Abstract
The goal of this study was to assess risk factors associated with HIV/AIDS progression. Between May 2007 and December 2014, 114 subjects were enrolled in Wuxi City and examined every 6 months. The pol gene sequence was amplified to ascertain the HIV-1 subtype. A Cox proportional hazards regression model was used to estimate the factors associated with HIV/AIDS progression. The median follow-up time for all 114 subjects was 26.70 months (IQR: 18.50–41.47), while the median progression time of the 38 progressed subjects was 24.80 months (IQR: 14.13–34.38). Overall, the CRF01_AE subtype was correlated with a significant risk of accelerated progression compared to non-CRF01_AE subtypes (HR = 3.14, 95%CI: 1.39–7.08, P = 0.006). In addition, a lower CD4 count (350–499) at baseline was associated with a risk of accelerated HIV/AIDS progression compared to higher CD4 count (≥500) (HR = 4.38, 95%CI: 1.95–9.82, P < 0.001). Furthermore, interaction analyses showed that HIV-1 subtypes interacted multiplicatively with transmission routes or CD4 count at baseline to contribute to HIV/AIDS progression (P = 0.023 and P < 0.001, respectively). In conclusion, the CRF01_AE subtype and a lower CD4 count at baseline tend to be associated with the faster progression of HIV/AIDS. Understanding the factors affecting HIV/AIDS progression is crucial for developing personalized management and clinical counselling strategies.
Introduction
Acquired immunodeficiency syndrome (AIDS) has been a major public health threat since its discovery in the United States in 1981. In general, the progression from human immunodeficiency virus (HIV) infection to AIDS development takes approximately 8–10 years1; however, this duration varies among individuals. Multiple factors have been found to contribute to the progression of HIV-1 infection, such as immunological, virological and host genetic factors2–6. In particular, the emergence of RNA sequencing technologies has provided a means for analysing the association between virological factors and HIV/AIDS progression.
The HIV-1 subtype has been associated with HIV/AIDS progression and has attracted much interest among researchers7–11. Unfortunately, no consensus has been reached in studies exploring the association between subtype and disease progression. Studies conducted in Tanzania and Uganda have suggested that subtype D is correlated with faster rates of CD4+ T-cell decline and disease progression than those observed in other subtypes and recombinant forms of the virus7, 8. However, a retrospective cohort study conducted during 1996 and 2007 revealed that Africans infected with B clade HIV-1 suffered from faster rates of HIV/AIDS progression compared with those infected with non-B clade subtypes10. Furthermore, a meta-analysis demonstrated that the trend of HIV/AIDS progression among different HIV-1 subtypes in a descending order was subtype C > D > AE > G > A9.
Though numerous prophylactic measures against HIV have been effectively employed in China12, they have not been able to efficiently terminate the pandemic. A systematic review and meta-analysis conducted in China showed that the pooled prevalence of CRF01_AE, subtype B, CRF07_BC, CRF08_BC, and subtype C was 44.54% (95% CI 40.81–48.30), 18.31% (95% CI 14.71–22.17), 16.45% (95% CI 13.82–19.25), 2.55% (95% CI 1.56–3.73), and 0.37% (95% CI 0.11–0.72), respectively13. Notably, the median time from the estimated date of seroconversion to the development of AIDS in the Chinese HIV-1 population was shorter compared with that in other countries14–16. Thus, understanding the factors that affect HIV/AIDS progression is vitally important for personalized disease management and clinical counselling. However, very few studies have explored the factors associated with HIV/AIDS progression among HIV-1 infected individuals in China. The aim of this study is to explore the natural history and molecular epidemiology of HIV-1 and to evaluate risk factors associated with HIV/AIDS progression.
