Abstract
Emerging evidence has indicated that microRNAs (miRNAs) are frequently dysregulated and are fundamental in the pathogenesis of hepatocellular carcinoma (HCC). However, the roles of miR-195 in HCC have not been well elucidated. In the present study, the expression of miR-195 was determined to be markedly downregulated in HCC tissues and cell lines, as compared with normal liver cells. Restoration of miR-195 expression resulted in significant inhibition of the proliferation and tumorigenicity of HCC cells in vitro and in vivo. Gene expression data and luciferase reporter assays revealed that miR-195 is able to directly inhibit the expression of astrocyte elevated gene 1 (AEG-1) through interaction with its 3′ untranslated region. Consistently, an inverse correlation between miR-195 and AEG-1 expression was observed in HCC tissues. Furthermore, the overexpression of AEG-1 was able to partially attenuate the miR-195-induced inhibition of cell growth and promotion of apoptosis. Taken together, these findings indicate that miR-195 functions as a tumor suppressor by inhibiting AEG-1. This pathway may provide new insights into the potential molecular mechanisms of HCC.
Keywords: microRNA, hepatocellular carcinoma, miR-195, AEG-1, tumor biology
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-associated mortality worldwide (1). Population-based studies have indicated that the incidence rate continues to approximate the mortality rate, indicating that HCC accounts for the mortality of a high proportion of patients who develop it (1). Excessive proliferation of cells is responsible for the initiation and progression of malignant tumors, including HCC (2). Therefore, identifying molecules that suppress proliferation and enhance apoptosis may provide novel targets for HCC therapies.
MicroRNAs (miRNAs) represent a large family of endogenous, non-protein-coding RNA molecules that are able to negatively regulate gene expression at the post-transcriptional level through translational repression and/or mRNA cleavage (3). miRNAs are involved in a variety of biological processes, including proliferation, differentiation, apoptosis and tumor metastasis, and may function as oncogenes or tumor suppressor genes by binding to the 3′ untranslated region (3′-UTR) of target mRNAs (3). Altered expression of various miRNAs, including miR-122, miR-375, miR-101 and miR-199a-3p, have been observed in cases of HCC, which indicates an involvement of miRNAs in the pathogenesis of HCC (4–7). More extensive investigations are required to elucidate the precise functions of miRNAs in hepatocarcinogenesis.
Previously, we identified a specific aberrant miRNA expression profile in HCC by comparison of miRNA expression profiles in cancerous hepatocytes and normal human hepatocytes. Among the aberrantly expressed miRNAs in HCC, miR-195 was of particular interest as its putative targets include astrocyte elevated gene 1 (AEG-1), which has been reported to serve critical roles in hepatocarcinogenesis (8). The aim of the present study was to clarify the functional significance of miR-195 in HCC and identify cancer-associated pathways that are regulated by miR-195. The results indicate that miR-195 is pathologically downregulated in HCC, and that restoration of miR-195 expression is able to inhibit proliferation as well as initiate apoptosis of liver cancer cells by targeting AEG-1, in vitro and in vivo.
Materials and methods
Tissue specimens and cell lines
In total, 36 pairs of HCC and adjacent non-tumor liver tissues were obtained from patients who were pathologically diagnosed with HCC and underwent hepatectomy at Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China) between September 2009 and September 2012. Tissue samples were immediately snap frozen in liquid nitrogen until use. The study was approved by the local Research Ethics Committee at Tongji Hospital of Huazhong University of Science and Technology, and written informed consent was obtained from the patients. The characteristics of the included patients are described in Table I.
Table I.
Clinical characteristics of patients with hepatocellular carcinoma included in the present study (n=36).
| Clinicopathological parameters | n (%) |
|---|---|
| Sex | |
| Male | 33 (91.7) |
| Female | 3 (8.3) |
| Age, years | |
| ≥40 | 33 (91.7) |
| <40 | 3 (8.3) |
| Serum alanine aminotransferase level, U/l | |
| <50 | 21 (58.3) |
| ≥50 | 15 (41.7) |
| Hepatitis B virus surface antigen | |
| Positive | 31 (86.1) |
| Negative | 5 (13.9) |
| Serum α-fetoprotein level, ng/ml | |
| <400 | 20 (55.6) |
| ≥400 | 16 (44.4) |
| Length of tumor, cm | |
| <5 | 3 (8.3) |
| ≥5 | 33 (91.7) |
| New American Joint Committee on Cancer stage | |
| Early (I–II) | 31 (86.1) |
| Late (IIIA-IIIB) | 5 (13.9) |
The cell lines used in this study included the immortalized human fetal liver cell line L02, and eight immortalized human liver cancer cell lines (HepG2, Huh7, PLC/PRF/5, SMMC-7721, SK-HEP-1, MHCC-97H, MHCC-97L and Hep 3B). All were maintained in Gibco Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 mg/ml streptomycin, and incubated at 37°C with 5% CO2.
