Abstract
Accumulating evidence suggests tumor protein 53 (p53) promotes correct cellular differentiation. Thus, mutant TP53 may be more frequent in tumors with irregular differentiation. This study investigated whether TP53 mutations were more frequent in diffuse large B cell lymphoma (DLBCL) that lacked the B cell lineage marker CD19. Sixteen CD19 negative and 78 CD19 positive DLBCL were sequenced for TP53 mutations. Twenty nine tumors had TP53 mutations and were associated with poorer survival. Mutant TP53 was more frequent in CD19 negative lymphomas (81% versus 21%, p < 0.0001). Analysis of other B cell markers revealed a lack of paired box 5 (PAX5) in CD19 positive lymphomas with mutant TP53 (50%), which was more frequent compared to tumors with wild-type TP53 (15%, p = 0.002). In summary, DLBCL lacking CD19 or PAX5 expression were more likely to have mutant TP53, suggesting irregular B cell marker phenotypes are associated with TP53 mutation.
Introduction
Normal functioning of p53 is critical for preventing cancers. We know this because mice with defects in Trp53 are highly tumor prone; in humans inherited TP53 mutations, Li-Fraumeni Syndrome, is characterized by multiple tumor types, and about half of all human cancers contain TP53 mutations (www.iarc.p53.fr) [reviewed in ref. 1]. The nature of TP53 mutations may provide clues as to the molecular events associated with the development of particular types of cancer2.
p53 is a transcription factor that prevents cancers forming by responding to DNA damage and initiating signaling pathways that repair DNA, permanently arrest cell division, or those that induce apoptosis to permanently remove damaged cells [reviewed in ref. 1]. As well as preventing cancer, normal functioning of p53 is required for many biological processes including cell differentiation. In hematopoietic cells, the introduction of wild-type p53 into transformed mouse pre-B cells induced B cell maturation3 and mice deleted for the proline-rich domain (PRD) of p53 develop B cell lymphomas comprised of incorrectly differentiated (CD19 negative) B cells4.
Diffuse large B cell lymphoma (DLBCL) is the most common and aggressive type of non-Hodgkin lymphoma, with TP53 mutations present in approximately 20% of tumors5, 6. A poorer prognosis is associated with DLBCL with mutant p535–10. Of interest, 10–12% of DLBCL have lost the B cell lineage marker CD1911, 12 and are also associated with a poor prognosis5, 6, 12. Given this, and the data from the PRD mutant mouse, we were interested to determine if mutant p53 might be driving abnormal B cell differentiation that pre-disposes to malignancy. This study investigated the prevalence of TP53 mutations in DLBCL with irregular expression of B cell markers including those that were CD19 negative.
Results
TP53 mutations are enriched in CD19 negative DLBCL
Ninety-four DLBCL were included in the study. The patient demographics and clinical information are provided in Table 1. Sixteen DLBCL were CD19 negative based on a low expression of CD19 positive cells by flow cytometry (a clear visible shift in fluorescence consistent with reduced expression) based on the initial diagnostic assessment. A lack of CD19 positive cells in these tumors was confirmed using immunohistochemistry (Fig. 1a). The CD19 negative DLBCL were characterized further for other B cell lineage and lymphoma markers including CD20, PAX5, CD138, BCL2, BCL6, BCL10 and MUM1 using immunohistochemistry (Table 2). All CD19 negative lymphomas were positive for CD20, the majority was positive for PAX5, and two were positive for BCL2 and BCL6. A CD138 negative status for all tumors indicated that the CD19 negative cohort used were not plasma cell tumors13.
Table 1.
Patient demographics and clinical characteristics of included individuals with DLBCL.
| CD19 negative (n = 16)* | CD19 positive (n = 78)** | ||
|---|---|---|---|
| Gender | Female | 37% | 40% |
| Male | 63% | 60% | |
| Age | <60 | 67% | 67% |
| ≥60 | 33% | 33% | |
| Treatment | CHOP | 100% | 100% |
| R-CHOP | 31% | 38% | |
| PFS at 5 years | 14% | 34% | |
| OS at 5 years | 21% | 54% |
OS, overall survival; PFS, progression free survival; follow-up data available for 14 individuals (*) and 50 individuals (**).
Figure 1.