Results
Demographic characteristics of subjects
Following the application of strict screening criteria, a total of 114 subjects were included in the analysis (Fig. 1). The majority of subjects were male (88.6%) and were of Han ethnicity (99.1%). The age of the subjects ranged from 15- to 74-years-old at the time of HIV-1 infection. The median CD4 count at baseline was 502.00 (IQR: 404.75–614.25). The median follow-up time was 26.70 months (IQR: 18.50–41.47), and the median progression time was 24.8 months (IQR: 14.13–34.38). The basic demographic data are summarized in Table 1. The distribution of HIV-1 subtypes in Wuxi was as follows: CRF01_AE (53.5%), B (14.9%), CRF07_BC/CRF08_BC (27.2%), and other subtypes (4.4%) (Fig. 2). Considering that the majority of the subjects were infected with CRF01_AE, we then separated the subjects into CRF01_AE and non-CRF01_AE groups. CRF01_AE was more prevalent in the MSM (men who have sex with men) group (65.7%, 44/67) than in the heterosexual group (36.2%, 17/47) (Fig. 2). There were significant differences between the subtypes with regard to gender, route of transmission, age at infection, and education status, all with P values less than 0.05 (Table 1).
Figure 1.
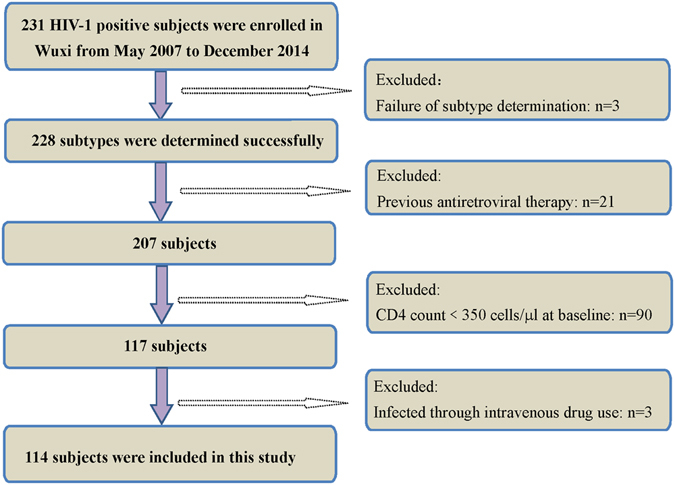
Flow chart of included and excluded study population.
Table 1.
Demographic characteristics of 114 HIV-1 participants classified by viral subtypes in Wuxi.
| Characteristics | All (n = 114) | CRF01_AE (n = 61) | Non-CRF01_AE (n = 53) | P |
|---|---|---|---|---|
| Gender (n, %) | 0.035 | |||
| Male | 101 (88.6) | 58 (95.1) | 43 (81.1) | |
| Female | 13 (11.4) | 3 (4.9) | 10 (18.9) | |
| Ethnicity (n, %) | 0.465 | |||
| Han | 113 (99.1) | 61 (100) | 52 (98.1) | |
| Minority | 1 (0.9) | 0 (0) | 1 (1.9) | |
| Route of transmission (n, %) | 0.002 | |||
| MSM | 67 (58.8) | 44 (72.1) | 23 (43.4) | |
| Heterosexual | 47 (41.2) | 17 (27.9) | 30 (56.6) | |
| Age at infection, yrs (n, %) | 0.007 | |||
| <30 | 59 (51.8) | 35 (57.4) | 24 (45.3) | |
| 30- | 19 (16.7) | 14 (22.9) | 5 (9.4) | |
| ≥40 | 36 (31.6) | 12 (19.7) | 24 (45.3) | |
| Marriage Status (n, %) | 0.300 | |||
| Single | 52 (45.6) | 31 (50.8) | 21 (39.6) | |
| Married | 37 (32.5) | 16 (26.2) | 21 (39.6) | |
| Divorced/widowed | 25 (21.9) | 14 (23.0) | 11 (20.8) | |
| Education (n, %) | 0.014 | |||
| Primary school or below | 12 (10.5) | 2 (3.3) | 10 (18.9) | |
| Junior/high school | 65 (57.0) | 35 (57.4) | 30 (56.6) | |
| College or above | 37 (32.5) | 24 (39.3) | 13 (24.5) | |
| CD4 count at baseline (n, %) | 0.260 | |||
| ≥500 | 59 (51.8) | 35 (57.4) | 24 (45.3) | |
| 350–499 | 55 (48.2) | 26 (42.6) | 29 (54.7) |
Figure 2.

