Abstract
Objective
To characterize the clinical and molecular epidemiology of multidrug-resistant (MDR) organisms in residents, in healthcare workers (HCWs), and on inanimate surfaces at a long-term care facility (LTCF).
Design
Point-prevalence study in 4 separate wards at a 600-bed urban LTCF that was conducted from October 31, 2006 through February 5, 2007.
Participants
One hundred sixty-one LTCF residents and 13 HCWs.
Methods
Nasal and rectal samples were obtained for culture from each resident, selected environmental surfaces in private and common rooms, and the hands and clothing of HCWs in each ward. All cultures were evaluated for the presence of MDR gram-negative bacteria, methicillin-resistant Staphylococcus aureus, and vancomycin-resistant enterococci. Clinical and demographic information were collected for each enrolled resident. Molecular typing was performed to identify epidemiologically related strains.
Results
A total of 37 (22.8%), 1 (0.6%), and 18 (11.1%) residents were colonized with MDR gram-negative bacteria, vancomycin-resistant enterococci, and methicillin-resistant S. aureus, respectively. MDR gram-negative bacteria were recovered from 3 (1.8%) of the 175 environmental samples cultured, all of which were obtained from common areas in LTCF wards. One (7.7%) of the 13 HCWs harbored MDR gram-negative bacteria. Molecular typing identified clonally related MDR gram-negative strains in LTCF residents. After multivariable analysis, length of hospital stay of at least 4 years, fecal incontinence, and antibiotic exposure for at least 8 days were independent risk factors associated with harboring MDR gram-negative bacteria among LTCF residents.
Conclusions
The prevalence of MDR gram-negative bacteria is high among LTCF residents and exceeds that of vancomycin-resistant enterococci and methicillin-resistant S. aureus. Common areas in LTCFs may provide a unique opportunity for person-to-person transmission of MDR gram-negative bacteria.
Multidrug-resistant (MDR) gram-negative bacteria pose an emerging threat in multiple clinical settings. Infections caused by MDR gram-negative bacteria are associated with worse clinical outcomes, higher rates of mortality, increased costs of care, and delayed receipt of appropriate antibiotics, compared with other types of bacteria.1-5 The elderly population and residents of long-term care facilities are consistently identified as being at increased risk of infection with MDR gram-negative bacteria.6-12 In addition, rates of MDR gram-negative bacterial infection are increasing among long-term care facility residents, both at their home facility and at hospital admission. A 2-year retrospective review of all clinical cultures from a 600-bed long-term care facility identified a substantial increase in the prevalence of MDR gram-negative bacteria (from 7% in 2003 to 13% in 2005).11 Another study that focused on bloodstream infections at hospital admission among older patients also identified an increase in the prevalence of MDR gram-negative bacteria (from 1% in 1999 to 16% in 2007).10
The prevalence of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) at long-term care facilities has been reported to be up to 20% among residents colonized with at least 1 MDR isolate.13-17 Recent evidence strongly suggests that the prevalence of MDR gram-negative bacteria in this population may exceed rates of MRSA and VRE.11,15,18 The increase in the prevalence of MDR gram-negative bacteria among older patients at long-term care facilities emphasizes the need to evaluate infection control strategies targeting this patient population at high risk of infection. This is especially important because existing guidelines primarily address MRSA and VRE and are extrapolated largely from acute care settings, leaving ambiguity with regard to the appropriate measures at long-term care facilities.19
Acquisition of MDR gram-negative bacteria may be endogenous, whereby antibiotic exposure leads to the emergence of antimicrobial resistance among previously susceptible bacteria.20 MDR gram-negative bacteria may also be acquired exogenously through patient-to-patient transmission.15 Previous studies at long-term care facilities have revealed that the hands of healthcare workers (HCWs) can be vehicles of transmission of gram-negative pathogens.21,22 Data on the role of the environment in the spread of antimicrobial-resistant pathogens at long-term care facilities are more limited. Environmental surfaces at long-term care facilities have demonstrated contamination with MRSA, VRE, and gram-negative pathogens.21-23 However, direct correlation of environmental contamination with patient colonization or infection has not been consistently demonstrated.15 Long-term care facilities differ from acute care settings in that they provide frequent opportunities for contact among residents through group activities, such as communal dining and use of common rooms. The potential for environmental contamination in communal spaces at long-term care facilities has not been fully explored; such contamination may provide a unique risk for transmission among long-term care facility residents. Because previous studies have not included communal spaces in their evaluation of environmental contamination with MDR gram-negative bacteria, we included these rooms in the sampling of environmental surfaces to evaluate the potential for transmission of MDR gram-negative bacteria in communal spaces.
