Abstract
Microglia play an important role in the pathology of CNS disorders, however, there remains significant uncertainty about the neuroprotective/degenerative role of these cells due to a lack of techniques to adequately assess their complex behaviour in response to injury. Advancing microscopy techniques, transgenic lines and well-characterized molecular markers, have made histological assessment of microglia populations more accessible. However, there is a distinct lack of tools to adequately extract information from these images to fully characterise microglia behaviour. This, combined with growing economic pressures and the ethical need to minimise the use of laboratory animals, led us to develop tools to maximise the amount of information obtained. This study describes a novel approach, combining image analysis with spatial statistical techniques. In addition to monitoring morphological parameters and global changes in microglia density, nearest neighbour distance, and regularity index, we used cluster analyses based on changes in soma size and roundness to yield novel insights into the behaviour of different microglia phenotypes in a murine optic nerve injury model. These methods should be considered a generic tool to quantitatively assess microglia activation, to profile phenotypic changes into microglia subpopulations, and to map spatial distributions in virtually every CNS region and disease state.
Subject terms: Cellular imaging, Neurodegeneration, Microglia, Retina
Introduction
Whilst small in size, microglia are of enormous importance for CNS health and repair. Fulfilling their neuroinflammatory role, these so-called ‘resident macrophages’ continuously probe the CNS environment for signs of distress, and migrate to sites of injury/infection/neurodegeneration where they recruit and activate additional cells, proliferate, phagocytose, clear debris and reorganize the CNS parenchyma. This microglial reactivity is a complex multistage activation process, which broadly encompasses their transformation from a ramified to an amoeboid morphology by first retracting the microglia processes and then extending dynamic protrusions, followed by cellular locomotion. Microglia dynamics are, however, sometimes very subtle, and go beyond their immune surveillance function. Indeed, microglia are believed to represent an important nexus between neurological and immunological activity in the CNS, and, for example, are also essential for synaptic homeostasis and regulation of neurotransmitters1–3.
Despite their central role in mounting a rapid, protective response against perturbations of CNS homeostasis, microglial activity has also been linked to various pathological conditions including neurodegenerative diseases. Indeed, while microglial reactivity may at first aid to the elimination of the source of distress and to the restoration of tissue homeostasis, persistent inflammatory stimuli (as present in e.g. Alzheimer’s, Parkinson’s diseases, amyotrophic lateral sclerosis) may ultimately overwhelm microglia and lead to a downward spiral of chronic inflammation that compromises neuronal survival4, 5. Altogether, in response to their ever-changing surroundings, microglia display a remarkable degree of phenotypic plasticity – both in terms of morphology, number and molecular markers – and may be skewed to either a neurotoxic or a neuroprotective role. Much remains to be investigated, however, in order to understand this dynamic process and to correlate microglia phenotypes and function.
Historically, microglia phenotypes have been described by categorizing cells based on their morphological features. In general, highly ramified cells were designated ‘resting’ microglia and amoeboid cells were called ‘activated’, with a range of intermediate activation states in between (e.g., ‘intermediately activated’, ‘bipolar’, ‘rod-like’, ‘hypertrophied’, ‘bushy’). No morphological classification standard exists, however, which complicates comparison of data from different studies, and prevents objective quantification of pathological status and therapeutic effects. Nevertheless, it is well-known that microglia morphological features and function are tightly coupled3, 6, and the development of unbiased and quantitative methods could therefore lead to improved insights into this form-function paradigm. Furthermore, microglia reactivity is a highly dynamic process, which is likely to be better described by a continuum of quantitative physiological measures – able to detect subtle changes in cell morphology and to capture the dynamics of microglia plasticity – rather than by a qualitative categorization3, 7. Over the last years, novel methodological approaches for adequate description of the reactive transformation of microglia have been emerging (e.g., refs 3, 8–12), yet correlating microglia morphology to function remains difficult and few easy-accessible user-friendly tools are presently available to adequately quantify morphological changes. In addition, microglia phenotypes appear to heterogeneous, both amongst CNS regions and within a single tissue, which emphasizes the need for comprehensive analysis of the entire microglia population.
In the current study, microglial reactivity was evaluated on whole-mounted mouse retinas, harvested four days after ONC. The ONC model has been extensively used to study retinal neurodegeneration, and the timing and pathological mechanisms leading to retinal ganglion cell death have been the subject of many studies, e.g. refs 13–16. Furthermore, a growing interest in the detailed course of microglial reactivity in this experimental glaucoma model has been emerging17–21. Aiming to address this recent focus, we designed an automated analysis algorithm that, based on Iba-1 (ionized calcium-binding adapter molecule 1) immunostaining of the retinal microglia, quantifies five parameters: cell density, nearest neighbour distance (NND) and regularity index, to describe microglia numbers and distribution; and cell soma size and roundness, to assess cell morphology. Together, in combination with a spatial statistics approach, this comprehensive set of measures allows us to extract greater information from whole-retinal histology than has previously been possible, to quantitatively describe the dynamic pathological state of microglia in the retina.
Results
Quantification of microglial activation by a novel comprehensive image analysis script
Retinal microglia were visualized by means of immunohistochemical staining for Iba-1, a commonly used macrophage/microglia-specific calcium binding protein and well-suited marker for morphological differentiation of ramified versus amoeboid microglia22. Automated detection and counting of Iba-1 immunopositive cells was validated by comparison to manual counts by three masked observers. Algorithm results correlated strongly with mean manual observations and subsequent Bland-Altman analysis23 revealed comparable inter-operator variability between manual observers and the algorithm described herein (Fig. 1, Table 1).
Figure 1.
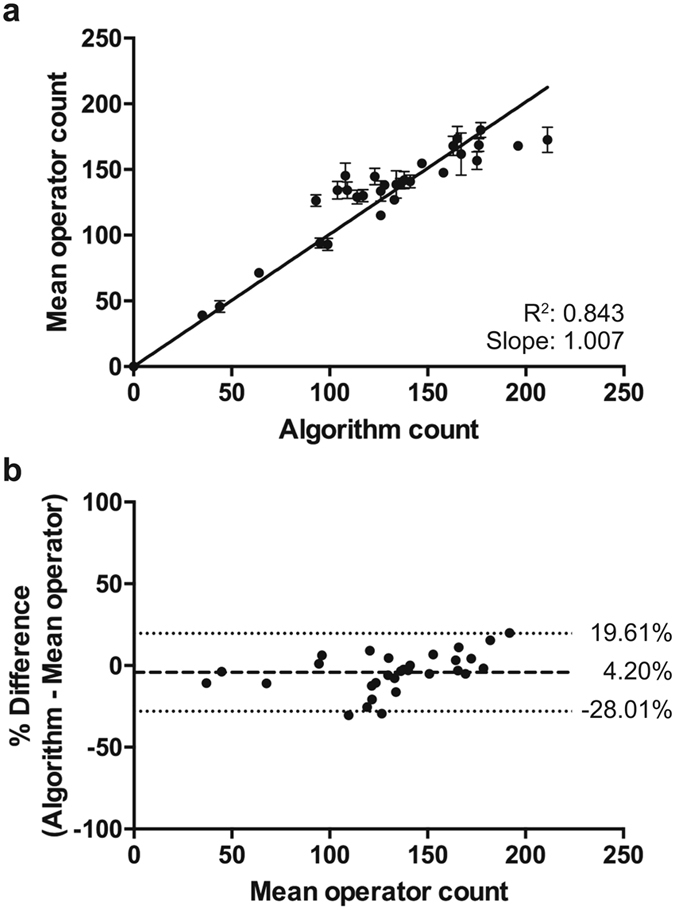
Algorithm optimization. (a) Linear fit of the mean operator counts versus the numbers of microglia detected by the automated algorithm, reveals a strong correlation (y = 1.007x; R2 = 0.843) and validates our algorithm. (b) This is confirmed by the Bland-Altman plot, which shows a bias of 4.20%. For comparison, bias for three observers was −8.35%, 4.33% and 3.50%. Data are depicted as mean ± 95% confidence intervals.