Distribution of HIV-1 subtypes in Wuxi.
Factors associated with HIV/AIDS progression
In the Cox proportional hazards regression model, the CRF01_AE subtype was correlated with a significantly accelerated rate of HIV/AIDS progression compared to non-CRF01_AE subtypes (HR = 3.14, 95%CI: 1.39–7.08, P = 0.006) (Table 2). In addition, a lower CD4 count (350–499) at baseline was also significantly associated with HIV/AIDS progression compared to a higher CD4 count (≥500) (HR = 4.38, 95% CI: 1.95–9.82, P < 0.001) (Table 2).
Table 2.
Factors associated with HIV/AIDS progression (defined as time for CD4+ T-cell count decrease to <350 cells/μL) using Cox proportional hazards model.
| Covariates | Progressor/Total | Median progression time (Months) | HR (95%CI) | Pª |
|---|---|---|---|---|
| Gender | ||||
| Male | 31/101 | 25.33 | 1 | |
| Female | 7/13 | 9.13 | 1.48 (0.51–4.31) | 0.475 |
| Route of transmission | ||||
| MSM | 18/67 | 25.26 | 1 | |
| Heterosexual | 20/47 | 24.25 | 1.33 (0.60–2.97) | 0.486 |
| Age at infection, yrs | ||||
| <30 | 15/59 | 26.93 | 1 | |
| 30- | 5/19 | 16.53 | 1.41 (0.50–3.99) | 0.519 |
| ≥40 | 18/36 | 24.78 | 1.76 (0.75–4.11) | 0.191 |
| Viral subtype | ||||
| Non-CRF01_AE | 16/53 | 26.13 | 1 | |
| CRF01_AE | 22/61 | 22.15 | 3.14 (1.39–7.08) | 0.006 |
| Education | ||||
| Primary school or below | 7/12 | 24.13 | 1 | |
| Junior/high school | 24/65 | 27.41 | 3.24 (0.92–11.36) | 0.067 |
| College or above | 7/37 | 19.93 | 1.72 (0.36–8.09) | 0.495 |
| CD4 count at baseline | ||||
| ≥500 | 8/59 | 26.06 | 1 | |
| 350–499 | 30/55 | 24.26 | 4.38 (1.95–9.82) | <0.001 |
Abbreviations: HR, hazard ratio; CI, confidence intervals. ªCox regression with adjustment for gender, route of transmission, age at infection, viral subtype, education and CD4 count at baseline where necessary.
Considering the significant differences in gender, route of transmission, age at infection and education between the CRF01_AE and non-CRF01_AE groups (P < 0.05), stratification analyses were performed to explore the association between the 4 factors above and HIV/AIDS progression. We found that HIV/AIDS progressed faster in male subjects infected with CRF01_AE compared to males infected with non-CRF01_AE subtypes (P = 0.046) (Fig. 3). In addition, MSM patients infected with CRF01_AE exhibited a significantly faster progression rate than MSM patients infected with non-CRF01_AE subtypes (P = 0.048) (Fig. 4), while no significant difference between HIV/AIDS progression in CRF01_AE and non-CRF01_AE subtypes was observed in heterosexuals (P = 0.405). Further, no significant association was found between HIV-1 subtype and progression in any of the age groups, nor for either of the education groups. In addition, since lower CD4 count at baseline was a risk factor for fast HIV/AIDS progression, in concordance with the well-known knowledge, we also performed the stratified analysis of CD4 count at baseline (divided into two groups: ≥500 and 350–499) to explore the association between HIV subtypes and progression and found that HIV/AIDS progressed borderline significantly faster in subjects with lower baseline CD4 count (350–499) infected with CRF01_AE compared to those with lower baseline CD4 count (350–499) infected with non-CRF01_AE subtypes (P = 0.055) (Fig. 5), however, no significant association was found between HIV-1 subtype and progression in the higher baseline CD4 count group (≥500).
Figure 3.

Kaplan-Meier survival curves for progression from estimated date of seroconversion to CD4 count <350 cells/μL in different subtypes of male group.