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol without any prior purification step. RNA concentrations were determined spectrophotometrically.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Complementary DNA (cDNA) was synthesized from 1 µg of total RNA using the First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). qPCR was performed in an Applied Biosystems® 7500 Real-Time PCR System (Thermo Fisher Scientific, Inc.) using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. The following thermocycling conditions were used: For RT, 37°C for 60 min and 70°C for 5 min; and for qPCR, 50°C for 2 min (UDG incubation), 95°C for 2 min, and 40 cycles of 95°C for 3 sec and 60°C for 30 sec. β-actin was used as an internal control. The primer sequences for AEG-1 and β-actin are listed in Table II. The expression levels of miR-195 were also analyzed by RT-qPCR using a Bulge-Loop™ miRNA qRT-PCR Primer Set (Guangzhou RiboBio Co., Ltd., Guangzhou, China) and normalized to U6. The catalog numbers of the primers for miR-195 and U6 were miRQ0000461-1-2 and MQP-0202 (both from Guangzhou RiboBio Co., Ltd.). The 2−ΔΔCq method was used for quantification (9). All reactions were performed in triplicate.
Table II.
Sequences of the primers used in SYBR Green reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence (5′-3′) |
|---|---|
| β-actin | |
| Forward | CATGTACGTTGCTATCCAGGC |
| Reverse | CTCCTTAATGTCACGCACGAT |
| AEG-1 | |
| Forward | AAATGGGCGGACTGTTGAAGT |
| Reverse | CTGTTTTGCACTGCTTTAGCAT |
AEG-1, astrocyte elevated gene 1.
miRNA mimics and plasmid transfection
An miR-195 mimic and its matched negative control (miR-NC) were purchased from Guangzhou RiboBio Co., Ltd. An AEG-1 expressing vector, pcDNA3.1-AEG-1 (which lacked the 3′-UTR of AEG-1) was prepared as previously described (6). Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for cell transfections according to the manufacturer's protocol. For each transfection, 50 nM miRNA mimics or siRNA were used, unless otherwise noted.
Cell proliferation assays
Huh7 or PLC/PRF/5 cells were plated in 96-well plates at a density of 3×103 cells per well and transfected with 50 nM miRNA mimics. In other words, complete growth medium with 3×103 cells was added to each well containing miRNA mimics-Lipofectamine 2000 complexes. Cell proliferation was determined using a Cell Counting Kit-8 (CCK-8; Promoter Biological Co., Ltd., Wuhan, China) at 0, 24, 48 and 72 h according to the manufacturer's recommendations. In each treatment group, at least three wells were measured for cell viability.
Cell apoptosis assays
Huh7 or PLC/PRF/5 cells were transiently transfected with miRNA-NC or miR-195 mimics and were harvested after 48 h by trypsinization. Double staining with fluorescein isothiocyanate (FITC)-annexin V and propidium iodide (PI) was carried out using an Annexin V-FITC/PI kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) according to the manufacturer's instructions. Cells were analyzed by flow cytometry (FACScan; BD Biosciences, Franklin Lakes, NJ, USA) equipped with CellQuest software version 3.3 (BD Biosciences). Cells were categorized as viable, dead, early apoptotic and late apoptotic, and the ratio of total apoptotic cells in the miR-195-transfected group relative to the total apoptotic cells in the miR-NC-transfected group was compared. Experiments were conducted in triplicate.
Analysis of clonogenicity in vitro
For the colony formation assay, 24 h after transfection, 1×102 transfected Huh7 cells were plated in each well of a fresh 24-well plate and maintained in DMEM containing 10% FBS for 2 weeks. Subsequently, the formed colonies were fixed with methanol and stained with 0.1% crystal violet in 20% methanol for 10 min.
For the anchorage-independent growth assay, transfected cells were trypsinized and suspended in complete medium plus 0.3% low-melting-temperature agarose (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). The mixture of cells and agar was then seeded onto a 0.5% agar base layer with complete medium at a density of 5×102 cells per well in 24-well plates. After 2 weeks, colonies containing >50 cells were counted. Triplicate wells were prepared for each group of transfectants and the experiment was repeated twice.
Target gene prediction for miR-195
Putative miR-195 target genes and their target miRNA-binding site seed regions were predicted using the target prediction programs TargetScan (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/microrna/home.do).
Western blot
Huh7 cells were harvested 48 h after transfection and protein lysates were prepared according to the manufacturer's recommendations. Cell lysates were prepared with 1 ml RIPA Lysis Buffer (P0013B; Beyotime Institute of Biotechnology, Haimen, China) with 10 µl phenylmethylsulfonyl fluoride (100 mM) (ST506; Beyotime Institute of Biotechnology). From each lysate, 30 µg proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and then electrophoretically transferred to polyvinylidene fluoride membranes. Following blocking with 5% nonfat dry milk for 1 h at room temperature, immunoblotting was performed at 4°C overnight with a diluted (1:2,000) polyclonal anti-AEG-1 antibody (#13860-1-AP; Proteintech, Wuhan, China), with a diluted (1:5,000) anti-GAPDH antibody (#10494-1-AP; Proteintech) used as an internal control. Subsequently, the membranes were incubated with secondary antibodies (1:4,000 dilution; #SA00001-2; Proteintech) for 1 h at room temperature. Band signals were visualized using an enhanced chemiluminescence kit (Pierce ECL Western Blotting Substrate; Thermo Fisher Scientific, Inc.).