Table 1.
Correlation analysis of automated detection versus manual counting of Iba-1 immunopositive cells.
| Slope | R2 | Pearson R | p-value | Bias (%) | 95% CI Upper | 95% CI Lower | |
|---|---|---|---|---|---|---|---|
| Observer 1 | 1.072 | 0.920 | 0.9787 | <0.0001 | −8.347 | 6.626 | −23.32 |
| Observer 2 | 0.963 | 0.873 | 0.9479 | <0.0001 | 4.328 | 21.77 | −13.12 |
| Observer 3 | 0.956 | 0.885 | 0.9620 | <0.0001 | 3.499 | −14.09 | 21.09 |
| Algorithm | 1.007 | 0.843 | 0.9376 | <0.0001 | 4.203 | 19.61 | −28.01 |
Analysis of cell number and distribution to measure microglial reactivity
The first set of parameters that was analysed in our data set comprised cell density, NND and regularity index, and aimed to assess microglia numbers and distribution. First, upon comparison of microglia cell numbers in retinas of naive mice versus retinas that underwent ONC (4 days post injury), overall cell density was found to dramatically increase by 43% (185 ± 5 cells/mm2 versus 264 ± 9 cells/mm2, respectively; P < 0.005) (Fig. 2a). This was also reflected in the NND, which decreased in the ONC retinas (40.8 ± 1.0 μm versus 34.2 ± 0.7 μm; P < 0.005) (Fig. 2b). A detailed analysis of microglia density and NND in function of the distance from the ONH, furthermore revealed that microglia density tends to be higher in the central retina for all treatment groups (Fig. 2c,d). In addition, the left-shift in the cumulative density graphs (Fig. 2c, arrow) and the larger NND at increasing distance from the ONH (Fig. 2d, arrows), are suggestive of a microglia redistribution from the periphery to the centre in response to the ONC injury. Notably, this microglial response appeared to be almost identical in ONC and contralateral eyes at 4 days post ONC, and – given that the overall microglia density did not change in the contralateral eyes – proposes that at least part of the increased central cell density in the ONC retinas is the result of relocating resident microglia. These spatial organizations are illustrated by the pseudocolour maps, giving a graphical representation of microglia NND (Fig. 2e).
Figure 2.
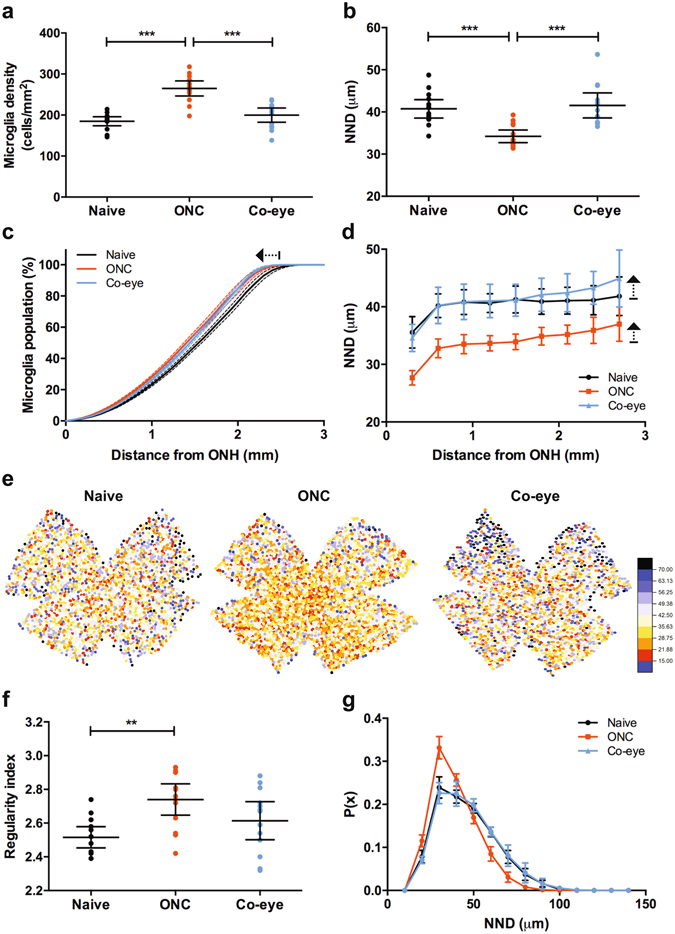
Microglia numbers and distribution. (a) A significant increase in overall microglia density is seen when comparing naive versus ONC retinas (4 days post injury). The microglia density of microglia is not significantly different between naive retinas and retinas from the contralateral eye of the ONC animals (one-way ANOVA with Tukey’s post hoc test). (b) Nearest neighbour distance (NDD) is significantly higher in ONC retinas, but not in the contralateral eye (one-way ANOVA with Tukey’s post hoc test). (c) Plotting cumulative microglia density, depicted as a percentage of the total microglia number, reveals that retinal microglia density is higher in the central regions of retina in ONC and contralateral eyes versus naive eyes. This can be observed as a left-shift of the ONC and Co-eye curves (arrow). (d) Distribution analysis of NND, in relation to the distance from the ONH, indicates that overall NND decreases in ONC eyes in comparison to naive eyes. However, NND disproportionally increases in the peripheral retina, a pattern that can also be discerned in the retinas of contralateral eyes (arrows). (e) Pseudocolor images of microglia NND/density in retinal whole-mounts. Red-to-blue pseudocolour representation of microglia distribution, ranging from small NND (15 μm, red tones) to large NND (70 μm, blue tones). (f) The regularity index of the microglia population increases upon ONC (one-way ANOVA with Tukey’s post hoc test). (g) A probability function of the NND points out that both the mean NND and NND standard deviation decrease after ONC, which explains the increase in regularity index. Data are depicted as mean ± 95% confidence intervals.
Second, the regularity index, which is defined as the ratio of the mean NND over the NND standard deviation and describes the regular spacing of cells, was seen to increase upon ONC (2.52 ± 0.03 versus 2.74 ± 0.04; P < 0.01) (Fig. 2e). Indeed, looking at the probability function of the NND, one can see that the mean NND shifts to the left (i.e., becomes smaller) and the distribution becomes more narrow (i.e., the standard deviation decreases) upon ONC (Fig. 2f). In other words, the NND in a population of activated microglia becomes smaller and more homogenous as the cells are more densely packed and their exclusion radii touch. The latter will be further explored in the following sections with Ripley’s K-statistics.
Intriguingly, analysis of microglia density, NND and regularity index in the contralateral eye of animals that underwent ONC, revealed that values tended to deviate from the naive eye. Although these differences were small and found not to be significant in this study, they are suggestive of a mild microglial activation in the contralateral eye in response to ONC (Fig. 2a–e).