Figure 4.
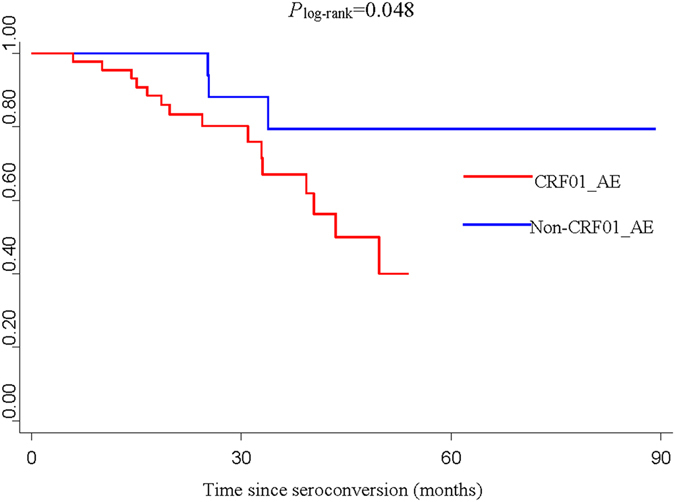
Kaplan-Meier survival curves for progression from estimated date of seroconversion to CD4 count <350 cells/μL in different subtypes of MSM group.
Figure 5.
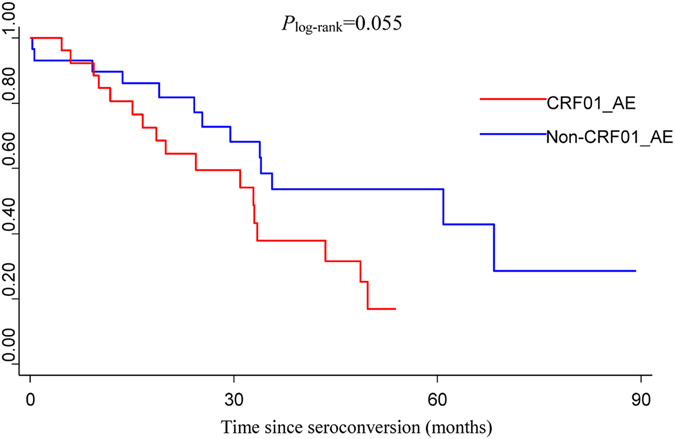
Kaplan-Meier survival curves for progression from estimated date of seroconversion to CD4 count <350 cells/μL in different subtypes of lower baseline CD4 count group (350–499).
Considering the significant difference in the route of disease transmission between the CRF01_AE and non-CRF01_AE groups, we investigated whether the effect of HIV-1 subtype on HIV/AIDS progression was modified by transmission route. Interaction analyses showed that HIV-1 subtypes interacted multiplicatively with transmission route to contribute to HIV/AIDS progression (interaction P = 0.023) (Table 3). In addition, since lower CD4 count at baseline was a risk factor for fast HIV/AIDS progression, in concordance with the well-known knowledge, we also investigated the interaction of HIV-1 subtypes and CD4 count at baseline. As expected, interaction analyses showed that HIV-1 subtypes interacted multiplicatively with CD4 count at baseline to contribute to HIV/AIDS progression (interaction P < 0.001) (Table 4).
Table 3.
The interaction between HIV-1 subtypes and transmission route on HIV/AIDS progression.
| Subtype | Transmission route | Progressor/Total | Median progression time (Months) | HR (95%CI) | P a |
|---|---|---|---|---|---|
| Non-CRF01_AE | MSM | 3/23 | 25.33 | 1 | |
| Non-CRF01_AE | Heterosexual | 13/30 | 26.93 | 1.72 (0.41–7.27) | 0.463 |
| CRF01_AE | MSM | 15/44 | 24.40 | 3.19 (0.89–11.36) | 0.074 |
| CRF01_AE | Heterosexual | 7/17 | 19.93 | 4.52 (1.11–18.47) | 0.036 |
| P for multiplicative interaction | 0.023 | ||||
a P value of interaction analysis between HIV-1 subtype and transmission route on HIV/AIDS progression with adjustment for gender, age at infection and baseline CD4 count.