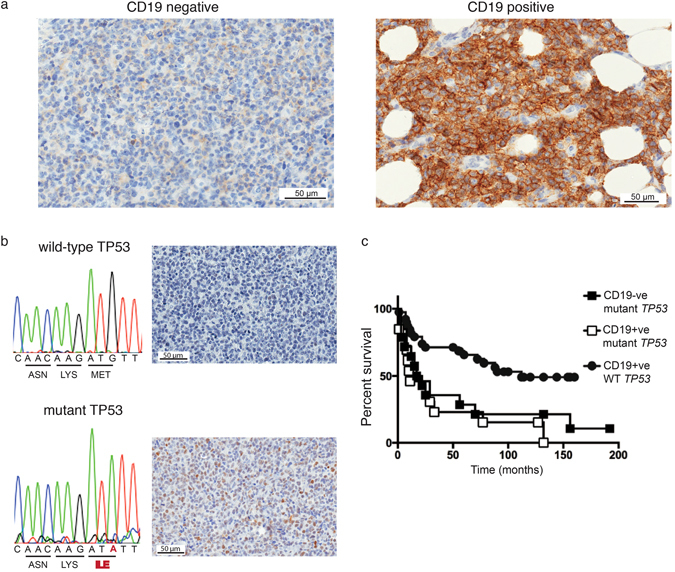
Tumor protein 53 mutations in CD19 negative DLBCL. (a) Left panel, CD19 negative lymphomas lacked CD19 staining using immunohistochemistry. Right panel, CD19 positive lymphomas showed strong CD19 staining. (b) Lymphomas were sequenced to identify TP53 mutations. Sequence analysis of a CD19 negative lymphoma with a M133I mutation is given in comparison to the wild-type TP53 sequence. Increased p53 by immunohistochemistry was used to predict lymphomas with TP53 mutations. The corresponding p53 immunohistochemistry staining for the lymphoma with the MI33I mutation and the lymphoma with wild-type TP53 are given. (c) Overall patient survival based on the CD19 and TP53 mutation statuses of lymphomas. TP53 mutations were identified by gene sequencing of tumor DNA and predicted by TP53 immunohistochemistry. WT, wild-type p53; TP53, tumor protein 53; −ve, negative; +ve, positive. p = 0.0003 (overall survival comparison between those with TP53 mutant and wild-type TP53 tumors).
Table 2.
Summary of the B-cell and lymphoma marker distribution amongst CD19 and TP53 subgroups.
| Marker | CD19 negative mutant TP53 (n = 13) | CD19 positive mutant TP53 (n = 16) | CD19 positive wild-type TP53 (n = 62) |
|---|---|---|---|
| BCL2 | 2/13 (15%) | 3/16 (19%) | 17/62 (27%) |
| BCL6 | 2/13 (15%) | 4/16 (25%) | 21/62 (34%) |
| BCL10 | 7/13 (54%) | 7/16 (44%) | 14/40 (35%) |
| CD20 | 13/13 (100%) | 16/16 (100%) | 61/62 (98%) |
| CD138 | 0/13 (0%) | 0/16 (0%) | 0/62 (0%) |
| MUM-1 | 2/13 (15%) | 6/16 (38%) | 16/40 (40%) |
| c-myc | 7/13 (54%) | 8/16 (50%) | 13/62 (21%) |
| PAX5 | 10/13 (77%) | 8/16 (50%) | 53/62 (85%) |
To determine if the irregular B-cell phenotype in CD19 negative lymphoma was associated with mutant TP53, all DLBCL were stained for p53 using immunohistochemistry and the TP53 gene was sequenced (Fig. 1b). We found that 13/16 (81%) CD19 negative lymphomas had mutations in TP53 whereas only 16/78 (21%) CD19 positive cases had TP53 mutations. Thus, a higher proportion of TP53 mutations occur in CD19 negative compared to CD19 positive lymphoma (p < 0.0001).
The lymphomas were further divided into 3 groups based on the CD19 status and the presence of TP53 mutations. Of interest, irrespective of the CD19 status, tumors with mutant TP53 had a poorer outcome compared to those with wild type TP53 (median survival of 17 and 112 months respectively, p = 0.0003, hazard ratio 2.9, 95% CI 1.7–6.5) (Fig. 1c).
Mutant TP53 is associated with poorer survival of DLBCL patients due in part to an association with increased function of the c-myc oncogene5, 7, 9. To determine if TP53 mutations were associated with increased c-myc expression in our cohort, all lymphomas were stained with an antibody to c-myc (Table 2)7. Consistent with the above, we found that c-myc positive tumors were present in 50% of CD19 positive lymphomas with TP53 mutations; 54% of CD19 negative lymphomas, but only 21% in tumors with wild-type TP53 (p = 0.008, TP53 mutant lymphoma (combined CD19 negative and positive cases) versus wild-type TP53 cases).