Analysis of cell shape to measure microglial reactivity
A more detailed analysis focusing on the microglia cell somata, revealed two additional parameters that were significantly affected upon ONC. First, compared to naive mice, average cell body size increased (53.1 ± 1.3 μm2 versus 74.3 ± 2.7 μm2; P < 0.005) (Fig. 3a). Second, microglial reactivity at 4 days post ONC was also characterized by a reduction in the roundness of the cell body (0.181 ± 0.006 versus 0.160 ± 0.006 in naive and ONC retinas, respectively; P < 0.05) (Fig. 3b). Distribution analyses furthermore revealed a wider, slightly right-shifted distribution for soma size and left-shifted distribution for roundness (Fig. 3c,d). Together, these data suggest that the number of microglia with a small, round cell body decreased after ONC and that these cells adopted a bigger, more irregular soma shape. This fits the general classification of microglia morphology, which describes the surveying ‘resting’ cells as small, round cells with elaborate ramifications, and the ‘activated’ microglia as bigger, more amoeboid cells with retracted processes. Of note, no signs of activation were found when analysing cell soma size and roundness in the retina of the contralateral eye of the ONC animals (Fig. 3a–d).
Figure 3.
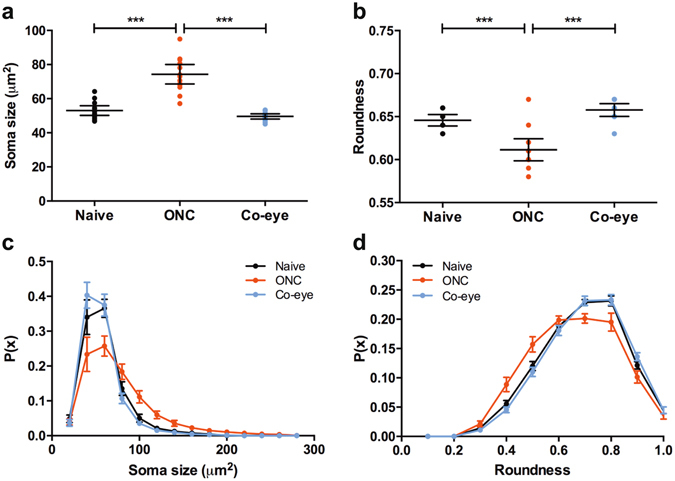
Microglia soma size and roundness. (a) Microglial activity following ONC is associated with an increase in average cell body area; while (b) roundness of the cell soma decreases (one-way ANOVA with Tukey’s post hoc test). These signs of microglial activation are not seen in the retina of the contralateral ONC eyes. (c) Distribution analysis of soma area shows a shift from small to larger cell body sizes after ONC; and (d) distribution analysis of circularity indicates that these cells are more likely to be more irregularly shaped (i.e., to have a lower roundness index). Data are depicted as mean ± 95% confidence intervals.
K-means clustering to gain a deeper insight into microglia subpopulations with different activation states
While the above measures provide an average value for microglia soma size and roundness for each retina, they fail to take into account the heterogeneity of microglia phenotypes within a single retina. Therefore, in order to define subpopulations of microglia with a ‘low’ versus ‘high’ activation status, we performed K-means clustering that divided the data into two main categories based on microglia soma size and roundness (Fig. 4a). We defined these clusters as ‘low activity’ and ‘high activity’ microglia, rather than ‘resting’ versus ‘activated’ cells, as we believe that this traditional classification fails to capture the diverse functions of microglia in both healthy and diseased CNS tissues. Microglia falling in the first category had small yet round somata and therefore resemble the typical morphological features of so-called resting microglia; microglia in the ‘high activity’ cluster displayed typical signs of microglial reactivity, i.e., an increased soma size and more irregular shape of their cell body, and thereby come close to the amoeboid-like morphology that is typically associated with activated microglia. This K-means clustering revealed that in a naive retina on average 65 ± 1% of the microglia fell into the ‘low activity’ and 35 ± 1% in the ‘high activity’ cluster (120 ± 3 versus 65 ± 4 cells/mm2, respectively). While the number of ‘low activity’ cells remained the same (126 ± 6 cells/mm2), the number of ‘high activity’ microglia doubled to 139 ± 8 cells/mm2 (P < 0.005) in the ONC eyes, resulting in a ratio of 49 ± 2% ‘low activity’ versus 51 ± 2% ‘high activity’ microglia (Fig. 4c,d). In the contralateral eye, no significant differences were seen, although the population of ‘low activity’ microglia tended to somewhat increase in comparison to naive retinas (from 65 ± 1% to 70 ± 1%) (Fig. 4c,d). Finally, graphical representations of the distribution of ‘low activity’ and ‘high activity’ microglia hint that there were no regional differences in microglia activation (Fig. 4c).
Figure 4.
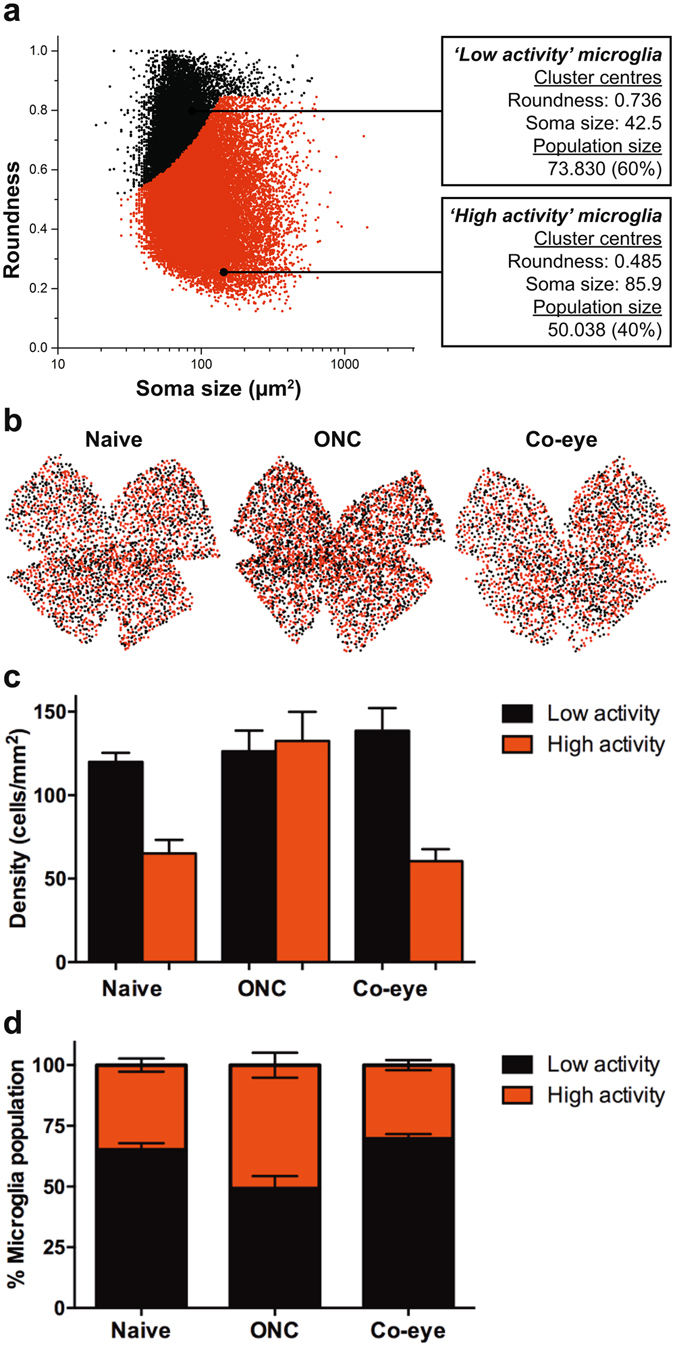
K-means clustering into ‘low activity’ and ‘high activity’ microglia subpopulations. (a) Two subpopulations of microglia were defined via K-means cluster analysis of a total of 123.868 cells (i.e., the sum of microglia from 14 naive retinas, 14 ONC retinas and 14 contralateral retinas), based on their roundness and soma size. (b) Images of representative retinal whole-mounts from naive, ONC and contralateral eyes, showing the distribution of ‘high activity’ (red) and ‘low activity’ (black) microglia. Each dot represents a microglia cell. (c) Absolute numbers of ‘low activity’ and ‘high activity’ microglia in the retina reveal that microgliosis after ONC merely presents as an increase in the number of ‘high activity’ microglia (one-way ANOVA with Tukey’s post hoc test). (d) This results in an increased proportion of ‘high activity’ versus ‘low activity’ microglia at day 4 post ONC, in comparison to naive retinas. No significant changes were observed in the contralateral eye. Data are depicted as mean ± 95% confidence intervals.