We conducted a point-prevalence study at a 600-bed long-term care facility to determine the prevalence of colonization with MDR gram-negative bacteria, relative to MRSA and VRE, and to evaluate potential sources of HCW and environmental contamination, with specific attention to communal rooms. To address these goals, we obtained samples for surveillance culture from long-term care facility residents, their environment, and HCWs. Pulsed-field gel electrophoresis (PFGE) was used to determine molecularly related strain types and potential epidemiological links between patients, inanimate surfaces, and HCWs.
Methods
The Institutional Review Board at Hebrew Senior Life and the Clinical Committee of Investigation at Beth Israel Deaconess Medical Center approved the conduct of this study.
Point-Prevalence Study
As part of an infection control initiative to determine the extent of colonization with antimicrobial-resistant bacteria, all residents from 4 of 15 wards at a 600-bed long-term care facility in metropolitan Boston were included in the study. A point-prevalence study was conducted for 1–2 days in each of these 4 wards over a 3-month period: ward A (starting on October 31, 2006), ward B (starting on November 21, 2006), ward C (starting on January 15, 2007), and ward D (starting on February 5, 2007). Wards were chosen to include a range of functional and cognitive abilities representative of patients housed at the study facility.
During each point-prevalence study, nasal and rectal specimens were obtained from residents by trained staff. All residents in each ward were eligible for study participation. Environmental samples were collected from each resident's room. Samples were obtained from the following sites over a standardized 5 × 5-cm area with use of Rodac plates: bed rail, light switch, and bathroom sink. To conserve resources, a single Rodac plate for each resident's room was used for all 3 tested inanimate surfaces. Samples from inanimate surfaces of the communal room in each ward were also obtained for culture. The communal rooms in each ward are used by the residents of that ward for dining and group activities at least 3 times daily, for a total of up to 8 hours per day. All residents, including those with limited mobility or with advanced dementia, are brought to these rooms regularly to encourage interaction between residents. For each communal room, samples were taken from 1 of the 6 tables in the dining area, the television, and the countertop used for food and beverage preparation with use of 3 separate Rodac plates.
Specimens were collected from HCWs who provided direct resident care in each ward. Samples were obtained from the normal working uniform worn by each HCW by taking an imprint of a 5 × 5-cm area over the umbilical region with use of Rodac plates. Hand specimens were obtained by finger impressions on plates.
Resident Data Collection
Demographic and clinical data were collected for all residents from clinical charts and the Minimum Data Set. The Minimum Data Set is a federally mandated, standardized assessment instrument completed for all residents of licensed nursing homes in the United States. Minimum Data Set data have been widely used and found to be reliable for research pur-poses.24,25 Clinical data included age; sex; fecal and urinary continence; presence of any pressure ulcer; and diagnosis of diabetes, dementia, congestive heart failure, chronic obstructive pulmonary disease, and chronic renal failure. The Global Deterioration Score, a measure of dementia stage, was determined from data obtained in interviews with the primary care providers. The Global Deterioration Score can range from 0 (normal) to 7 (end-stage dementia).26 The Activities of Daily Living score was obtained from Minimum Data Set data. The Activities of Daily Living score describes the ability to conduct tasks independently and can range from 0 (completely dependent) to 6 (fully independent). The Activities of Daily Living score is calculated from 6 activities, with 1 point assigned for ability to complete each task independently: bathing, dressing, toileting, transferring, continence, and feeding.27 The Charlson Comorbidity Index was used to provide a composite score of comorbid conditions.28 Data on antibiotic exposure during the 12-month period before study enrollment were collected from pharmacy records. Dates and duration of acute care hospital stays during the 12 months before study enrollment were obtained from existing medical records.