An analysis of BCL10 and MUM1 was performed to determine if CD19 negative lymphomas had a germinal center (GC) or a non-germinal center-like phenotype14. Seventy-two cases were stained for BCL10 and MUM1: all CD19 negative lymphoma, and 56/78 CD19 positive lymphoma. Fifty-four percent (7/13) of the CD19 negative lymphoma cohort with TP53 mutations (Table 2) was positive for BCL10 compared to 38% of CD19 positive lymphoma. Fifteen percent (n = 2/13) of the CD19 negative lymphoma cohort with TP53 mutations (Table 2) was positive MUM-1 compared to 39% of the CD19 positive lymphoma cohort. The different frequencies of BCL10 and MUM1 positive cases were not significant between CD19 negative and positive lymphoma.
Other B-cell irregularities are found in DLBCL with mutant TP53
To determine if TP53 mutant DLBCL had other aberrantly expressed lineage markers, CD20 and PAX5 immunohistochemistry results were evaluated in CD19 positive lymphomas with TP53 mutations in comparison to CD19 positive lymphomas with wild-type TP53 (Table 2). No evidence was found that CD20 was lost on CD19 positive lymphomas with mutant TP53; however, 50% (n = 8/16) were negative for PAX5 compared with only 15% (n = 9/62) of DLBCL with wild-type TP53 (p = 0.002). This result suggests aggressive lymphomas with mutant TP53 have other irregular B-cell marker phenotypes. No differences were found between these lymphomas for BCL2, BCL6, and CD138 markers.
CD19 negative lymphomas have an atypical distribution of TP53 mutations
The nature of TP53 mutations may provide clues as to the molecular events associated with the development of some cancer types2. Here we investigated the mutation spectrum in CD19 negative and CD19 positive DLBCL. Mutations detected were analyzed using the International Agency for Research on Cancer (IARC) database (http://www-p53.iarc.fr/). Overall, the mutation type was similar between the CD19 negative and CD19 positive cohorts with missense mutations being most frequent accounting for 85% and 100% of cases, respectively (Fig. 2). A nonsense mutation and a splice site mutation were identified in two CD19 negative DLBCL at Q52X and IVS4-2A > G, respectively.
Figure 2.
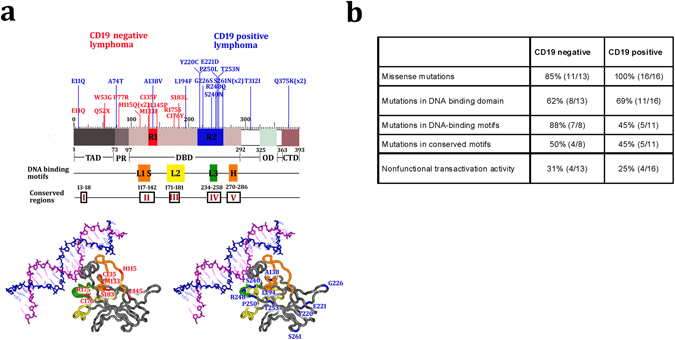
Atypical distribution of TP53 mutations in CD19 negative compared to CD19 positive B cell lymphomas. (a) Top panel: TP53 mutation distribution column plot in CD19 negative (red columns) and CD19 positive (blue columns) DLBCL and their relation to p53 protein structure. TP53 mutations in DBD regions 1 and 2 (R1 and R2) targeted for comparison using RNAseq TCGA data are boxed in red and blue, respectively. The DNA binding loops of p53: LSH, L2 and L3 are boxed in orange, yellow and green, respectively. The location of highly conserved regions of the DNA binding domain are marked (I to V). Numbers indicate amino acid position. TAD, transactivation domain; PRR, proline-rich region; DBD, DNA-binding (core) domain; OD, tetramerization domain; CTD, regulatory domain at the extreme C-terminus. Bottom panel: position of TP53 mutations within the 3D structure of p53 DNA-binding domain in CD19 negative (bottom left; highlighted in red) and CD19 positive (bottom right; highlighted in blue) DLBCL. Protein Data Bank code 1TSR file; chain B visualized by cn3d version 4.327. (b) Structural and IARC mutation information for TP53 mutants in CD19 negative and CD19 positive DLBCL (http://www-p53.iarc.fr/).