Ripley’s K-statistics and Dixon’s χ2-test to resolve microglia territories and spatial dynamics
In a spatial statistics analysis, using L(r) and H(r) derivatives of Ripley’s K-function, we investigated the topographical organization of retinal microglia. The negative values found for L(r) and H(r) indicate that microglia tend to disperse rather than cluster together in foci (Fig. 5a,b). Calculating the second minimum of the derivative of H(r) yielded the average size of the microglia territories under the different experimental conditions (Fig. 5b). In naive and ONC eyes, the average microglia domain radius was 58.6 ± 1.4 μm and 44.9 ± 0.6 μm, respectively, indicating that microglia activation coincides with a reduction in size of the microglia territories (P < 0.005) (Fig. 5c). Notably, in the contralateral eyes, microglia also somewhat decreased their domain radius (54.6 ± 1.5 μm), yet this effect was not significant. Running cross-K (K ij) and self-K functions (K ii and K jj) (where i stands for ‘low activity’ microglia and j for ‘high activity’ microglia) furthermore revealed that, in response to ONC, both ‘low activity’ and ‘high activity’ microglia reduce their domain radii to a similar extent (Fig. 5d). Domain radii are thus modulated independent of activation status.
Figure 5.
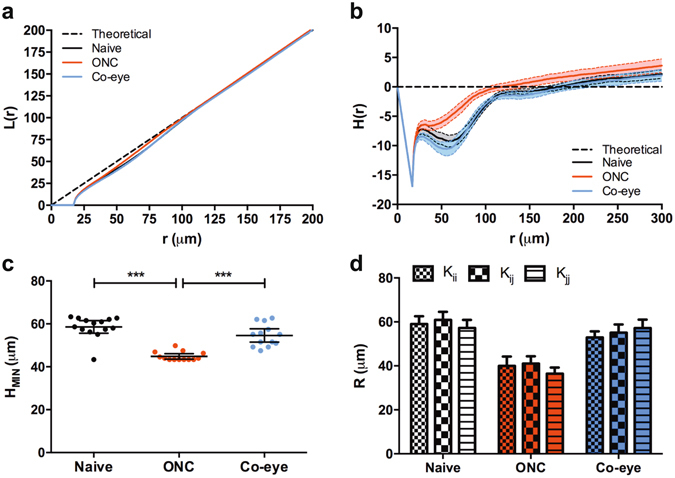
Ripley’s K-statistics, analysing spatial clustering of microglia subtypes. (a) For all treatment groups, L(r) and H(r) derivatives of Ripley’s K-function are negative for small values of r, indicating that retinal microglia have a dispersed topographical organisation. (b) Upon ONC, defined as the minimum of H(r), the domain radius of microglia decreases in size (one-way ANOVA with Tukey’s post hoc test). (c) Comparison of cross-K (K ij) and self-K functions (K ii and K jj) shows that ‘low activity’ and ‘high activity’ microglia subpopulations evenly decrease their domain radius after ONC injury (two-way ANOVA with Tukey’s post hoc test). Data are depicted as mean ± 95% confidence intervals.
Finally, Dixon’s χ2-test for testing spatial segregation was applied, focusing on the identity (i.e., ‘low activity’ or ‘high activity’) of the nearest neighbour of each microglia rather than the distance between them. A Dixon’s χ2-test was applied to the distribution of ‘low activity’ or ‘high activity’ microglia versus simulated random distributions, to examine whether microglia had a tendency to cluster beyond the exclusion radii identified above. A summary of the number of ‘low activity’ and ‘high activity’ microglia and the χ2-statistics, degrees of freedom, and p-values is provided in Table 2. The complete results are available as a Supplementary Table. Investigation of these test statistics revealed that microglia from both subpopulations have a mild tendency towards self-association. Upon ONC, however, a large increase in ‘high activity’ microglia clustering was observed. Indeed, across the 14 naive retinas included in this analysis, the aggregate C = 47.81 with 28 degrees of freedom yielded a p-value of 0.0112. Dixon’s χ2-test thus points to a tendency for ‘low activity’ and ‘high activity’ microglia to spatially segregate differently that what is expected under random labelling. The p-values of squared-summed z ii = 25.00 (P = 0.0346) and z jj = 26.55 (P = 0.0220) with 14 degrees of freedom indicated that both microglia subtypes tend to self-associate. At four days post ONC, the aggregate C = 143.57 with 28 degrees of freedom had a p-value < 0.000001, indicative of a very strong deviation from random labelling and from the naive condition. The square-summed z ii = 23.72 with 14 degrees of freedom and p-value = 0.0495 corresponded to our observations in the naive retina, suggesting that ‘low activity’ microglia did not behave differently in naive versus ONC retinas. In contrast, the squared summed z jj = 104.80 with 14 degrees of freedom and P < 000001 proposes that a large increase in ‘high activity’ microglia clustering is responsible for the increase in aggregate C versus naive retina. Spatial segregation of retinal microglia in the contralateral eye was similar to the naive retina. In summary, ‘high activity’ microglia preferentially self-associate, which could be explained by their cytokine-mediated cross-talk leading to activation of neighbouring cells and/or recruitment of additional microglia/macrophages.
Table 2.
Summary of Dixon’s χ2-test results across retinas from naive, ONC and contralateral eyes, each with N i ‘low activity’ microglia and N j ‘high activity’ microglia.
| Condition | Ni | Nj | Overall | Group-specific | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ΣC | df | p-value | df | P | df | p-value | |||||
| Naive | 25208 | 13713 | 47.81 | 28 | 0.0112 | 25.00 | 14 | 0.0346 | 26.55 | 14 | 0.0220 |
| ONC | 24242 | 25631 | 143.57 | 28 | 0.000001 | 23.72 | 14 | 0.0495 | 104.80 | 14 | 0.00001 |
| Co-eye | 24380 | 10694 | 49.33 | 26 | 0.0038 | 24.77 | 13 | 0.0248 | 15.09 | 13 | 0.3017 |
Discussion
Given that changes in microglia activation can be elucidated from changes in population size and morphology, independent of any molecular markers of activation, quantitative analysis these parameters can be used to assess the pathologic state of a given tissue. In this study, we exemplified this approach by describing microglia activation in a mouse ONC model, based on a five-parameter morphological analysis. Initially, we performed a global analysis of these activity markers in the entire retina and calculated average values, using traditional parametric statistical methods. Next, non-parametric cluster analysis and spatial segregation statistics were applied to this data set, in order to investigate the heterogeneity of microglia subpopulations and their spatial organization within the retina.