Definition of MDR Gram-Negative Bacteria
Multidrug resistance among gram-negative bacteria was defined as resistance to 3 or more of the following antimicrobials or antimicrobial classes: extended-spectrum penicillins (ampicillin-sulbactam or piperacillin-tazobactam), third-generation cephalosporins (ceftazidime or ceftriaxone), aminoglycosides (gentamicin), fluororquinolones (ciprofloxacin), and carbepenems (meropenem).10
Microbiological Methods
MDR gram-negative bacteria and VRE were identified from rectal swabs, and MRSA was identified from nasal and rectal swabs. For the isolation of MDR gram-negative bacteria, 2 McConkey media supplemented with 2 μg of ciprofloxacin per mL or 2μg of ceftazidime per mL were used to minimize the recovery of pansusceptible gram-negative bacteria. Species identification and susceptibility testing were performed as described elsewhere.20 MRSA and VRE were isolated in accordance with standard procedures.29,30 PFGE was performed for all MDR gram-negative bacteria, MRSA, and VRE isolates. Indistinguishable or closely related strains, defined as strains differing by no more than 3 bands, were considered to be clonally related according to the criteria of Tenover et al.31
Statistical Analysis
Continuous variables were dichotomized at the mean or at clinically relevant intervals. Categorical variables and dichotomized continuous variables were analyzed using the χ2 test. Statistical significance was defined as 2-sided P ≤ .05. Variables that were statistically significant in univariate analysis were included in a stepwise logistic regression model. Potential interaction between relevant variables was also analyzed (Stata, version 10.0; Stata).
Results
A total of 161 (95.3%) of 169 eligible residents were enrolled in the study: 39 (98%) of 40 residents in ward A, 37 (93%) of 40 in ward B, 39 (98%) of 40 in ward C, and 46 (94%) of 49 in ward D. Eight residents refused cultures or were not on site at the time of specimen collection. The mean age among all enrolled residents was 88.5 years (range, 57–103 years); 82.6% were female, and 94.4% were white. The mean Activities of Daily Living score was 1.1 (range, 0–6), the mean Charlson score was 2.5 (range, 0–6), and the mean Global Deterioration Score was 5.3 (range, 1–7). The mean duration of antibiotic therapy during the 12 months before study enrollment was 16.2 days (range, 0–142 days). Twenty-one residents (13.0%) were hospitalized during the 12 months before study enrollment, and the mean number of hospitalizations was 0.18 (range, 0–4). Among all enrolled residents, the following comorbid conditions were present: fecal incontinence (120 residents [74.5%]), pressure ulcer (19 [11.8%]), urinary incontinence (134 [83.2%]), diabetes (38 [23.6%]), chronic obstructive pulmonary disease (7 [4.3%]), congestive heart failure (35 [21.7%]), and chronic renal failure (15 [9.3%]).
A total of 175 environmental samples were obtained for culture from residents' rooms, including 34 single rooms, 63 double rooms, and 1 triple room; 12 environmental samples were taken from the common rooms in each of the 4 wards. Thirteen HCWs who provided direct care in each participating ward had samples obtained for culture (4 in ward A, 3 in ward B, 3 in ward C, and 3 in ward D).
MDR Gram-Negative Bacteria Isolates
A total of 47 MDR gram-negative isolates were identified among 37 (23.0%) of the 161 enrolled residents. Nine residents (5.6%) were colonized with at least 1 different MDR gram-negative species; 7 (4.3%) were colonized with 2 different MDR gram-negative species, and 2 (1.2%) were colonized with 3 different MDR gram-negative species. The type and frequency of MDR gram-negative species are shown in Table 1. Resistance to individual antimicrobials or classes was as follows: ampicillin-sulbactam (100%), ceftazidime (40.4%), ceftriaxone (29.8%), ciprofloxacin (97.9%), gentamicin (55.3%), meropenem (18.8%), and piperacillin-tazobactam (12.8%). Three- and 4-drug resistance were present in 41 (87.2%) and 6 (12.8%) MDR gram-negative isolates, respectively. The most common 3-drug coresistance pattern among all MDR gram-negative species was ampicillin-sulbactam, ciprofloxacin, and gentamicin (among 22 isolates [46.8%]) (Table 1).