Table 4.
The interaction between HIV-1 subtypes and CD4 count at baseline on HIV/AIDS progression.
| Subtype | CD4 count at baseline | Progressor/Total | Median progression time (Months) | HR (95%CI) | P a |
|---|---|---|---|---|---|
| Non-CRF01_AE | ≥500 | 3/24 | 26.93 | 1 | |
| Non-CRF01_AE | 350–499 | 13/29 | 25.33 | 3.18 (0.90–11.28) | 0.073 |
| CRF01_AE | ≥500 | 5/35 | 24.37 | 2.09 (0.47–9.27) | 0.331 |
| CRF01_AE | 350–499 | 17/26 | 19.93 | 10.99 (2.95–41.01) | <0.001 |
| P for multiplicative interaction | <0.001 | ||||
a P value of interaction analysis between HIV-1 subtype and CD4 count at baseline on HIV/AIDS progression with adjustment for gender, age at infection and transmission route.
Discussion
In the present study, in order to understand the natural history and disease progression of HIV-1, we studied HIV/AIDS progression in subjects infected with different subtypes of HIV-1 in Wuxi, Jiangsu by measuring the length of time between the estimated date of seroconversion to the time of a decrease in CD4 count to <350 cells/μL. The subtype distribution in our study was 53.5% CRF01_AE, 14.9% B, 27.2% CRF07_BC/CRF08_BC and 4.4% other subtypes, which was consistent with previous reports17, 18, indicating that recombinant forms of the virus might be on the rise worldwide, which have already been associated with faster disease progression as seen in Cuba19 and Brazil20. Most importantly, our study demonstrated that CRF01_AE is associated with faster HIV/AIDS progression, which is in agreement with a previous study conducted in China14. In another study, Ng et al. (2011) showed that CRF01_AE-infected seroconverters experienced a faster rate of CD4+ T-cell decline and thus required earlier cART initiation compared to non-CRF01_AE patients21. It remains unclear as to why CRF01_AE was associated with rapid CD4 decline. However, some studies have shown that a high proportion of X4 tropism is observed in the CRF01_AE subtype, and numerous studies have demonstrated that X4 tropism is associated with increased rates of CD4 count decline and progression to advanced immunosuppression22–24. In addition, it has been postulated that the decline of the host immune system after HIV-1 infection may allow X4 viruses to evolve and replicate freely25, which may explain the increased rate of disease progression in CRF01_AE patients.
In our study, we found that a lower CD4 count (350–499 cells/μL) at baseline was a risk factor for fast HIV/AIDS progression, in concordance with previous studies26. CD4+ T cells are crucial in forming an immune response to foreign antigens, and they are the primary target cells of the HIV-1 virus. The continuous loss of CD4+ T cells could eventually result in the loss of ability to mount an effective immune response to pathogens, as well as the deaths of subjects in the terminal stage of HIV infection27.
The impact of transmission route on disease progression has long been a subject of debate28. In our study, no significant relationship between transmission route and HIV-1 progression was observed (P = 0.486); this finding is inconsistent with a previous study conducted in China by Li et al., which revealed that the transmission route in MSM subjects is an independent risk factor for the progression of HIV infection to AIDS14. There are several possible reasons for the difference in our results. First, differences in the eligibility criteria of the studied populations may have affected the outcome; we included subjects with CD4 > 350 cells/μL at baseline and defined HIV/AIDS progression as CD4 < 350 cells/μL, whereas the inclusion criteria used by Li et al. was a CD4 count <350 cells/μL and they defined HIV/AIDS progression as CD4 < 200 cells/μL. Second, the difference may have resulted from the relatively short follow-up time in our study.