There was however, a striking difference in spatial distribution of TP53 mutations seen in CD19 negative compared to CD19 positive lymphomas. Notably, the TP53 mutations identified in CD19 negative lymphomas were mainly located within the N-terminus of p53 and in the proximal region of the central core DNA-binding domain (DBD) with exons 4–5 most often mutated (Fig. 2a). Of the mutations located in the DBD: 88% (7/8) were located in DNA-binding motifs (DBM), four of these mutations were located in Loop 1 (L1) of the Loop-sheet-helix/LSH motif that interacts with the DNA major groove, and three mutations were in Loop 2 (L2). No mutations were located in L3 (codons 237–250) that enhances the binding affinity of p53 with the DNA helix (Fig. 2a and b)15. Four mutations were localized to highly conserved areas: two mutations in area II (codons 117–142) and two mutations in area III (codons 171–181). In contrast, in the CD19 positive cohort the majority of mutations were located in the distal part of the DBD, exons 6–7 (Fig. 2a). Only 45% (5/11) of mutations were inside the DBM with three located in L3 (Fig. 2a and b).
A comparison of the loss of function predictions using the IARC database did not reveal any differences in regard to mutations identified in CD19 positive and CD19 negative cases (Fig. 2b; http://www-p53.iarc.fr)2.
Spatial distribution of TP53 mutations alters expression of differentiation genes
Given the differences in mutation spectrum between CD19 positive and negative lymphomas and a tendency towards altered lineage marker expression, we asked whether different mutations might affect a generalized alteration in expression of differentiation genes. To test this, RNAseq data for tumors from The Cancer Genome Atlas (TCGA) with mutations analogous to those found in the CD19 negative lymphomas (region (R) 1 highlighted in Fig. 2a) were compared with mutations found in CD19 positive lymphomas (R2 highlighted in Fig. 2a). We found that 429 genes were differentially regulated (fold change cut off ≥1.5 or ≤−1.5 and p ≤ 0.05). These differentially expressed genes enriched for 13 GO-biological processes (p < 0.05). Interestingly, one of these over represented GO-biological processes included in developmental processes (GO:0032502), which encompasses genes involved in cell differentiation. To verify the involvement of these differentially expressed genes in cell differentiation geneset enrichment analysis was performed. We found that there was a significant enrichment for genes involved in differentiation (geneSetTest p = 0.002).
Discussion
In this study, we showed TP53 mutations were present in the majority of DLBCL with an irregular CD19 B-cell marker phenotype. These results suggest aggressive lymphomas with an irregular B-cell marker phenotype are enriched for TP53 mutations. Based on gene expression differences TP53 mutations present in CD19 negative lymphomas were associated with differentiation processes that could contribute to irregular B cell lineage marker expression. In mice without the p53 proline domain irregular B-cell phenotypes develop with age suggesting early changes to p53 lead to irregular B-cells4. Although this evidence is consistent with wild-type p53 playing a role in aiding correct B cell differentiation, in the current study we do not know when the TP53 mutations occurred and thus were unable to determine if the TP53 mutations were directly responsible for the irregular B-cell phenotype.
Growing evidence suggests that the pattern of TP53 mutations varies amongst cancer types and may contribute to the tumor phenotype6. TP53 mutations in CD19 negative lymphomas were distributed in the N-terminus of p53 in the proximal part of the DBD, and were more likely to be in the DBM in L1 and L2 regions. Previous studies including those in DLBCL have emphasized that missense mutations in the DBMs have a worse prognosis compared to missense mutations outside these motifs6, 16. Loss of transactivation capacity of p53 has been reported as the main determinant of the poor prognostic value of TP53 in a number of tumors including those with inherited TP53 mutations2, 16, 17. In CD19 positive lymphomas TP53 mutations were distributed predominantly in the DBD distal to those in CD19 negative lymphomas and included mutations in L3. The location of mutations in CD19 positive lymphomas were more similar to that reported for TP53 mutations in cancer in general (http://www-p53.iarc.fr/).
A limitation of the current study is that with the restricted number of CD19 negative lymphomas other factors, such as the treatment received, were not factored into the survival analyses. However, other studies of lymphomas have associated TP53 mutations or loss of CD19 as factors associated with poorer patient survival5, 6, 12. The finding of this study, that CD19 negative lymphomas are enriched for TP53 mutations, suggests TP53 mutations may be the underlying factor associated with poorer patient outcome.