Table 1. Coresistance Patterns Among Multidrug-Resistant Gram-Negative Bacterial Isolates Recovered from Long-Term Care Facility Residents.
| Coresistance profile | Total isolates, % | No. (%) of isolates | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Providencia stuartii (n = 12) | Proteus mirabilis (n = 4) | Morganella morganii (n = 9) | Klebsiella pneumoniae (n = 11) | Escherichia coli (n = 10) | Citrobacter freundii (n = 1) | |||
| ESP, ciprofloxacin, and gentamicin | 22 | 5 (41.7) | 4 (100)a | 4 (44.4) | 0 (0) | 9 (90.0) | 0 (0) | |
| ESP, ciprofloxacin, and meropenem | 3 | 2 (16.7) | 0 (0) | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) | |
| ESP, ciprofloxacin, and third-generation cephalosporinb | 15 | 1 (8.3) | 0 (0) | 3 (33.3) | 10 (90.9)c | 1 (10)d | 0 (0) | |
| ESP, cephalosporin, and meropenem | 1 | 0 (0) | 0 (0) | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) | |
| ESP, cephalosporin, and ciprofloxacin-meropenem | 2 | 1 (8.3) | 0 (0) | 0 (0) | 1 (9.1)e | 0 (0) | 0 (0) | |
| ESP, ciprofloxacin, and gentamicin-meropenem | 3 | 3 (25.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| ESP, cephalosporin, ciprofloxacin, and gentamicin | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100)f | |
Note. ESP, extended-spectrum penicillin (ampicillin-sulbactam or piperacillin-tazobactam).
Two (50%) of these isolates were resistant to both ampicillin-sulbactam and piperacillin-tazobactam.
Ceftazidime or ceftriaxone.
Two isolates were resistant to both ampicillin-sulbactam and piperacillin-tazobactam, and all 10 were resistant to both ceftazidime and ceftriaxone.
This isolate was resistant to both ampicillin-sulbactam and piperacillin-tazobactam.
This isolate was resistant to both ceftazidime and ceftriaxone.
This isolate was resistant to both ampicillin-sulbactam and piperacillin-tazobactam and to both ceftazidime and ceftriaxone.
MDR gram-negative strains with indistinguishable or closely related PFGE patterns were identified in multiple residents in the same ward and in residents in different wards. Among the most prevalent MDR gram-negative species, Klebsiella pneumoniae and Providencia stuartii were the most clonally related, with 4 and 2 unique strains identified among 11 and 12 isolates, respectively. Among MDR Escherichia coli isolates, less clonality was detected, with 7 unique strains identified among 10 isolates. Table 2 shows the epidemiological links between MDR gram-negative bacteria isolates recovered from 2 or more residents.
Table 2. Multidrug-Resistant Gram-Negative Strains Recovered From 2 or More Residents of a Long-Term Care Facility, by Unit of Residence.
| Species, pulsed-field gel electrophoresis strain | No. of isolates | ||||
|---|---|---|---|---|---|
|
| |||||
| Total | Ward A | Ward B | Ward C | Ward D | |
| Klebsiella pneumoniae: KP1 | 9 | 6 | 0 | 0 | 3 |
| Morganella morganii | |||||
| MM1 | 3 | 0 | 3 | 0 | 0 |
| MM2 | 3 | 1 | 2 | 0 | 0 |
| MM3 | 2 | 1 | 1 | 0 | 0 |
| Escherichia coli | |||||
| EC1 | 2 | 0 | 0 | 1 | 1 |
| EC2 | 3 | 0 | 0 | 0 | 3 |
| Proteus mirabilis: PM1 | 2 | 0 | 2 | 0 | 0 |
| Providencia stuartii: PS1 | 12 | 8 | 3 | 0 | 1 |
MDR gram-negative bacteria were isolated from 3 environmental specimens (1.7%) and 1 specimen (7.7%) from an HCW. Two environmental isolates, recovered from the countertop and television of the communal room in ward C, and the 1 MDR gram-negative isolate recovered from an HCW who provided care in ward C, were identified as MDR Enterobacter cloacae. All 3 isolates had indistinguishable PFGE patterns (Figure). Colonization with the same strain type of MDR E. cloacae was not detected in any resident. MDR Stenotrophomonas maltophilia was recovered from the third MDR environmental isolate, which was obtained from the sink of a resident's private room in ward A.