To the best of our knowledge, although the data are not new, this is the first study of the natural course of HIV-1 infection in antiretroviral-naive subjects in Jiangsu, China. Interestingly, this is the first report of transmission routes interacting multiplicatively with HIV-1 subtypes to contribute to HIV/AIDS progression. Moreover, our study may have important implications for personalized disease management and clinical counselling in other Asian countries since the CRF01_AE strain is dominant in Asia. However, our study has limitations that need to be addressed. First, we defined CD4 < 350 cells/μL as a time-to-event endpoint in HIV/AIDS progression, and we could not exclude the possibility that the standards of baseline CD4 count and progression CD4 count were so approach that the judgment of HIV/AIDS progression might be not specific enough. Further study measuring viral load and the clinical symptoms of AIDS should be conducted. Second, in the current sample size (114 subjects), the statistical power is approximately 49.8% in detecting an effect size of 1.50 with an α-level of 0.05 for the association between subtype and HIV/AIDS progression. Nevertheless, we could not exclude the possibility that a small sample size may result in low statistical power. Therefore, further studies with larger sample sizes are needed to confirm our findings. Third, the differing rates of HIV/AIDS progression in HIV-infected individuals are complex and involve various factors, with viral and host factors being the most impactful; thus, further studies are needed to extensively explore additional factors (e.g., viral load and host genetics)8, 15, 29, 30. Fourth, we inferred HIV-1 subtype by sequencing the pol region, which could result in the misclassification of recombinants as pure subtypes. It is also possible that some of the reported recombinants are actually HIV-1 dual/multiple infections, which have been associated with faster disease progression31, 32. In addition, we only sequenced the pol region and have not sequenced env region at present, thus no data are available about the co-receptor use by HIV-1. Therefore, whole genome sequencing is the ideal method for inferring subtype and performing co-receptor use analysis. Finally, we observed that CRF01_AE is associated with faster HIV/AIDS progression. However, we were unable to elucidate the biological mechanisms underlying this phenomenon; thus, further functional evaluations are necessary.
Conclusions
This study revealed that subjects carrying the CRF01_AE subtype may suffer a faster loss of CD4 cells and earlier HIV/AIDS progression compared with those carrying a non-CRF01_AE subtype. Regular surveillance of HIV-1 subtypes and CD4 count would be beneficial in monitoring HIV/AIDS progression, as well as for improving personalized disease management and clinical counselling. Further studies incorporating subjects with different ethnic backgrounds combined with functional evaluations are warranted to verify the relationship between HIV-1 subtypes and HIV/AIDS progression.
Methods
Ethics Statement
This study was reviewed and approved by the Institutional Review Board at the Human Medical Research Ethics Committee of the Center for Disease Control and Prevention (CDC) in Wuxi, and written informed consent was obtained from every subject. The methods were carried out in accordance with the approved guidelines.
Study population
A total of 231 subjects were enrolled in the study between May 2007 and December 2014 in Wuxi with follow-up intervals of 6 months. CD4 count was determined at each visit. Follow-up started in May 2007, and the last follow-up was concluded in December 2014. Subjects were included if they met the following eligibility criteria: (1) CD4 count was higher than 350 cells/μL at baseline; (2) subject was antiretroviral-naive; (3) HIV was contracted through sexual infection; and (4) the subjects attended subsequent follow-ups from the estimated date of seroconversion (Fig. 1). HIV/AIDS progression was defined as a CD4 count less than 350 cells/μL. Ultimately, a total of 114 individuals were included in this study. Clinical data, including basic demographic information and CD4 counts, were collected from medical charts at the Wuxi CDC. Basic demographic information of the subjects included age at HIV-1 infection, gender, ethnicity, marital status, education status and route of HIV-1 transmission.
Viral RNA extraction, PCR amplification and sequencing
All blood samples were processed at the Shanghai Municipal CDC for population-based nucleotide sequencing of plasma HIV-1 RNA encoding regions of reverse transcriptase (RT) and protease (PR) genes. HIV-1 RNA was extracted from 200 µl of stored plasma specimen using the QIAmp Viral RNA Mini kit (Qiagen, Valencia, CA, USA) per the manufacturer’s instructions. RT-PCR and nested polymerase chain (nPCR) amplification for the pol region were performed according to previously published methods17, 33. Subtypes were determined using the Recombinant Identification Program (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html).
