populations were compared. The Fisher’s exact test using SPSS software version 15.0 (SPSS, Inc., Chicago, IL, USA) was conducted to determine whether the ratio of mating types (MAT1-1-1:MAT1-2-1) did not deviate from a theoretical ratio of 1:1.
Karyological analysis
Ascospores were stained with DAPI (4′-6-diamidino-2-phenilindole) to visualize nuclei. Pools of purified ascospores were incubated for 15 min in the staining reagent (4 mM DAPI, 100 mM Tris-HCl pH 7.5, and 20% glycerol) directly on the microscope slide. Nuclei were observed under fluorescent light using an Olympus Fluoview FV1000 laser-scanning microscope.
Electronic supplementary material
Acknowledgements
The authors thank Dr. Jianping Xu (Department of Biology, McMaster University), Dr. Bang Feng and Dr. Jing Yang (Kunming Institute of Botany, CAS), Dr. Yu-Guang Fan (Changbai Mountain Academy of Sciences), Dr. Jean-Michel Bellanger (Centre d’Ecologie Fonctionnelle et Evolutive, CNRS), Prof. Xinsheng He (Southwest University of Science and Technology), Mr. Michael Loizides, Mr. Philippe Clowez, Mr. Hao Hu, Mr. Kaihua He, and Mr. Di Luan for their help during the authors’ study. Thanks also are due to the anonymous reviewers and Dr. Timothy Y. James (Department of Ecology and Evolutionary Biology, University of Michigan) for their constructive comments and suggestions on the manuscript. This work was supported by the National Basic Research Program of China (973 Program, No. 2014CB138305), the National Natural Science Foundation of China (No. 31300022), West Light Foundation of The Chinese Academy of Sciences and the CAS/SAFEA International Partnership Program for Creative Research Teams, Scientific and Technological Research Program of Chongqing Municipal Education Commission (No. KJ1703052).
Author Contributions
Z.L.Y. and X.H.D. conceived, designed and coordinated this study. X.H.D. designed, performed, analyzed the experiments, interpreted all of results, contributed to the preparation of the figures and wrote the paper. X.H.D., Q.Z. and F.R. collected samples. All authors reviewed the results and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01682-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kuo, M. Morels. University of Michigan Press, Ann Arbor. 923 (2005).
- 2.Du XH, Zhao Q, Xu J, Yang ZL. High inbreeding, limited recombination and divergent evolutionary patterns between two sympatric morel species in China. Scientific Reports. 2016;6:22434. doi: 10.1038/srep22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masaphy S. Biotechnology of morel mushrooms: successful fruiting body formation and development in a soilless system. Biotechnology Letters. 2010;32:1523–1527. doi: 10.1007/s10529-010-0328-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Q, Xu ZZ, Cheng YH, Qi SW, Hou ZJ. Bionic cultivation of Morchella conica. Southwest China Journal of Agricultural and Science. 2009;22:1690–1693. [Google Scholar]
- 5.Lee SC, Ni M, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiology and Molecular Biology Reviews. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalgleish H, Jacobson K. A first assessment of genetic variation among Morchella esculenta (Morel) populations. Journal of Heredity. 2005;96:396–403. doi: 10.1093/jhered/esi045. [DOI] [PubMed] [Google Scholar]
- 7.Pagliaccia D, et al. Development of molecular markers and preliminary investigation of the population structure and mating system in one lineage of black morel (Morchella elata) in the Pacific Northwestern USA. Mycologia. 2011;103:969–982. doi: 10.3852/10-384. [DOI] [PubMed] [Google Scholar]
- 8.Volk, T. J. & Leonard, T. J. Experimental studies on the morel. I. Heterokaryon formation between monoascosporous strains of Morchella. Mycologia 523–531 (1989).
- 9.Martin SH, Steenkamp ET, Wingfield MJ, Wingfield BD. Mate-recognition and species boundaries in the ascomycetes. Fungal Diversity. 2013;58:1–12. doi: 10.1007/s13225-012-0217-2. [DOI] [Google Scholar]
- 10.Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora. BioEssays. 1990;12(2):53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 11.Heitman J. Evolution of sexual reproduction: A view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biology Reviews. 2015;29(3–4):108–117. doi: 10.1016/j.fbr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart JE, Kawabe M, Abdo Z, Arie T, Peever TL. Contrasting codon usage patterns and purifying selection at the mating locus in putatively asexual Alternaria fungal species. PloS One. 2011;6:e20083. doi: 10.1371/journal.pone.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan J, Torriani SFF, McDonald BA. Significant divergence in pathogenicity between MAT1-1 and MAT1-2 isolates in the wheat pathogen Mycosphaerella graminicola. Fungal Genetics and Biology. 2007;44:339–346. doi: 10.1016/j.fgb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhang GZ, Liang Y. Improvement of fruiting body production in Cordyceps militaris by molecular assessment. Archives of Microbiology. 2013;195:579–585. doi: 10.1007/s00203-013-0904-8. [DOI] [PubMed] [Google Scholar]
- 15.O’Donnell K, et al. Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genetics and Biology. 2011;48:252–265. doi: 10.1016/j.fgb.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Du XH, Zhao Q, O’Donnell K, Rooney AP, Yang ZL. Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China. Fungal Genetics and Biology. 2012;49:455–469. doi: 10.1016/j.fgb.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 17.O’Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Pőggeler S. Phylogenetic relationships between mating-type sequences from homothallic and heterothallic ascomycetes. Current Genetics. 1999;36:222–231. doi: 10.1007/s002940050494. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y. J. et al. Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: Implications for its evolution and conservation. BMC Evolutionary Biology9(290), doi:10.1186/1471-2148-9-290 (2009). [DOI] [PMC free article] [PubMed]
- 20.Woudenberg JC, Gruyter JDE, Crous PW, Zwoers LH. Analysis of the mating-type loci of co-occurring and phylogenetically related species of Ascochyta and Phoma. Molecular Plant Pathology. 2012;13(4):350–362. doi: 10.1111/j.1364-3703.2011.00751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faretra, F. & Pollastro, S. Genetic studies of the phytopathogenic fungus Botryotinia fuckeliana (Botrytis cinerea) by analysis of ordered tetrads. Mycol. Res100, 620–624 (1996).