Figure.
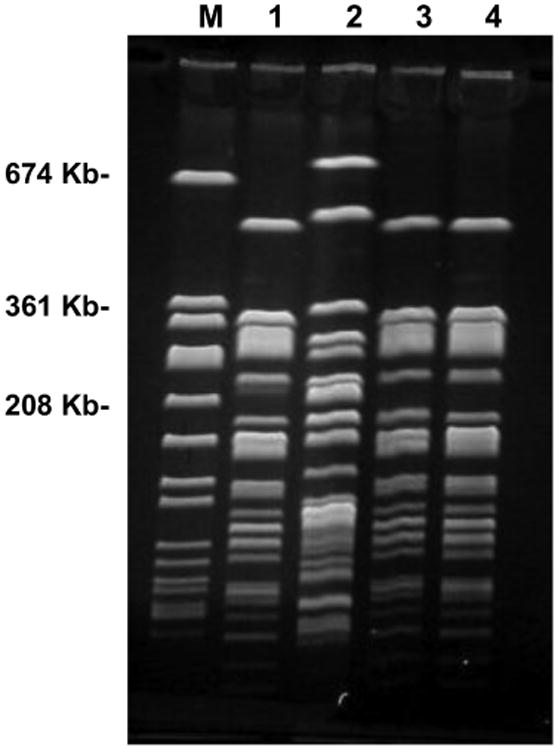
Pulsed-field gel electrophoresis of multidrug-resistant (MDR) Enterobacter cloacae isolates. Staphylococcus aureus 519 size marker (lane M), MDR E. cloacae from the counter top of ward C (lane 1), non-MDR E. cloacae from the table surfaces of ward C (lane 2), MDR E. cloacae from the television of ward C (lane 3), MDR E. cloacae from the hands of healthcare workers in ward C (lane 4). MDR E. cloacae strains shown in lanes 1, 3, and 4 are indistinguishable.
MRSA Isolates
MRSA colonization was identified in 18 residents (11.2%). MRSA was recovered by culture of nasal and rectal specimens from 18 and 4 residents, respectively. All MRSA isolates had indistinguishable PFGE patterns (Table 3).
Table 3. Multidrug-Resistant Gram-Negative Bacteria by Site of Isolation: Total Number of Isolates Identified and Pulsed-Field Gel Electrophoresis Strain Type.
| Species | No. of isolates (no. of strains) | ||
|---|---|---|---|
|
| |||
| Patients (n = 161) | Environmental (n = 163) | Healthcare Workers (n = 13) | |
| Klebsiella pneumoniae | 11 (3) | 0 (0) | 0 (0) |
| Morganella morganni | 9 (4) | 0 (0) | 0 (0) |
| Providencia stuartii | 12 (1) | 0 (0) | 0 (0) |
| Proteus mirabilis | 4 (3) | 0 (0) | 0 (0) |
| Eschericia coli | 10 (7) | 0 (0) | 0 (0) |
| Citrobacter species | 1 (1) | 0 (0) | 0 (0) |
| Enterobacter cloacae | 0 (0) | 2 (1) | 1 (1) |
| Stenotrophomonas maltophilia | 0 (0) | 1 (1) | 0 (0) |
| MRSA | 18 (1) | 2 (1) | 0 (0) |
| VRE | 1 (1) | 0 (0) | 0 (0) |
Note. MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant enterococci.
MRSA was isolated from 2 environmental specimens (1.2%) collected from 2 residents' private rooms. These MRSA strains were indistinguishable from the MRSA strain that colonized the resident in the contaminated room. MRSA was not recovered from any HCW.
VRE Isolates
VRE was recovered from only 1 resident (0.6%). VRE was not isolated from residents' rooms, common areas, or any HCW.
The distribution of MDR gram-negative bacteria, MRSA, and VRE isolates and strain type by site of isolation are shown in Table 3.