Statistical analysis
Baseline characteristics were analysed by using the χ2 test for categorical variables. The Cox proportional hazards regression model was used to estimate risk factors associated with the time of CD4 count decrease to <350 cells/μL. P < 0.05 was considered statistically significant. The SPSS 17.0 statistical package and STATA 13 were used for all analyses.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81373060), the Six Major Human Resources Project of Jiangsu Province (WSN-015), Preventive Medicine Project of Jiangsu Province (Y2013071) and the Science and Technology Program of Nantong City (MS12015125).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
M.C., W.Z. and Xun Zhuang: study design, data collection, interpretation of results, critical revision of manuscript, and writing of the manuscript; Xuan Zhang, X.M., B.Z., Y.Y. and Y.T. prepared the samples. W.J., X.H., L.Z., L.J., T.T. and Y.L. helped with the interpretation of data. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Minjie Chu and Wuhong Zhang contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saina MC, et al. Comparison of HIV-1 nef and gag Variations and Host HLA Characteristics as Determinants of Disease Progression among HIV-1 Vertically Infected Kenyan Children. PLoS One. 2015;10:e0137140. doi: 10.1371/journal.pone.0137140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantaleo G, Fauci AS. Immunopathogenesis of HIV infection. Annu Rev Microbiol. 1996;50:825–54. doi: 10.1146/annurev.micro.50.1.825. [DOI] [PubMed] [Google Scholar]
- 3.Nascimento-Brito S, et al. HIV-1 Tropism Determines Different Mutation Profiles in Proviral DNA. PLoS One. 2015;10:e0139037. doi: 10.1371/journal.pone.0139037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivera-Rivera, Y., Vazquez-Santiago, F. J., Albino, E., Sanchez, M. D. & Rivera-Amill, V. Impact of Depression and Inflammation on the Progression of HIV Disease. J Clin Cell Immunol7, doi:10.4172/2155-9899.1000423 (2016). [DOI] [PMC free article] [PubMed]
- 5.Lin N, et al. Humoral Immune Pressure Selects for HIV-1 CXC-chemokine Receptor 4-using Variants. EBioMedicine. 2016;8:237–47. doi: 10.1016/j.ebiom.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pananghat AN, et al. IL-8 Alterations in HIV-1 Infected Children With Disease Progression. Medicine (Baltimore) 2016;95:e3734. doi: 10.1097/MD.0000000000003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiwanuka N, et al. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J Acquir Immune Defic Syndr. 2010;54:180–4. doi: 10.1097/QAI.0b013e3181c98fc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasan A, et al. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42:843–52. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- 9.Pant Pai N, Shivkumar S, Cajas JM. Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996–2010. J Acquir Immune Defic Syndr. 2012;59:382–8. doi: 10.1097/QAI.0b013e31824a0628. [DOI] [PubMed] [Google Scholar]
- 10.Keller M, Lu Y, Lalonde RG, Klein MB. Impact of HIV-1 viral subtype on CD4+ T-cell decline and clinical outcomes in antiretroviral naive patients receiving universal healthcare. Aids. 2009;23:731–7. doi: 10.1097/QAD.0b013e328326f77f. [DOI] [PubMed] [Google Scholar]
- 11.Touloumi G, et al. Impact of HIV-1 subtype on CD4 count at HIV seroconversion, rate of decline, and viral load set point in European seroconverter cohorts. Clin Infect Dis. 2013;56:888–97. doi: 10.1093/cid/cis1000. [DOI] [PubMed] [Google Scholar]
- 12.Laga M, Piot P. Prevention of sexual transmission of HIV: real results, science progressing, societies remaining behind. Aids. 2012;26:1223–9. doi: 10.1097/QAD.0b013e32835462b8. [DOI] [PubMed] [Google Scholar]
- 13.Yuan R, Cheng H, Chen LS, Zhang X, Wang B. Prevalence of different HIV-1 subtypes in sexual transmission in China: a systematic review and meta-analysis. Epidemiol Infect. 2016;144:2144–53. doi: 10.1017/S0950268816000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. CRF01_AE subtype is associated with X4 tropism and fast HIV progression in Chinese patients infected through sexual transmission. Aids. 2014;28:521–30. doi: 10.1097/QAD.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 15.Rangsin R, et al. The natural history of HIV-1 subtype E infection in young men in Thailand with up to 14 years of follow-up. Aids. 2007;21(Suppl 6):S39–46. doi: 10.1097/01.aids.0000299409.29528.23. [DOI] [PubMed] [Google Scholar]