A comprehensive view on retinal microglia activation following optic nerve injury
Together, the results from this study can be integrated in order to model how microglia number, distribution, and morphology change in relation to microglia activation (Fig. 6). This shows that, at four days post ONC, microglial activation in the retina leads to, first, microglia proliferation/macrophage infiltration, which is visible as an increase in the density of Iba-1+ cells; and, second, to a more closely packed yet regularly spaced organization, as evidenced by the measures of nearest neighbour distance and regularity index. More in-depth analysis of microglia topographic organization via Ripley’s K-statistics confirms that their exclusion radius reduces upon ONC. This plasticity of microglia territories appears to be conserved in all treatment groups, suggesting that this may constitute an intrinsic cell-cell interaction mechanism that ensures efficient microglia surveillance covering the entire retina. As the radii of these territories were found to be similar for ‘low activity’ and ‘high activity’ microglia, this phenomenon appears to be independent from microglia size/morphology (i.e., the territory covered by microglia processes), but may instead be the result of chemotactic signalling24. In addition, microglia activation is associated with an increase in cell body size and a decrease in its roundness, reflecting the morphological transition from a ramified ‘resting’ to an activated amoeboid-like phenotype. Based on these two measures, we performed a K-means clustering to segment microglia populations into two subtypes, i.e., ‘high activity’ and ‘low activity’ microglia. Although this clustering is somewhat artificial, by categorizing microglia into either of the two limits of their spectra of phenotypes, it can be used to gain more insight into the diverging behaviour of ‘high activity’ and ‘low activity’ microglia, provided that the limitations of this approach are considered. Indeed, re-evaluation of microglia density for each of the clusters pointed out that the increase in microglia density after ONC is merely an increase in the number of ‘high activity’ microglia. Finally, Dixon’s χ2 spatial statistics disclosed that these ‘high activity’ microglia are more often surrounded by other ‘high activity’ microglia, which might be a reflection of their cell-cell communication that is designed to mount a self-propagating inflammatory response (Fig. 6).
Figure 6.
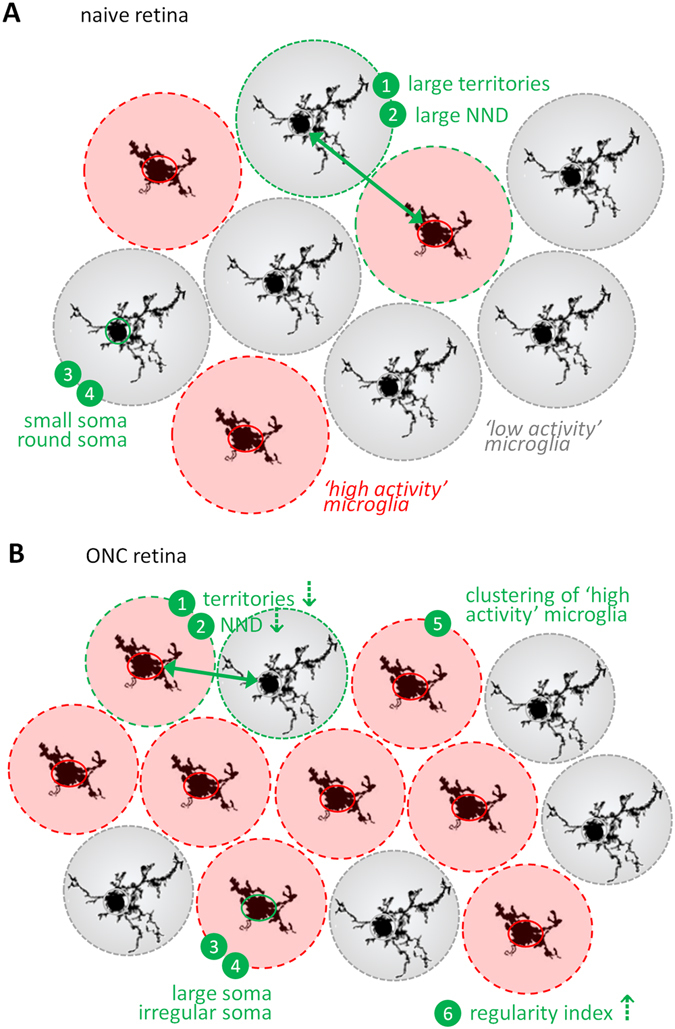
Summary figure integrating all findings from this study. (a) In a naive retina, microglia typically have small somata and large territories, corresponding to a ‘low activity’ status (grey). They are regularly interspersed with a minority of ‘high activity’ cells (red). (b) At day 4 post ONC, microglia density has increased and the majority of the microglia has adopted a ‘high activity’ phenotype, characterized by a reduction in their territory, enlargement of their cell body and a more irregular cell shape. Furthermore, although ‘low activity’ and ‘high activity’ microglia are regularly spaced, ‘high activity’ microglia tend to self-associate.
Overall, these findings agree with other studies of microglial activity in mouse ONC models of glaucomatous retinal neurodegeneration, which revealed proliferation of microglia taking place within the first week post ONC, as well as shape simplification and enlargement of their soma size20, 21. Despite following a different time course, identical changes in microglia appearance are also seen in other experimental rodent glaucoma models17–19, 21, 25–30. Notably, although not statistically significant, our findings hinted towards a low-grade immune response in the contralateral eyes, thereby corroborating previous findings in rodent glaucoma models30–32.
Microscopic spatial statistics: a novel, generic approach to study microglia dynamics
The major strength of our approach, wherein its innovation lies in comparison to the traditional morphological categorization of microglia, is that it allows for the detection of changes in cell morphology in a very large number of cells. Indeed, rather than focusing on a detailed, manual/qualitative/semi-quantitative analysis of a limited number of cells in a small region of interest, we performed a population analysis including all microglia in the retina. As such, and by focusing on cell soma shape and size rather than on skeleton analysis, we choose high-throughput and moderate computing over detailed, single-cell analysis and avoid the need for high-resolution, high-magnification images. This overall approach is in contrast to fractal and Sholl analyses, the two most commonly used alternative analysis methods3, 10, 33, 34. Our results prove that this strategy allows to gain advanced insights into microglia activation dynamics, especially by combining conventional statistics on population averages with clustering analyses of these large data sets. Because of the very large sample sizes, combined with K-means, Ripley’s K-statistics and Dixon’s χ2 spatial statistics, our method is able to capture the heterogeneous morphological transformations that go with microglia activation dynamics and may – in a future approach – aid to link the continuum of these morphological subpopulations to physiological differences. Indeed, as was also recently suggested by Verdonck et al.12 – who used a similar K-means approach to analyse microgliosis in the brain following endotoxemia – expanding the number of parameters in the clustering analysis (e.g., additional morphological features, gene expression signatures,…) could refine the classification of microglia subtypes and lead to a better understanding of morphology-function relations. Furthermore, microscopic spatial statistical approaches can be used to shed light on how the spatiotemporal distribution of microglia subtypes changes during disease progression by interacting with themselves and other cell types.