CD19 positive lymphomas with TP53 mutations may have other irregularities in the B-cell phenotype. CD19 positive lymphomas with mutant p53 were more likely to have lost PAX5 expression. Loss of PAX5 contributes to leukemia and lymphoma with haplo-insufficiency of PAX5 occurring in 32% of B-progenitor acute lymphoblastic leukemia18. Loss of PAX5 could be expected to create a more aggressive tumor. In vitro silencing of PAX5 in mantle cell lymphoma led to decreased TP53 expression and a more aggressive and drug resistant cell19. Mouse models lacking PAX5 in mature B-cells were prone to aggressive progenitor B-cell lymphomas20. CD19 is a downstream target of PAX5, so it is not immediately obvious how lymphomas can maintain CD19 expression without PAX5 function21–23. The analysis of PAX5 was limited to an immunohistochemistry analysis and although reduced there may be sufficient PAX5 present in the CD19 positive lymphomas classified as PAX5 negative to allow CD19 expression.
Overall our study demonstrates that TP53 mutations are enriched in DLBCL with irregular expression of CD19 and PAX5. These findings suggest that the association between loss of B cell marker expression and TP53 mutations may be relevant to those with relapsed disease following treatment with B cell targeted therapies. This has recently been shown in a case study, where a patient that experience a relapse post CD19 targeted therapy had CD19 negative disease at relapse24.
Materials and Methods
Case Selection
CD19 negative DLBCL (as determined by flow cytometry to have <10% positive cells as part of the initial diagnosis) cases were selected from biopsies taken between 1998–2015. Seventy-eight CD19 positive DLBCL were also selected from biopsies taken over the same period for comparison. Exclusion criteria included plasmablastic DLBCL and recurrent biopsies. Follow-up data was available for 64 individuals. This study was contacted with ethical approval from the New Zealand Health and Disability Ethics Committee and all methods were performed in accordance with the relevant guidelines and regulations. All study participants provided written and informed consent for study participation and acquisition of relevant clinical history.
Immunohistochemistry
For immunohistochemical staining 4 µm sections from paraffin embedded tissues were subjected to heat mediated epitope-retrieval, and DAB chromogenic detection methods. B cell lymphoma 2, 6, and 10 (BCL2, BCL6, and BCL10), CD19, CD20, melanoma associated antigen (mutated) 1 (MUM1), PAX5, p53, and v-myc avian myelocytomatosis viral oncogene homolog (MYC) staining was performed. The primary antibodies and criteria for determining positive and negative tumors are given in Supplementary Table S1. Positive cells were counted at 400x magnification with at least 10 fields selected. The percent positive cells were calculated from the total number of cells counted (at least 500) by two blinded examiners.
TP53 mutation analysis
Tumor DNA was extracted from paraffin embedded tissues using the NucleoSpin FFPE RNA/DNA kit (Macherey-Nagel, Duren, Germany) according to the manufacturer’s instructions with each extraction including 3–10, 10 µm sections. TP53 exons 1–11 including the intron-exon boundaries were amplified by PCR and sequenced by direct sequencing. Primer sequences are given in Supplementary Table S2.
Statistical analyses
The frequency of TP53 mutations amongst CD19 negative and CD19 positive lymphomas was compared using the Chi-square test. Kaplan Meier survival curves were compared between groups using the Log-rank test using SPSS version 22 software, with p values corrected for multiple comparisons using the Bonferroni correction. p < 0.05 was considered statistically significant.
Retrieval of mutation and RNAseq data from TCGA datasets and differential gene expression analysis
Tumors containing missense and nonsense TP53 mutations common to the position of the amino acids substituted in the CD19 negative and positive DLBCL respectively, were identified using cBioPortal (http://www.cbioportal.org/). Level 3 RNAseq data, processed using the RNAseq by Expectation-Maximization (RSEM) method25 and normalized within samples to a fixed upper quartile, was downloaded on October 20th 2015 (Supplementary Table S3). Detailed description of the processing protocol can be found in the TCGA open access FTP download directories (https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/). A list of differentially expressed genes in tumors with TP53 mutations in the DBD (highlighted as R1 in Fig. 2a) common to the amino acid positions substituted in CD19 negative DLBCL (n = 6) versus TP53 mutations in the DBD (highlighted as R2 in Fig. 2a) in CD19 positive DLBCL (n = 9) was generated using the limma-voom pipeline26. Nucleotide positions R175, C176, L194, Y220, R248 have been documented to be natural mutation hot-spots and hence were excluded from this analysis. Differentially expressed genes with a fold change of 1.5 and p value < 0.05 were used for pathway analyses. Pathway analyses were performed using PANTHER (www.pantherdb.org). A customized list of genes involved in differentiation of different cell lineages was downloaded from genesetdb (http://genesetdb.auckland.ac.nz/haeremai.html) (Supplementary Table S4). The genesetTest function from the “LIMMA” package (https://bioconductor.org/packages/release/bioc/html/limma.html) was used to analyze genes enriched within the customized differentiation geneset. All data analyses and visualizations were performed using the R statistical framework (https://www.r-project.org/).