Risk Factors for Colonization with MDR Gram-Negative Bacteria
Demographic and clinical characteristics of residents with and without MDR gram-negative bacteria colonization are compared in Table 4. In bivariate analysis, MDR gram-negative bacteria colonization was associated with the presence of pressure ulcers, an Activities of Daily Living score of zero, fecal incontinence, and a duration of stay of at least 4 years. MDR gram-negative bacteria colonization was also associated with exposure to antibiotics for at least 8 days during the 12 months before study enrollment. These variables were included in the multivariable model. In multivariable analysis, duration of stay for at least 4 years (odds ratio [OR], 2.43; 95% confidence interval [CI], 1.06–5.56; P = .036), fecal incontinence (OR, 4.93; 95% CI, 1.13–21.57; P = .034), and antibiotic exposure (OR, 3.6; 95% CI, 1.51–8.66; P = .004) were independently associated with MDR gram-negative bacteria colonization. Interaction effects between these variables were not present (P ≥ .05).
Table 4. Demographic and Clinical Characteristics of Residents With and Without Multidrug-Resistant (MDR) Gram-Negative Bacterial Colonization.
| Variable | No. (%) of residents | ||
|---|---|---|---|
|
| |||
| Unadjusted OR (95% CI) | Colonized with MDR gram-negative bacteria (n = 37) | Not colonized with MDR gram-negative bacteria (n = 124) | |
| Age >85 years | 24 (64.9) | 93 (75) | 0.62 (0.26–1.49) |
| Female sex | 32 (86.5) | 101 (81.5) | 0.68 (0.19–2.06) |
| White | 35 (94.6) | 117 (94.4) | 1.05 (0.19–10.77) |
| Mean duration of stay ≥4 years | 21 (56.8) | 42 (33.9) | 2.56 (1.13–5.82)a |
| Urinary catheter | 1 (2.7) | 4 (3.2) | 0.83 (0.02–8.78) |
| Pressure ulcer | 8 (21.6) | 11 (8.9) | 2.83 (0.89–8.51)a |
| Urinary incontinence | 34 (91.9) | 100 (80.6) | 2.72 (0.75–14.91) |
| Fecal incontinence | 34 (91.9) | 86 (69.4) | 5.00 (1.42– 26.82)a |
| Dementia | 33 (89.2) | 103 (83.1) | 1.68 (0.51–7.20) |
| Diabetes mellitus | 12 (32.4) | 26 (21) | 1.81 (0.72–4.34) |
| GDS score >5 | 26 (70.3) | 72 (58.1) | 1.7 (0.73–4.17) |
| ADL score of 0 | 11 (29.7) | 63 (50.8) | 2.44 (1.04–5.95)a |
| Charlson Comorbidity Index >2 | 30 (81.1) | 84 (67.7) | 2.04 (0.79–5.96) |
| Total number of hospitalizations ≥1b | 5 (13.5) | 16 (12.9) | 1.05 (0.28–3.32) |
| Any antibiotic prescription in previous 12 months | 27 (73) | 77 (62) | 1.64 (0.69–4.16) |
| ≥8 Days of antibiotic treatment | 26 (70.2) | 56 (45.2) | 2.87 (1.23–6.99)a |
Note. ADL, Activity of Daily Living; CI, confidence interval; GDS, Global Deterioration Score; OR, odds ratio.
P < .05.
Total number of admissions to an acute care facility during the 12 months before study enrollment.
Discussion
This study describes the prevalence of MDR gram-negative bacteria at a long-term care facility and characterizes important aspects of transmission of MDR gram-negative bacteria. A total of 22.9% of residents were colonized with MDR gram-negative bacteria, which greatly exceeded the 11% and 0.4% of residents colonized with MRSA and VRE, respectively. Other studies have also documented a greater prevalence of MDR gram-negative bacteria than of VRE and MRSA at long-term care facilities.11,17-18 The reasons for these different rates remains unclear.