Luciferase assays
A pMIR-REPORT miRNA Expression Reporter Vector System (Thermo Fisher Scientific, Inc.) was used to verify the precise target of miR-195. The wild-type sequences of the AEG-1 3′-UTR (wt-AEG-1) and those with mutated miR-195 target sites (mut-AEG-1) were synthesized and ligated between the SpeI and HindIII restriction sites of the pMIR-REPORT Luciferase vector to establish the pLUC-wt-AEG-1 and pLUC-mut-AEG-1 vectors. The sequences of oligonucleotides are described in Table III. In 96-well plates, Huh7 cells were transiently cotransfected with 100 ng of pLUC-wt-AEG-1 or pLUC-mut-AEG-1, 13 ng of internal transfection efficiency control pMIR-REPORT β-galactosidase plasmid, as well as miR-195 mimics or miR-NC to a 50 nM final concentration, using 0.3 µl Lipofectamine 2000. At 24 h after transfection, luciferase and β-galactosidase activities were measured using a Dual-Light System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Table III.
Sequences of 55-mer double-stranded oligonucleotides containing the predicted miRNA binding sites.
| Targeted gene | Sequence (5′-3′) |
|---|---|
| wt-AEG-1 | |
| Forward | CTAGTGAGCAGAAATTTGGAAGGCTATTCAGTGCTGCTTAGTGTAGCAGCTAATA |
| Reverse | AGCTTATTAGCTGCTACACTAAGCAGCACTGAATAGCCTTCCAAATTTCTGCTCA |
| mut-AEG-1 | |
| Forward | CTAGTGAGCAGAAATTTGGAAGGCTATTCATATTCTTGTAGTGTAGCAGCTAATA |
| Reverse | AGCTTATTAGCTGCTACACTACAAGAATATGAATAGCCTTCCAAATTTCTGCTCA |
Nucleotides predicted to pair with the corresponding seeding sequences of miR-195 are shown in bold and underlined. Italic letters indicate overhangs of restriction enzyme sites. AEG-1, astrocyte elevated gene 1; wt, wild type; mut, mutant type.
Lentivirus-based miR-195 overexpression
In order to elucidate the role of miR-195 in vivo, a recombinant lentivirus (LV-miR-195) was constructed to generate stable gain-of-function of miR-195 in hepatoma cells. The recombinant lentivirus LV-miR-195 was prepared as previously described (10). The negative control lentivirus was designated LV-miR-NC.
Tumorigenesis assay in nude mice
BALB/c athymic nude mice (male; age, 4–6 weeks; weight, 16–20 g) were purchased from Beijing HFK Bioscience Co., Ltd. (Beijing, China) and were bred in pathogen-free conditions. The mice were provided with sterile water and food, and were kept in the following environment: Room temperature, 26–28°C; relative humidity, 40–60%; and daily light cycle, 10 h light:14 h dark. All animal experiments were approved by the Tongji Medical College Institutional Animal Care and Use Committee. Nude mice were randomly divided into two groups (n=6 for the LV-miR-NC group, and n=6 for the LV-miR-195 group), and 6×106 cells in phosphate-buffered saline were inoculated subcutaneously into the flanks of the nude mice. From the 10th day after inoculation, the length (L) and width (W) of the tumors were measured with vernier calipers every 4 days. Tumor volume (V) was calculated with the formula V = (L × W2) × 0.5. Mice were sacrificed at the 30th day after inoculation.
Statistical analysis
Data are presented as the mean ± standard error from three separate experiments performed in triplicate, unless otherwise noted. Error bars indicate standard error. A Student's t-test was employed to evaluate the differences between groups, and P<0.05 was considered to indicate a statistically significant difference.
Results
miR-195 is downregulated in HCC cell lines and tissues
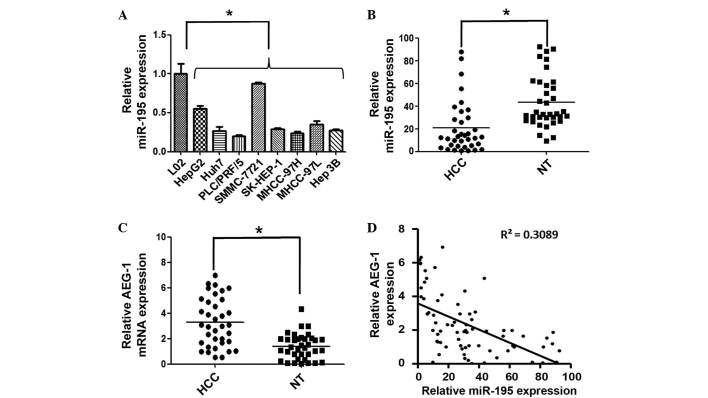
Previously, we identified a specific aberrant miRNA expression profile in HCC by comparison of miRNA expression profiles in cancerous hepatocytes with normal primary human hepatocytes, and identified miR-195 as one of the most downregulated miRNAs in HCC (6). In the present study, RT-qPCR was performed to further verify this result. First, the expression levels of miR-195 were detected in eight human hepatoma cell lines, including HepG2, Huh7, PLC/PRF/5, SMMC-7721, SK-HEP-1, MHCC-97H, MHCC-97L and Hep 3B, as well as in the normal human liver cell line L02. Generally, a marked reduction in the expression of miR-195 was observed in the cancer cell lines compared with the normal liver cell line (Fig. 1A). In addition, among the eight HCC cell lines, the expression levels of miR-195 in HepG2, Huh7, PLC/PRF/5, MHCC-97H, MHCC-97L, SK-HEP-1 and Hep 3B were markedly lower than that of SMMC-7721 (Fig. 1A). This result informs the selection of cell model for use in further research of miR-195 in HCC.