- 16.Time from HIV-1 seroconversion to AIDS and death before widespread use of highly-active antiretroviral therapy: a collaborative re-analysis. Collaborative Group on AIDS Incubation and HIV Survival including the CASCADE EU Concerted Action. Concerted Action on SeroConversion to AIDS and Death in Europe. Lancet355, 1131–7 (2000). [PubMed]
- 17.He X, et al. A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One. 2012;7:e47289. doi: 10.1371/journal.pone.0047289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, et al. The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990 s. Aids. 2013;27:1793–802. doi: 10.1097/QAD.0b013e328360db2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouri V, et al. CRF19_cpx is an Evolutionary fit HIV-1 Variant Strongly Associated With Rapid Progression to AIDS in Cuba. EBioMedicine. 2015;2:244–54. doi: 10.1016/j.ebiom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarosso LF, et al. Short communication: HIV type 1 subtype BF leads to faster CD4+ T cell loss compared to subtype B. AIDS Res Hum Retroviruses. 2014;30:190–4. doi: 10.1089/aid.2012.0243. [DOI] [PubMed] [Google Scholar]
- 21.Ng OT, et al. Increased rate of CD4+ T-cell decline and faster time to antiretroviral therapy in HIV-1 subtype CRF01_AE infected seroconverters in Singapore. PLoS One. 2011;6:e15738. doi: 10.1371/journal.pone.0015738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters L, et al. The impact of HIV tropism on decreases in CD4 cell count, clinical progression, and subsequent response to a first antiretroviral therapy regimen. Clin Infect Dis. 2008;46:1617–23. doi: 10.1086/587660. [DOI] [PubMed] [Google Scholar]
- 23.Weiser B, et al. HIV-1 coreceptor usage and CXCR4-specific viral load predict clinical disease progression during combination antiretroviral therapy. Aids. 2008;22:469–79. doi: 10.1097/QAD.0b013e3282f4196c. [DOI] [PubMed] [Google Scholar]
- 24.Hamlyn E, et al. Increased levels of CD4 T-cell activation in individuals with CXCR4 using viruses in primary HIV-1 infection. Aids. 2012;26:887–90. doi: 10.1097/QAD.0b013e328351e721. [DOI] [PubMed] [Google Scholar]
- 25.Philpott SM. HIV-1 coreceptor usage, transmission, and disease progression. Curr HIV Res. 2003;1:217–27. doi: 10.2174/1570162033485357. [DOI] [PubMed] [Google Scholar]
- 26.Mellors JW, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–54. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 27.Pattanapanyasat K, Thakar MR. CD4+ T cell count as a tool to monitor HIV progression & anti-retroviral therapy. Indian J Med Res. 2005;121:539–49. [PubMed] [Google Scholar]
- 28.Lin PH, et al. Slow immunological progression in HIV-1 CRF07_BC-infected injecting drug users. Emerg Microbes Infect. 2013;2:e83. doi: 10.1038/emi.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baeten JM, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195:1177–80. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 30.Tscherning C, et al. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–8. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 31.Ssemwanga D, et al. Multiple HIV-1 infections with evidence of recombination in heterosexual partnerships in a low risk Rural Clinical Cohort in Uganda. Virology. 2011;411:113–31. doi: 10.1016/j.virol.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottlieb GS, et al. Dual HIV-1 infection associated with rapid disease progression. Lancet. 2004;363:619–22. doi: 10.1016/S0140-6736(04)15596-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhong P, et al. Genetic diversity and drug resistance of human immunodeficiency virus type 1 (HIV-1) strains circulating in Shanghai. AIDS Res Hum Retroviruses. 2007;23:847–56. doi: 10.1089/aid.2006.0196. [DOI] [PubMed] [Google Scholar]