- 22.Robertson, S. J., Bond, L. and Read, N. D. (1998). Homothallism and heterothallism in Sordaria brevicollis. Mycological Research102, 1215–1223 (1996).
- 23.Zheng P, et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biology. 2011;12:R116. doi: 10.1186/gb-2011-12-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon, C. S., Gessner, R. V. & Romano, M. A. Population genetics and systematics of the Morchella esculenta complex. Mycologia 227–235 (1990).
- 25.Marra RE, Milgroom MG. The mating system of the fungus Cryphonectria parasitica: selfing and self-incompatibility. Heredity. 2001;86:134–143. doi: 10.1046/j.1365-2540.2001.00784.x. [DOI] [PubMed] [Google Scholar]
- 26.Rubini A, et al. Tuber melanosporum: mating type distribution in a natural plantation and dynamics of strains of different mating types on the roots of nursery-inoculated host plants. New Phytologist. 2011;189:723–735. doi: 10.1111/j.1469-8137.2010.03493.x. [DOI] [PubMed] [Google Scholar]
- 27.Debets AJM, Griffiths AJF. Polymorphism in hetgenes prevents resource plundering in Neurospora crassa. Mycological Research. 1998;102:1343–1349. doi: 10.1017/S095375629800639X. [DOI] [Google Scholar]
- 28.Leslie JF. Fungal vegetative compatibility. Annual Review of Phytopathology. 1993;31:127–151. doi: 10.1146/annurev.py.31.090193.001015. [DOI] [PubMed] [Google Scholar]
- 29.Rubini A, Riccioni C, Belfiori B, Paolocci F. Impact of the competition between mating types on the cultivation of Tuber melanosporum: Romeo and Juliet and the matter of space and time. Mycorrhiza. 2014;24(Suppl 1):S19–S27. doi: 10.1007/s00572-013-0551-6. [DOI] [PubMed] [Google Scholar]
- 30.Choi GH, et al. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics. 2012;90:113–127. doi: 10.1534/genetics.111.133983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iotti M, et al. Self/nonself recognition in Tuber melanosporum is not mediated by a heterokaryon incompatibility system. Fungal Biology. 2012;116(2):261–275. doi: 10.1016/j.funbio.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Glass NL, Jacobson DJ, Patrick KTS. The genetics of hyphal fusion and vegetative incompatibility filamentous ascomycete fungi. Annual Review of Genetics. 2000;34:165–186. doi: 10.1146/annurev.genet.34.1.165. [DOI] [PubMed] [Google Scholar]
- 33.Carris LM, Peever TL, McCotter SW. Mitospore stages of Disciotis, Gyromitra and Morchella in the inland Pacific Northwest USA. Mycologia. 2015;107(4):729–744. doi: 10.3852/14-207. [DOI] [PubMed] [Google Scholar]
- 34.Paolocci F, Rubini A, Riccioni C, Arcioni S. Reevaluation of the life cycle of Tuber magnatum. Applied and Environmental Microbiology. 2006;72:2390–2393. doi: 10.1128/AEM.72.4.2390-2393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selosse MA, Taschen E, Giraud T. Do black truffles avoid sexual harassment by linking mating type and vegetative incompatibility? New Phytologist. 2013;199(1):10–13. doi: 10.1111/nph.12329. [DOI] [PubMed] [Google Scholar]
- 36.Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. Journal of Clinical Microbiology. 2005;43:556–564. doi: 10.1128/JCM.43.2.556-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan J, Kema GHJ, Waalwijk C, McDonald BA. Distribution of mating type alleles in the wheat pathogen Mycosphaerella graminicola over spatial scales from lesions to continents. Fungal Genetics and Biology. 2002;36:128–136. doi: 10.1016/S1087-1845(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 38.Cooke DEL, et al. Phenotypic and genotypic diversity of Phytophthora infestans populations in Scotland (1995–1997) Plant Pathology. 2003;52:181–192. doi: 10.1046/j.1365-3059.2003.00817.x. [DOI] [Google Scholar]
- 39.Couch BC, et al. Origins of host-specific populations of the blast pathogen Magnaporthe oryzaein crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics. 2005;170:613–630. doi: 10.1534/genetics.105.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke DL, Woodlee GL, McClell CM, Seymour TS, Wickes BL. The Cryptococcus neoformans STE11 gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Molecular Microbiology. 2001;40:200–213. doi: 10.1046/j.1365-2958.2001.02375.x. [DOI] [PubMed] [Google Scholar]
- 41.Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research30(14), 3059–3066. PMID:12136088 (2002). [DOI] [PMC free article] [PubMed]
- 42.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. in. Nucleic acids symposium series. 1995;41:95–98. [Google Scholar]
- 43.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 44.Swofford, D.L. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4 (2003).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