Figure 1.
miR-195 expression is downregulated and is inversely correlated with AEG-1 expression in HCC. (A) RT-qPCR was used to measure the expression levels of miR-195 in eight human liver cancer cell lines (HepG2, Huh7, PLC/PRF/5, SMMC-7721, SK-HEP-1, MHCC-97H, MHCC-97L and Hep 3B) and a normal human hepatic cell line (L02). U6 was used as an internal control and the mean miR-195 expression level in L02 liver cells was designated as 1. (B) The expression of miR-195 was detected in 36 paired HCC and tumor-adjacent NT samples by RT-qPCR. miR-195 was downregulated in HCC tissues compared with the adjacent NTs. The central horizontal line represents the mean value. (C) AEG-1 mRNA was detected in 36 paired HCC and NT samples by RT-qPCR. β-actin was used as internal control and the central horizontal line represents the mean value. (D) miR-195 expression is inversely correlated with AEG-1 expression (R2=0.308; P<0.01, Pearson's correlation). *P<0.05. miR-195, microRNA-195; AEG-1, astrocyte elevated gene 1; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NT, normal tissue.
The expression level of miR-195 was further examined in 36 paired HCC and adjacent noncancerous liver tissues. Compared with the corresponding noncancerous tissues, the mean relative expression level of miR-195 was significantly decreased in HCC samples (P<0.05; Fig. 1B). Additionally, the expression of miR-195 was inversely correlated with the expression of AEG-1, which is a potential target for miR-195 (Fig. 1C and D). These results indicated that the aberrant expression of miR-195 may be involved in the process of hepatocarcinogenesis and may provide insight into pathogenesis of HCC.
miR-195 decreases liver cancer cell growth and induces apoptosis
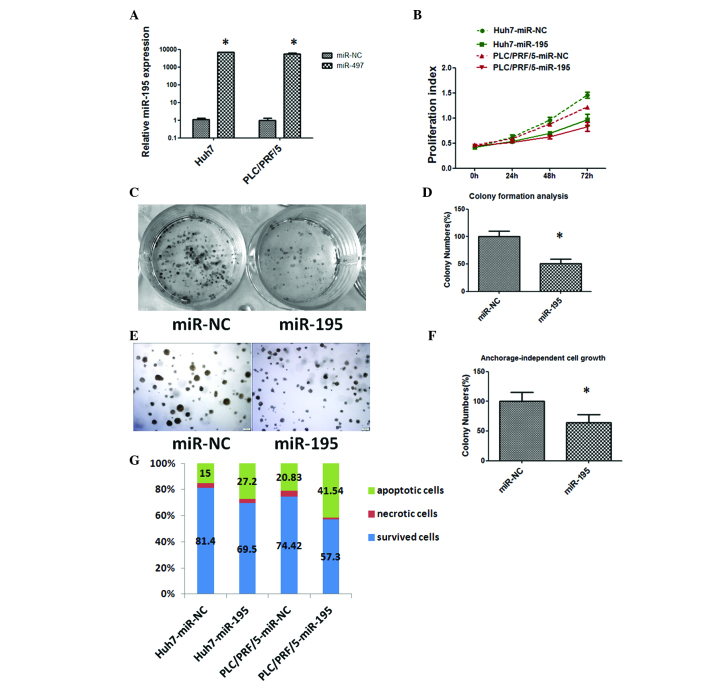
Sustained cell growth and proliferation is considered to contribute to cancer-associated mortality by disrupting the balance of growth promotion and growth limitation. To determine the function of miR-195 in HCC, a gain-of-function analysis was performed using miR-195 mimics or miRNA-NC transfectants. As shown in Fig. 2A, the increased expression of miR-195 was verified by RT-qPCR in Huh7 and PLC/PRF/5 cells transfected with miR-195 mimics.
Figure 2.
Tumor suppressive effects of miR-195 in hepatocellular carcinoma. (A) Relative expression of miR-195 in Huh7 and PLC/PRF/5 cells was detected by reverse transcription-quantitative polymerase chain reaction at 48 h following transfection of the cells with 50 nM miR-195 mimics or miR-NC. The average miRNA expression level from the miR-NC group was designated as 1. (B) Overexpression of miR-195 suppressed the proliferation of Huh7 and PLC/PRF/5 cells. (C) miR-195 overexpression decreased the colony-forming ability of Huh7 cells. (D) A histogram indicated that miR-195 inhibited colony formation. The percentage of colony numbers of the miR-NC group was designated as 100%. (E) Restoration of the expression of miR-195 inhibited the anchorage-independent cell growth ability of Huh7 cells. (F) A histogram indicated that miR-195 inhibited anchorage-independent cell growth. The percentage of colony numbers of the miR-NC group was designated as 100%. (G) miR-195 induced apoptosis. Apoptosis was detected by Annexin V/propidium iodide staining combined with flow cytometry in Huh7 and PLC/PRF/5 cells at 48 h subsequent to transfection. Apoptotic evaluation was determined by the percentage of apoptotic cells out of the total cell number. *P<0.05. miR-195, microRNA-195; miR-NC, negative control microRNA.