Electronic supplementary material
Acknowledgements
Ms Sarah Bowie, Ms Janine Neill and Ms Amanda Fisher are thanked for their technical assistance on the project. Southern Community Laboratories are thanked for their assistance with the pathology material. The Health Research Council of New Zealand funded this project.
Author Contributions
M.K., I.R., R.E., and T.S. performed, analyzed, and designed the experiments related to the immunohistochemistry, sequencing, and p53 mutant data. S.M. performed, analyzed, and designed the bioinformatics analyses, N.H., B.S., A.R., M.K., and T.S. performed the histopathological analyses; N.H., B.S., A.R., M.B., A.B. designed the research study, contributed essential tools, and analysed the data. M.K., S.M., M.B., A.W. and T.S. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01800-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature reviews. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human mutation. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 3.Shaulsky G, Goldfinger N, Peled A, Rotter V. Involvement of wild-type p53 in pre-B-cell differentiation in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:8982–8986. doi: 10.1073/pnas.88.20.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slatter TL, et al. p53-mediated apoptosis prevents the accumulation of progenitor B cells and B-cell tumors. Cell death and differentiation. 2010;17:540–550. doi: 10.1038/cdd.2009.136. [DOI] [PubMed] [Google Scholar]
- 5.Xu-Monette ZY, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120:3986–3996. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young KH, et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood. 2008;112:3088–3098. doi: 10.1182/blood-2008-01-129783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, et al. p53 expression is a strong marker of inferior survival in de novo diffuse large B-cell lymphoma and may have enhanced negative effect with MYC coexpression: a single institutional clinicopathologic study. American journal of clinical pathology. 2014;141:593–604. doi: 10.1309/AJCPPHMZ6VHF0WQV. [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa A, et al. Mutations of the p53 gene as a prognostic factor in aggressive B-cell lymphoma. The New England journal of medicine. 1997;337:529–534. doi: 10.1056/NEJM199708213370804. [DOI] [PubMed] [Google Scholar]
- 9.Stefancikova L, et al. Prognostic impact of p53 aberrations for R-CHOP-treated patients with diffuse large B-cell lymphoma. International journal of oncology. 2011;39:1413–1420. doi: 10.3892/ijo.2011.1170. [DOI] [PubMed] [Google Scholar]
- 10.Leroy K, et al. p53 gene mutations are associated with poor survival in low and low-intermediate risk diffuse large B-cell lymphomas. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2002;13:1108–1115. doi: 10.1093/annonc/mdf185. [DOI] [PubMed] [Google Scholar]
- 11.Tedder TF. CD19: a promising B cell target for rheumatoid arthritis. Nature reviews. Rheumatology. 2009;5:572–577. doi: 10.1038/nrrheum.2009.184. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M, et al. Clinicopathologic significance of loss of CD19 expression in diffuse large B-cell lymphoma. International journal of hematology. 2007;85:41–48. doi: 10.1532/IJH97.06148. [DOI] [PubMed] [Google Scholar]
- 13.Uckun FM, et al. Developmental hierarchy during early human B-cell ontogeny after autologous bone marrow transplantation using autografts depleted of CD19 + B-cell precursors by an anti-CD19 pan-B-cell immunotoxin containing pokeweed antiviral protein. Blood. 1992;79:3369–3379. [PubMed] [Google Scholar]
- 14.Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England journal of medicine. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 15.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 16.Olivier M, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 17.Kato S, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 19.Teo AE, et al. Differential PAX5 levels promote malignant B-cell infiltration, progression and drug resistance, and predict a poor prognosis in MCL patients independent of CCND1. Leukemia. 2016;30:580–593. doi: 10.1038/leu.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 21.Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14:779–790. doi: 10.1016/S1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 22.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/MCB.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 24.Evans AG, et al. Evolution to plasmablastic lymphoma evades CD19-directed chimeric antigen receptor T cells. British journal of haematology. 2015;171:205–209. doi: 10.1111/bjh.13562. [DOI] [PubMed] [Google Scholar]
- 25.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law CW, Chen Y, Shi W, Smyth G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/S0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