To conclude, the spatial statistics approach described herein addresses some of the current needs of modern biological research, by providing a means to fully explore the quantity and complexity of information contained in microscopic images35, 36. Our method is efficient, objective, and sensitive, and helps to reveal subtleties in microglia behaviour that are otherwise undetectable to the human eye. Importantly, the workflow described here comes with very few prerequisites and can therefore be considered a generic approach applicable to the entire CNS. Virtually any tissue of any species in any pathophysiological state can be assessed, as long as microglia are labelled with a sufficient signal-to-noise ratio. The current approach may thus also be applied to tissues stained with alternative microglia/macrophage markers (e.g. CD11b, isolection B4, F4/80) or with transgenically labelled cells in reporter lines (e.g. CX3CR1-GFP mice, Tg(coro1a:eGFP; lyz:Dsred) zebrafish)17, 22, 37, and aid to the maximization of information extracted from any study looking into microglia activation. As such, besides dramatically improving the quality of the data, with this microscopic spatial statistics approach we also react to the increasing scrutiny of the scientific, ethical and economic use of procedures involving animals.
Methods
Animals
All studies were conducted in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the European Communities Council Directive of 22 September 2010 (2010/63/EU) and the Belgian legislation (KB of 29 May 2013), and were approved by the KU Leuven institutional ethical committee (P025-2013). Wild type mice (C57Bl6/J; Charles River Laboratories), aged 2 to 4 months, were kept under a 12/12 h light-dark cycle and had ad libitum access to food and water.
Surgical procedures
Intraorbital optic nerve crush (ONC) was performed as described15 in 14 animals. Briefly, a temporal incision was made in the conjunctiva and the exposed optic nerve was then crushed 1 mm from the globe with a cross-action forceps (no. 11262-30, Fine Science Tools) for 5 seconds. Funduscopy was performed before and after the procedure to assess integrity of retinal perfusion. Animals were sacrificed at 4 days post ONC.
All surgical procedures were performed under general anaesthesia with xylazine (i.p. 10 mg/kg body weight, XYL-M) and ketamine (i.p. 70 mg/kg body weight, Nimatek). After the procedure, an antibiotic ointment (tobramycin 3 mg/g, Tobrex) was applied to avoid infection and corneal desiccation. Naive eyes from a separate cohort of untreated animals served as controls (N = 14).
Immunohistochemistry for Iba-1 on retinal whole-mounts
In order to label microglia (and potential macrophages infiltration from the circulation), an Iba-1 immunostaining was performed on retinal whole-mounts30. Mice were deeply anaesthetized (i.p. 30 mg/kg sodium pentobarbital, Nembutal) and sacrificed by cervical dislocation. Eyes were dissected and fixed for 1 hour in 4% PFA. Next, retinas were whole-mounted and again fixed for 1 hour in 4% PFA. Whole-mounted retinas were frozen for 15 minutes at −80 °C and rabbit anti-Iba-1 (Wako Chemicals, #019-19741) (1:1000), diluted in 10 mM phosphate-buffered saline (PBS) containing 2% Triton X-100 and 2% goat pre-immune serum, was applied overnight. The next day, a secondary goat anti-rabbit IgG antibody conjugated to an Alexa fluorophore-488 (Life Technologies) (1:200) was applied for 2 hours. Retinal whole-mounts were rinsed with PBS with 0.5% Triton X-100 in between steps, and mounted using mowiol mounting medium (10% mowiol 4–88, 40% glycerol, 0.1% 1,4-diazabicyclo-[2,2,2]-octane in 0.2 M Tris-HCl [pH 8.5]). Mosaic z-stack images (step size 3 μm; 20–30 stacks per image, comprising the retinal nerve fibre layer till the photoreceptor layer; 20x objective; 1 pixel = 1.24 μm) of the entire whole-mount were taken with a laser confocal scanning microscope (FV1000, Olympus), controlled with FluoViewer 4.2 software (Olympus), and a maximum intensity projection was made for further analysis.
Automatic quantification of Iba-1 labelled microglia in retinal whole-mounts
(Step 1) Image analysis was performed using Fiji (ImageJ, v1.51 h, 64 bit Windows) software38 with MorphoLibJ (v1.3.1) integrated library and plugin39. Quantification of Iba-1 labelled microglia in retinal whole-mounts was achieved using a multi-step algorithm. First, segmentation of Iba-1 labelled microglia was achieved by applying a grey scale attribute opening filter (area minimum: 25 pixels; connectivity: 8) to an 8-bit maximum projection of the retinal whole-mount. An opening morphological filter (1-pixel radius octagon) was then used effectively to separate microglia soma from processes, before a maximum entropy threshold was used to segment microglia soma from image background.
(Step 2) Quantification of microglia soma was completed using the Fiji (ImageJ) Analyse Particles function with a particle size threshold of 10 pixels, to exclude small pixel noise and extract information regarding microglial soma area, centre of mass and roundness (equation 1).
| 1 |
where A is the area of the microglia soma and M is the length of the major axis, derived from the longest axis of an ellipse fit to each microglia soma. In order to plot the above measures as a function of the distance from the ONH, the Euclidean distance of each microglia centroid, relative to the centre of the optic nerve head (ONH), was determined using a technique previously described for retinal ganglion cells (equation 2)40.
| 2 |
where d is the microglia Euclidian distance, (a, b) is the coordinate centre of the ONH (determined manually) and (x, y) is the centroid of each microglia within a retina. Results were presented as percentage frequency distributions (bin size: 50 µm) to allow averaging of multiple retinas and comparisons between treatment groups.
(Step 3) Nearest neighbour distance (NND) was determined using a script for Fiji (ImageJ) developed by Y. Mao41 and only particles with a NND < 14 pixels were considered to be microglia, in order to exclude large process artefacts from subsequent analysis. Regularity index (RI) was calculated using equation 3:
| 3 |
where XNND is the average NND of a population and δNND is the standard deviation of that population42.
(Step 4) Whole-mount retinal area was determined as described previously40. Briefly, a low intensity threshold (0–5) was applied to each 8-bit image by operators masked to treatment groups. The pixel area of this selection was measured using Fiji (ImageJ) and converted to mm2, and from this selection the global microglia density was determined by dividing the total microglia population by total retinal area. The boundary of each retinal area were exported as a list of xy-coordinates and used in conjunction with microglia centroid coordinates to generate spatial point processes with retinal area boundaries.
A detailed description of the microglia segmentation process, as well as a link to the Fiji source code, is included as a supplementary file. The Fiji source code is also publicly available via http://bio.kuleuven.be/df/lm/research/methods. The data sets generated and analysed during the current study are available from the corresponding author on reasonable request.
Parametric statistical analyses
For parametric statistics, normal distribution and parallel equal variance between groups was tested via D’Agostino-Pearson (omnibus K2) and Brown-Forsythe tests, respectively. A significance level of α < 0.05 was considered significant (*P < 0.05, **P < 0.01, ***P < 0.005). All data are described as mean ± SEM in the text. GraphPad Prism 5 (GraphPad Software) was used for all analyses, including Bland-Altman plots.
K-means clustering and colour mapping
Soma area and roundness data for each microglia identified in this study were pooled before normalization with 90% winsorizing of each population to remove extreme values43. K-means clustering, using the protocol described by Hartigan and Wong44, was determined using the K-means function in the R stats software package (v3.3.1)45, with two centres corresponding to ‘low’ and ‘high’ activity microglia populations. After ascribing clusters, microglia populations were separated by retina, and colour maps of microglia centroids (x, y coordinates) plotted with ‘low’ activity microglia were identified as black and ‘high’ activity microglia in red.