Sustained cell growth is a notable hallmark of cancer. The CCK-8 assay showed statistically significant inhibition of HCC cell proliferation in miR-195 mimic transfectants in comparison with miR-NC transfectants at 48 and 72 h after transfection (Fig. 2B). Subsequently, the capacity of the cells for colony formation and anchorage-independent growth was evaluated in Huh7 cells transfected with miR-195 mimics or miR-NC. As shown in Fig. 2C, D, E and F, miR-195 mimic-transfected cells formed markedly fewer and smaller colonies compared with miR-NC transfectants.
Evading apoptosis is another property of tumor cells that facilitates their limitless growth. Cell apoptosis in miR-195-transfected Huh7 and PLC/PRF/5 cells was assessed by flow cytometry, revealing that the fraction of apoptotic cells was significantly increased in miR-195 transfectants compared with that in miR-NC transfectants (P<0.05; Fig. 2G). These data indicate a growth-inhibitory role of miR-195 in HCC.
miR-195 directly targets AEG-1 and blocks the carcinogenic effect of AEG-1 in HCC cells
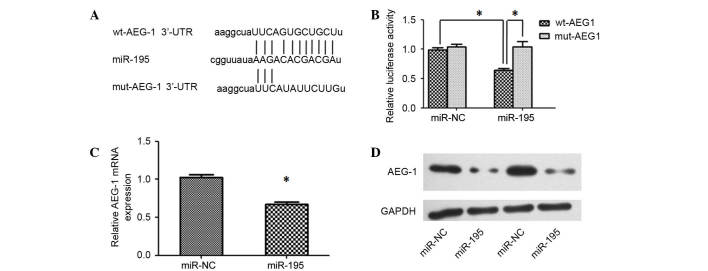
To unravel the mechanism underlying miR-195-mediated disruption of proliferation, positive regulators of carcinogenesis were searched using miRNA target prediction software (TargetScan and microRNA.org). Among the predicted target genes, AEG-1 was determined to be of interest as it is reported to have important roles in hepatocarcinogenesis (8). RT-qPCR was performed to analyze the expression levels of AEG-1 in HCC tissues and adjacent noncancerous liver tissues, and the results demonstrated that AEG-1 was upregulated in HCC tissues (Fig. 1C). Furthermore, an inverse correlation between the expression levels of miR-195 and AEG-1 was identified (Fig. 1D). A luciferase reporter analysis, which was performed to determine whether AEG-1 mRNA contained a target site for miR-195, revealed that the cotransfection of miR-195 significantly inhibited the activity of the luciferase reporter construct containing the wild-type 3′-UTR of AEG-1, whereas this effect was abrogated when the predicted 3′-UTR binding site was mutated (Fig. 3A and B). Additionally, the expression levels of AEG-1 mRNA and protein were significantly reduced in Huh7 cells transfected with miR-195 mimics compared with those transfected with miR-NC (Fig. 3C and D). These results indicate that the effect of miR-195 is partially due to its specific and direct interaction with the putative binding sites in the 3′-UTRs of AEG-1.
Figure 3.
miR-195 inhibits AEG-1 expression by targeting its 3′-UTR. (A) Putative binding sequence between the miR-195 and wild-type (wt-AEG-1) or mutant-type (mut-AEG-1) 3′-UTR of AEG-1. Mutations were generated in the complementary site that binds to the seed region of miR-195. (B) miR-195 inhibited the activity of a luciferase reporter that contained the wild-type 3′-UTR of AEG-1 (wt-AEG-1). miR-NC or miR-195 mimic was co-transfected with luciferase reporter plasmid that contained either the wild-type (wt) or mutant (mut) 3′-UTR of AEG-1. β-gal signal was used as an internal control to calibrate the differences in the transfection and harvest efficiencies. The luciferase activity of each sample was normalized to the β-gal signal. *P<0.05. (C) AEG-1 mRNA was downregulated in Huh7 cells transfected with miR-195 mimic. mRNA relative expression was measured by reverse transcription-quantitative polymerase chain reaction at 48 h after transfection of miR-NC or miR-195 mimic. The average mRNA expression from miR-NC group is designated as 1. *P<0.05 vs. miR-NC. (D) Expression of miR-195 reduced the protein levels of AEG-1 in Huh7 cells. HCC cells that were transfected with miR-NC or miR-195 mimic for 48 h were analyzed via western blot assay. GAPDH was used as an internal control. miR-195, microRNA-195; AEG-1, astrocyte elevated gene 1; 3′-UTR, 3′ untranslated region; wt-, wild type; mut-, mutant.
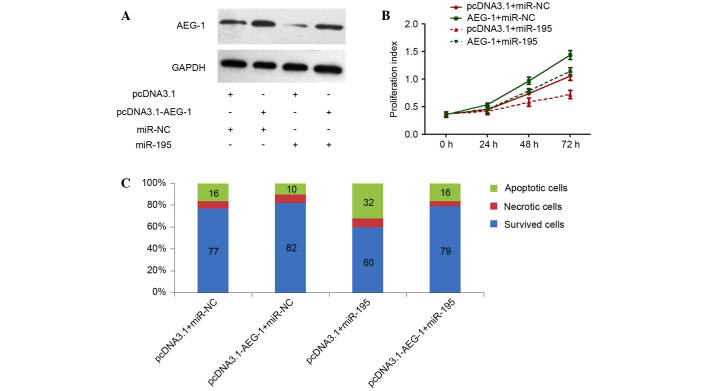
In our previous studies, we have silenced AEG-1 by RNAi to explore the role of this gene in HCC (6), demonstrating that specific knockdown of AEG-1 could inhibit cell proliferation and clonogenicity, and also induce apoptosis (6), which phenocopied the effects of miR-195 overexpression in HCC cells. In the present study, whether AEG-1 could counteract the proliferation-inhibiting and apoptosis-promoting functions of miR-195 was investigated. An AEG-1 recombinant plasmid was constructed, which encoded the entire AEG-1 coding sequence but lacked the 3′-UTR of AEG-1 mRNA. Cotransfection of the AEG-1 plasmid and miR-195 in Huh7 cells efficiently restored AEG-1 expression with enhanced expression of miR-195 (Fig. 4A), and partially reversed the antitumor effects induced by miR-195 (Fig. 4B and C). Taken together, these results suggest that miR-195 suppressed AEG-1 expression by binding directly to the 3′-UTR of AEG-1, and that the negative regulation of AEG-1 by miR-195 may contribute partially to the antitumor effects of miR-195 involved in HCC.
Figure 4.
Rescue of miR-195 ectopic expression effects by simultaneous overexpression of AEG-1. (A) Expression of AEG-1 protein in Huh7 cells. Proteins were extracted and analyzed by western blot at 48 h after co-transfection with miR-NC or miR-195 mimic and pcDNA3.1-AEG-1 or pcDNA3.1 in Huh7 cells. GAPDH was used as an internal control. (B) Cell proliferation detected in Huh7 cells at 0, 24, 48 and 72 h after transfection. (C) Cell apoptosis detected by Annexin-V/propidium iodide combined labeling flow cytometry in Huh7 cells at 48 h after transfection. Apoptotic evaluation was determined by the percentage of the apoptotic cell number out of the total cell number. miR-195, microRNA-195; AEG-1, astrocyte elevated gene 1; miR-NC, negative control microRNA.
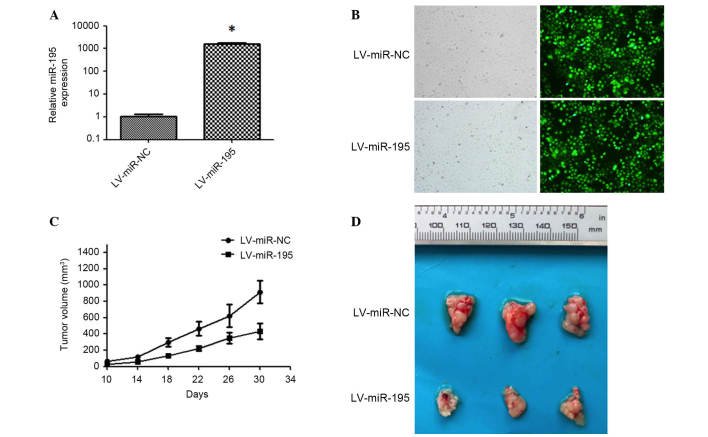
miR-195 inhibits tumor growth of hepatoma xenografts in nude mice
To investigate the potential antitumor activity of miR-195 in vivo, the recombinant lentiviral vector LV-miR-195 and its matched negative control, LV-miR-NC, were constructed and transduced into Huh7 cells to induce the expression of miR-195 (Fig. 5A and B). These lentiviral-transfected Huh7 cells were then injected subcutaneously into athymic nude male mice (n=6 in each control and experimental group). Tumor sizes in the two groups were measured every 4 days from the 10th day after inoculation, and the results indicated that tumor growth in the LV-miR-195-transfected group was significantly reduced (Fig. 5C). At 30 days after inoculation, all mice were sacrificed and the tumor volumes in the miR-195 group were 47.4% of that of the control (P<0.05; Fig. 5D). These results suggest that miR-195 inhibits tumorigenesis in vivo.
Figure 5.
Overexpression of miR-195 impairs the growth of hepatoma xenografts in vivo. (A) Relative expression of miR-195 in Huh7 cells transfected with lentivirus LV-miR-195 or LV-miR-NC was analyzed by reverse transcription-quantitative polymerase chain reaction. The average miRNA expression in LV-miR-NC group was designated as 1. *P<0.05. (B) Fluorescence microscopy was applied to detect the transfection efficiency of the lentivirus in Huh7 cells (right panels). The images in the left panels show the cells in the same field under normal white light. (C) Dynamic growth curves of xenografted tumors. Huh7 cells transfected with LV-miR-NC or LV-miR-195 were injected subcutaneously into nude mice. From the 10th day after injection, tumor sizes were measured every 4 days and tumor growth curves were obtained. Data are presented as the mean tumor volume ± standard error (n=6). (D) Mice were sacrificed and representative images of tumor volumes were captured at 30 days after injection. miR-195, microRNA-195; AEG-1, astrocyte elevated gene 1; miR-NC, negative control microRNA.