Ripley’s K- and L-functions
Microglia centroids from each retina were used in conjunction with ‘low’ or ‘high’ activity designations to create spatial point processes bounded by retinal area. To determine the extent of microglia distribution deviation from spatial homogeneity, Ripley’s K-functions46 were applied using the Kest function from the R spatstat software package47, to compare microglia spatial point patterns and assess deviation from compete spatial randomness. Initially, K-functions for unmarked spatial point processes (equation 4) were drawn as previously described48.
| 4 |
where N is the number of cells within distance r of another point, λ is the average cell density per unit area and P i is the ith cell and the sum taken over n cells.
Compete spatial randomness was estimated using a random Poisson distribution K(r) = πr 2 where r > 0. Deviations from this estimation are indicative of incidence of clustering or dispersion. To facilitate data analysis, the variance stabilized Ripley’s K-function L(r) was calculated using the protocol proposed by Besag49 (equation 5).
| 5 |
Using this transformation, under compete spatial randomness, r is linear and L(r) > r is indicative of clustering and L(r) < r is indicative of dispersal. L(r) can be further normalized to produce the H-function50, such that the expected value of r under compete spatial randomness is zero (equation 6).
| 6 |
This transformation is useful for the analysis of microglia spatial point processes. Using an adaptation of the work of Kiskowski et al.48, the radii of microglia territories (exclusion radii) can be estimated as the second minima – as microglia distribution is a dispersal rather than clustering process – of H(r), excluding the first minima that are an artefact of the algorithms NND exclusion radii. To determine whether the exclusion radii of ‘low activity’ (Kii), ‘high activity’ (Kjj) and intergroup (Kij) microglia populations changed upon ONC injury, changes in the spatial behaviour of ‘high’ and ‘low activity’ microglia populations across marked point processes were assessed, using the self-K and cross-K functions of the R spatstat software package for each retina.
Dixon’s χ2-test
As microglia population clustering could occur independent of the existence of exclusion radii previously identified, a Dixon χ2-test was employed to evaluate spatial segregation of ‘low activity’ and ‘high activity’ microglia51. Using the dixon software package in R52, nearest neighbour contingency tables were constructed for microglia populations for each retina and results summed across multiple retina (Naive, ONC and Co-eye treatment groups) to produce a process-wide test statistics. Deviation from random distribution of microglia populations was assessed using an overall test of segregation between (C) and within (z ii and z jj) groups, compared to results expected under random labelling, according to the methods described by Dixon et al.51.
Electronic supplementary material
Acknowledgements
The authors want to thank Marijke Christiaens and Véronique Brouwers for their excellent technical assistance. This research was supported by the KU Leuven Research Council (BOF-OT/10/033), the Hercules Foundation (AKUL-09-038), and the Research Foundation Flanders (G.05311.10). BMD and MFC were supported by the Wellcome Trust Healthcare Innovation Challenge fund and Medical Research Council confidence in concept award; MSN and LDG are postdoctoral research fellows of the Research Foundation Flanders.
Author Contributions
B.M.D. designed the study, performed the analyses and interpreted the data; M.S.N. performed the experiments; F.M.C. and L.M. designed the study and interpreted the data; L.D.G. designed the study, interpreted the data and wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01747-8
References
- 1.Nayak D, Roth TL, McGavern DB. Microglia Development and Function. Annl Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wake H, Moorhouse AJ, Nabekura J. Functions of microglia in the central nervous system–beyond the immune response. Neuron glia biol. 2011;7:47–53. doi: 10.1017/s1740925x12000063. [DOI] [PubMed] [Google Scholar]
- 3.Karperien A, Ahammer H, Jelinek HF. Quantitating the subtleties of microglial morphology with fractal analysis. Frontiers Cell Neurosci. 2013;7:3. doi: 10.3389/fncel.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang Y, Le W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol Neurobiol. 2015 doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 5.London, A., Cohen, M. & Schwartz, M. Microglia and monocyte-derived macrophages: functionally distinct populations that act in concert in CNS plasticity and repair. Front Cell Neurosci7, doi:10.3389/fncel.2013.00034 (2013). [DOI] [PMC free article] [PubMed]
- 6.Gomez-Nicola D, Perry VH. Microglial Dynamics and Role in the Healthy and Diseased Brain: A Paradigm of Functional Plasticity. Neuroscientist. 2015;21:169–184. doi: 10.1177/1073858414530512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Streit WJ. Microglial Response to Brain Injury: A Brief Synopsis. Toxicol Pathol. 2000;28:28–30. doi: 10.1177/019262330002800104. [DOI] [PubMed] [Google Scholar]
- 8.Zanier ER, Fumagalli S, Perego C, Pischiutta F, De Simoni M-G. Shape descriptors of the “never resting” microglia in three different acute brain injury models in mice. Intensive Care Med Exp. 2015;3:1–18. doi: 10.1186/s40635-015-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Megjhani M, et al. Population-scale three-dimensional reconstruction and quantitative profiling of microglia arbors. Bioinformatics. 2015;31:2190–2198. doi: 10.1093/bioinformatics/btv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison HW, Filosa JA. A quantitative spatiotemporal analysis of microglia morphology during ischemic stroke and reperfusion. J Neuroinflammation. 2013;10:1–20. doi: 10.1186/1742-2094-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hovens IB, Nyakas C, Schoemakers RG. A novel method for evaluating microglial activation using ionized calcium-binding adaptor protein-1 staining: cell body to cell size ratio. Neuroimmunol Neuroinflamm. 2014;1:82–88. doi: 10.4103/2347-8659.139719. [DOI] [Google Scholar]
- 12.Verdonk F, et al. Phenotypic clustering: a novel method for microglial morphology analysis. J Neuroinflammation. 2016;13:153. doi: 10.1186/s12974-016-0614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agudo M, et al. Time course profiling of the retinal transcriptome after optic nerve transection and optic nerve crush. Mol Vis. 2008;14:1050–1063. [PMC free article] [PubMed] [Google Scholar]
- 14.Tezel G, Yang X, Yang J, Wax MB. Role of tumor necrosis factor receptor-1 in the death of retinal ganglion cells following optic nerve crush injury in mice. Brain Res. 2004;996:202–212. doi: 10.1016/j.brainres.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 15.De Groef L, et al. Differential visual system organization and susceptibility to experimental models of optic neuropathies in three commonly used mouse strains. Exp Eye Res. 2016;145:235–247. doi: 10.1016/j.exer.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Migallon MC, Valiente-Soriano FJ, Nadal-Nicolas FM, Vidal-Sanz M, Agudo-Barriuso M. Apoptotic Retinal Ganglion Cell Death After Optic Nerve Transection or Crush in Mice: Delayed RGC Loss With BDNF or a Caspase 3 Inhibitor. Invest Ophthalmol Vis Sci. 2016;57:81–93. doi: 10.1167/iovs.15-17841. [DOI] [PubMed] [Google Scholar]