Discussion
Accumulating evidence has indicated that miRNAs are aberrantly expressed in numerous types of human cancer (11). Furthermore, miRNA expression signatures of HCC have been reported by a number of researchers (6,12,13). Our group also reported 37 differentially expressed miRNAs between cancerous hepatocytes and normal primary human hepatocytes based on expression profiles (6). miR-195 was one of the aberrantly expressed miRNAs that was significantly downregulated in HCC. In the current study, miR-195 was further confirmed to be significantly suppressed in HCC tissues. These results are in agreement with those previously reported (14,15). The detection of miR-195 expression in various cell lines in the present study indicated that miR-195 was also significantly suppressed in the 8 hepatoma cell lines compared with the normal liver cell line. These results provide reference for the selection of cell models to be used in further research of miR-195 in HCC.
The mechanism underlying miR-195 dysregulation in HCC remains elusive. In our previous study, the preliminary data and bioinformatics analysis suggested that DNA methylation may play an important and complex role in the regulation of miR-195 expression in HCC, which may provide insight into the mechanism underlying miR-195 dysregulation in HCC (16).
Sustaining proliferation and evading apoptosis are hallmarks of malignant tumors, and the aberrant expression of miRNAs has been implicated in the regulation of various cellular processes that are often dysregulated during the initiation and progression of HCC (2,11). For instance, the upregulation of miR-221 in HCC augments cell proliferation, colony formation and invasion, and increases the number of cells in S phase, as well as inhibiting cell apoptosis (17). miR-199a/b-3p was reported to be downregulated in HCC and restoration of its expression could target tumor-promoting P21 (RAC1)-activated kinase 4 (PAK4) to suppress HCC growth through inhibiting the PAK4/Raf/MEK/ERK pathway, in vitro and in vivo (4). In the present study, it was demonstrated that miR-195 could inhibit proliferation and colony-formation by HCC cells, as well as induce apoptosis in vitro. Furthermore, miR-195 suppressed tumorigenesis of Huh7 subcutaneous xenografts in a nude mouse tumor model. This research indicate that miR-195 functions as a tumor suppressor in HCC, in vivo and in vitro.
AEG-1 gene is always amplified in patients with HCC and serves key role in regulating hepatocarcinogenesis (8). The molecular mechanisms mediating the oncogenic properties of AEG-1 involve proliferation, apoptosis, metastasis, angiogenesis and chemoresistance. AEG-1 is a downstream gene of the oncogenic Ha-Ras pathway, being transcriptionally regulated by c-Myc upon Ha-Ras and phosphoinositide 3-kinase (PI3K) activation (8). Overexpression of AEG-1 activates the PI3K/Akt, nuclear factor-κB, and Wnt/β-catenin signaling pathways (8). Our data also documented that AEG-1 was overexpressed in HCC tissues compared with the corresponding normal liver tissues. Furthermore, an inverse correlation was observed between miR-195 and AEG-1 expression in HCC tissues for the first time. miR-195 was shown to suppress AEG-1 expression by directly binding to the 3′-UTR of AEG-1. To the best of our knowledge, AEG-1 as a direct target gene of miR-195 has not been reported previously. Furthermore, restoration of AEG-1 could partially reverse the antitumor effects induced by miR-195. Theoretically, downregulation of miR-195 should, at least partially, contribute to the increased expression of AEG-1 in HCC as an alternative mechanism.
The function of miR-195 has already been investigated in several distinct cancer types, such as non-small cell lung cancer (18,19), bladder cancer (20,21), breast cancer (22,23), colorectal cancer (24) and HCC (25–27). To date, 17 genes, including PCMT1, SRC-3, CCND1, CCNE1, CDC25A, CCND3, CDK4, CDK6, E2F3, BTRC, IKKα, TAB3, VEGF, VAV2, CDC42, LATS2 and BCL-w, have been identified as targets of miR-195 in HCC (14,15,25–30). miR-195 has also been implicated in various biological processes of HCC, including cell cycle (14,28), proliferation (25,29), apoptosis (26), drug resistance (30), angiogenesis (15) and metastasis (15,25). The reported targets of miR-195 and our observations suggest that miR-195 may regulate multiple signaling pathways, and that loss of miR-195 could lead to tumor progression in HCC.
In summary, the present study investigated the potential role of miR-195 in HCC tumorigenesis and found that miR-195 negatively regulates oncogenic AEG-1 and inhibits hepatoma cell proliferation and tumor growth in vitro and in vivo. These data suggest an important role of miR-195 in the molecular etiology of HCC and indicate a potential application of miR-195 in cancer therapy.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 81472832, 81302112 and 81372663).
References
- 1.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Hung CH, Chiu YC, Chen CH, Hu TH. MicroRNAs in hepatocellular carcinoma: Carcinogenesis, progression, and therapeutic target. Biomed Res Int. 2014;2014:486407. doi: 10.1155/2014/486407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Nakao K, Miyaaki H, Ichikawa T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J Gastroenterol. 2014;49:589–593. doi: 10.1007/s00535-014-0932-4. [DOI] [PubMed] [Google Scholar]
- 6.He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, Liu C, Kabar I, Schmidt HH, Zhang X, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590–598. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar D. AEG-1/MTDH/LYRIC in liver cancer. Adv Cancer Res. 2013;120:193–221. doi: 10.1016/B978-0-12-401676-7.00007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak and Schmittgen: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8:re3. doi: 10.1126/scisignal.2005825. [DOI] [PubMed] [Google Scholar]
- 12.Tao J, Ji J, Li X, Ding N, Wu H, Liu Y, Wang XW, Calvisi DF, Song G, Chen X. Distinct anti-oncogenic effect of various microRNAs in different mouse models of liver cancer. Oncotarget. 2015;6:6977–6988. doi: 10.18632/oncotarget.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek S, Cho KJ, Ju HL, Moon H, Choi SH, Chung SI, Park JY, Choi KH, do Kim Y, Ahn SH, et al. Analysis of miRNA expression patterns in human and mouse hepatocellular carcinoma cells. Hepatol Res. 2015;45:1331–1340. doi: 10.1111/hepr.12510. [DOI] [PubMed] [Google Scholar]
- 14.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Zhao N, Li S, Fang JH, Chen MX, Yang J, Jia WH, Yuan Y, Zhuang SM. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2 and CDC42. Hepatology. 2013;58:642–653. doi: 10.1002/hep.26373. [DOI] [PubMed] [Google Scholar]
- 16.He XX, Kuang SZ, Liao JZ, Xu CR, Chang Y, Wu YL, Gong J, Tian DA, Guo AY, Lin JS. The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol Biosyst. 2015;11:532–539. doi: 10.1039/C4MB00563E. [DOI] [PubMed] [Google Scholar]
- 17.He XX, Guo AY, Xu CR, Chang Y, Xiang GY, Gong J, Dan ZL, Tian DA, Liao JZ, Lin JS. Bioinformatics analysis identifies miR-221 as a core regulator in hepatocellular carcinoma and its silencing suppresses tumor properties. Oncol Rep. 2014;32:1200–1210. doi: 10.3892/or.2014.3306. [DOI] [PubMed] [Google Scholar]
- 18.Guo H, Li W, Zheng T, Liu Z. MiR-195 targets HDGF to inhibit proliferation and invasion of NSCLC cells. Tumour Biol. 2014;35:8861–8866. doi: 10.1007/s13277-014-2153-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Wang Y, Lan H, Li J. MiR-195 inhibits the growth and metastasis of NSCLC cells by targeting IGF1R. Tumour Biol. 2014;35:8765–8770. doi: 10.1007/s13277-014-2140-5. [DOI] [PubMed] [Google Scholar]
- 20.Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa M, Enokida H. The microRNA expression signature of bladder cancer by deep sequencing: The functional significance of the miR-195/497 cluster. PLoS One. 2014;9:e84311. doi: 10.1371/journal.pone.0084311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fei X, Qi M, Wu B, Song Y, Wang Y, Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012;586:392–397. doi: 10.1016/j.febslet.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian J, Weng Y. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep. 2013;30:877–889. doi: 10.3892/or.2013.2532. [DOI] [PubMed] [Google Scholar]
- 23.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Qian L, Li X, Yan J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony-formation and invasion through targeting CARMA3. Mol Med Rep. 2014;10:473–478. doi: 10.3892/mmr.2014.2178. [DOI] [PubMed] [Google Scholar]
- 25.Ding J, Huang S, Wang Y, Tian Q, Zha R, Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al. Genome-wide screening reveals that miR-195 targets the TNF-α/NF-κB pathway by down-regulating IκB kinase alpha and TAB3 in hepatocellular carcinoma. Hepatology. 2013;58:654–666. doi: 10.1002/hep.26378. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Yu J, Yin J, Xiang Q, Tang H, Lei X. MiR-195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Pharmazie. 2012;67:645–651. [PubMed] [Google Scholar]
- 27.Amer M, Elhefnawi M, El-Ahwany E, Awad AF, Gawad NA, Zada S, Tawab FM. Hsa-miR-195 targets PCMT1 in hepatocellular carcinoma that increases tumor life span. Tumour Biol. 2014;35:11301–11309. doi: 10.1007/s13277-014-2445-4. [DOI] [PubMed] [Google Scholar]
- 28.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang HL, Yu H, Ma X, Xu D, Lin GF, Ma DY, Jin JZ. MicroRNA-195 regulates steroid receptor coactivator-3 protein expression in hepatocellular carcinoma cells. Tumour Biol. 2014;34:6955–6960. doi: 10.1007/s13277-014-1933-x. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, Yin J, Yu J, Xiang Q, Liu Y, Tang S, Liao D, Zhu B, Zu X, Tang H, Lei X. MiRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol Rep. 2012;27:250–257. doi: 10.3892/or.2011.1472. [DOI] [PubMed] [Google Scholar]