- 17.Bosco A, Romero CO, Ambati BK, Vetter ML. In vivo dynamics of retinal microglial activation during neurodegeneration: confocal ophthalmoscopic imaging and cell morphometry in mouse glaucoma. J Vis Exp. 2015;11:52731. doi: 10.3791/52731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebneter A, Casson RJ, Wood JPM, Chidlow G. Microglial Activation in the Visual Pathway in Experimental Glaucoma: Spatiotemporal Characterization and Correlation with Axonal Injury. Invest Ophthalmol Vis Sci. 2010;51:6448–6460. doi: 10.1167/iovs.10-5284. [DOI] [PubMed] [Google Scholar]
- 19.Bosco A, Steele MR, Vetter ML. Early microglia activation in a mouse model of chronic glaucoma. J Comp Neurol. 2011;519:599–620. doi: 10.1002/cne.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mac Nair CE, Schlamp CL, Montgomery AD, Shestopalov VI, Nickells RW. Retinal glial responses to optic nerve crush are attenuated in Bax-deficient mice and modulated by purinergic signaling pathways. J Neuroinflammation. 2016;13:93. doi: 10.1186/s12974-016-0558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohl SG, Schmeer CW, Witte OW, Isenmann S. Proliferative response of microglia and macrophages in the adult mouse eye after optic nerve lesion. Invest Ophthalmol Vis Sci. 2010;51:2686–2696. doi: 10.1167/iovs.09-4537. [DOI] [PubMed] [Google Scholar]
- 22.Das Sarma S, Chatterjee K, Dinda H, Chatterjee D, Das Sarma J. Cytomorphological and Cytochemical Identification of Microglia. ISRN Immunology. 2013;2013:10. doi: 10.1155/2013/205431. [DOI] [Google Scholar]
- 23.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 24.McLarnon JG. Microglial chemotactic signaling factors in Alzheimer’s disease. Am J Neurodegener Dis. 2012;1:199–204. [PMC free article] [PubMed] [Google Scholar]
- 25.Galindo-Romero C, et al. Effect of Brain-Derived Neurotrophic Factor on Mouse Axotomized Retinal Ganglion Cells and Phagocytic Microglia. Invest Ophthalmol Vis Sci. 2013;54:974–985. doi: 10.1167/iovs.12-11207. [DOI] [PubMed] [Google Scholar]
- 26.Trost A, et al. Time-dependent retinal ganglion cell loss, microglial activation and blood-retina-barrier tightness in an acute model of ocular hypertension. Exp Eye Res. 2015;136:59–71. doi: 10.1016/j.exer.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Chiu K, Yeung S-C, So K-F, Chang RC-C. Modulation of morphological changes of microglia and neuroprotection by monocyte chemoattractant protein-1 in experimental glaucoma. Cell Mol Immunol. 2010;7:61–68. doi: 10.1038/cmi.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Hoz R, et al. Rod-Like Microglia Are Restricted to Eyes with Laser-Induced Ocular Hypertension but Absent from the Microglial Changes in the Contralateral Untreated Eye. PLoS One. 2013;8:e83733. doi: 10.1371/journal.pone.0083733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Gracia P, et al. Automatic Counting of Microglial Cells in Healthy and Glaucomatous Mouse Retinas. PLoS One. 2015;10:e0143278. doi: 10.1371/journal.pone.0143278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojas B, et al. Microglia in mouse retina contralateral to experimental glaucoma exhibit multiple signs of activation in all retinal layers. J Neuroinflammation. 2014;11:133. doi: 10.1186/1742-2094-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallego BI, et al. IOP induces upregulation of GFAP and MHC-II and microglia reactivity in mice retina contralateral to experimental glaucoma. J Neuroinflammation. 2012;9:92. doi: 10.1186/1742-2094-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodeutsch N, Siebert H, Dermon C, Thanos S. Unilateral injury to the adult rat optic nerve causes multiple cellular responses in the contralateral site. J Neurobiol. 1999;38:116–128. doi: 10.1002/(SICI)1097-4695(199901)38:1<116::AID-NEU9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 33.Soltys Z, et al. Quantitative morphological study of microglial cells in the ischemic rat brain using principal component analysis. J Neurosci Methods. 2005;146:50–60. doi: 10.1016/j.jneumeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Soltys Z, Ziaja M, Pawlinski R, Setkowicz Z, Janeczko K. Morphology of reactive microglia in the injured cerebral cortex. Fractal analysis and complementary quantitative methods. J Neurosci Res. 2001;63:90–97. doi: 10.1002/1097-4547(20010101)63:1<90::AID-JNR11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Meijering E, Carpenter AE, Peng H, Hamprecht FA, Olivo-Marin J-C. Imagining the future of bioimage analysis. Nat Biotech. 2016;34:1250–1255. doi: 10.1038/nbt.3722. [DOI] [PubMed] [Google Scholar]
- 36.Davis BM, et al. Towards maximising information extraction from rodent models of ocular disease. Cell Death Dis. 2016;7:e2270. doi: 10.1038/cddis.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Yan B, Shi Y-Q, Zhang W-Q, Wen Z-L. Live Imaging Reveals Differing Roles of Macrophages and Neutrophils during Zebrafish Tail Fin Regeneration. J Biol Chem. 2012;287:25353–25360. doi: 10.1074/jbc.M112.349126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat Meth9, 676–682, http://www.nature.com/nmeth/journal/v9/n7/abs/nmeth.2019.html (2012). [DOI] [PMC free article] [PubMed]
- 39.Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with Image. J. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 40.Davis BM, et al. Automatic quantitative analysis of experimental primary and secondary retinal neurodegeneration: implications for optic neuropathies. Cell Death Discov. 2016;2:16031. doi: 10.1038/cddiscovery.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao, Y. Nearest Neighbor Distances Calculation with ImageJ, <https://icme.hpc.msstate.edu/mediawiki/index.php/Nearest_Neighbor_Distances_Calculation_with_ImageJ> (2016).
- 42.Wassle H, Grunert U, Rohrenbeck J. Immunocytochemical staining of AII-amacrine cells in the rat retina with antibodies against parvalbumin. J Comp Neurol. 1993;332:407–420. doi: 10.1002/cne.903320403. [DOI] [PubMed] [Google Scholar]
- 43.Tukey, J. W. The Future of Data Analysis. 1–67, doi:10.1214/aoms/1177704711 (1962).
- 44.Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. J R Stat Soc Ser C Appl Stat. 1979;28:100–108. doi: 10.2307/2346830. [DOI] [Google Scholar]
- 45.R Core Team. R: A language and environment for statistical computing. (Foundation for Statistical Computing, 2013).
- 46.Ripley, B. D. Statistical Inference for Spatial Processes. (Cambridge University Press, 1989).
- 47.Baddeley, A., Rubak, E. & Turner, R. Spatial Point Patterns: Methodology and Applications with R. (CRC Press, 2015).
- 48.Kiskowski MA, Hancock JF, Kenworthy AK. On the Use of Ripley’s K-Function and Its Derivatives to Analyze Domain Size. Biophys J. 2009;97:1095–1103. doi: 10.1016/j.bpj.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Besag JE. Comments on Ripley’s Paper. J R Stat Soc. 1977;39:193–195. [Google Scholar]
- 50.Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 51.Dixon P. Testing Spatial Segregation Using a Nearest-Neighbor Contingency Table. Ecology. 1994;75:1940–1948. doi: 10.2307/1941598. [DOI] [Google Scholar]
- 52.De la Cruz, M. In Introduccion al Analisis Espacial de Datos en Ecologia y Ciencias Ambientales: Metodos y Aplicaciones (eds Maestre, F. T., Escudero, A. & Bonet, A.) (Asociacion Espanola de Ecologia Terrestre, Universidad Rey Juan Carlos y Caja de Ahorros del Mediterraneo, 2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