The results of this study provide insight into the possible transmission mechanisms of MDR gram-negative bacteria in long-term care facilities. In acute care settings, the hands of HCWs and the environment in patient rooms are frequently contaminated and have been linked to horizontal transmission of gram-negative bacteria.32-37 Studies at long-term care facilities have also identified contamination of environmental surfaces and the hands of HCWs with gram-negative pathogens. The dissemination of a single clone of gram-negative bacteria at a long-term care facility has also been described.21,38 Consistent with these previous reports, in the present study, molecular characterization of the MDR gram-negative bacteria strains recovered from the long-term care facility residents revealed that the 2 most prevalent pathogens, K. pneumoniae and P. stuartii, exhibited dissemination of a single dominant strain between multiple residents, including those residing in different wards; this pattern suggests exogenous acquisition.
In the present study, we also identified an identical strain of MDR gram-negative bacteria on the hands of an HCW and on environmental surfaces in a common room. These findings suggest that common rooms in long-term care facilities may provide opportunities for resident-to-resident spread of MDR gram-negative bacteria. In contrast to the acute care setting, common rooms are a unique feature of long-term care facilities; daily interaction between residents is encouraged in these areas. Infection control strategies may need to specifically address this setting in long-term care facilities.
Risk factors for MDR gram-negative bacteria colonization, including prolonged duration of stay, fecal incontinence, and antibiotic exposure, were identified in the present study. Other studies involving long-term care facility populations have identified similar risk factors for colonization with antimicrobial-resistant bacteria, including poor functional status, low Activities of Daily Living score, and advanced dementia.17,39-40 A common feature of these risk factors is the requirement for frequent contact with HCWs for assistance with toileting, eating, repositioning, and bathing needs. These interactions provide ample opportunity for both acquisition and spread of MDR gram-negative bacteria to other residents through HCWs and through environmental contamination. Similarly, prolonged duration of stay in a long-term care facility would increase the likelihood of acquisition of MDR gram-negative bacteria from other colonized residents.
The present study has several limitations. First, environmental and HCW contamination were likely to have been underestimated, because samples were obtained from only a small percentage of inanimate surfaces and only samples from hands and clothes of HCWs were collected for culture. Second, although our study provides data suggestive of transmission of MDR gram-negative bacteria through HCWs or environmental surfaces, a direct link in the form of an identical strain of MDR gram-negative bacteria between patients and HCWs or environmental surfaces was not identified. However, we believe that the context of these results is important. Although very low rates of MDR gram-negative bacteria contamination were found among HCWs, we obtained only a single sample from each provider. Residents at long-term care facilities interact with HCWs countless times throughout the day. Therefore, even low rates of HCW contamination would result in important opportunities for transmission of MDR gram-negative bacteria. Third, the contribution of endogenous acquisition in the spread of MDR gram-negative bacteria was not assessed and would have provided a more complete characterization of the molecular epidemiology of MDR gram-negative bacteria and mechanisms of acquisition. Finally, the definition of MDR gram-negative bacteria has not been standardized. In the present study, multidrug resistance in gram-negative bacteria was defined as resistance to at least 3 antimicrobials or major antimicrobial groups frequently used in clinical practice. A different definition of multidrug resistance in gram-negative bacteria may have provided different data.
In conclusion, residents of long-term care facilities have high rates of colonization with MDR gram-negative bacteria that are in excess of rates of MRSA and VRE colonization. The present study provides evidence of resident-to-resident spread of MDR gram-negative bacteria at long-term care facilities, with common rooms providing unique opportunities for cross-transmission of these antimicrobial-resistant bacteria. Infection control strategies appropriate for long-term care facilities that are aimed at limiting the exogenous transmission of MDR gram-negative bacteria within the long-term care facility population should be further evaluated to reduce the burden of MDR gram-negative bacteria in this vulnerable population.
Acknowledgments
We thank Lata Venkataraman for technical support.
Financial support. This study was supported in part by the National Institutes of Health–funded Harvard–Beth Israel Deaconess Medical Center T32 fellowship program (Public Health Service grant 5 AG023480-04 to E.O.).
Footnotes
Potential conflicts of interest. All authors report no conflicts of interest relative to this study.
References
- 1.Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006;42:S82–89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 2.Daxboeck F, Budic T, Assadian O, Reich M, Koller W. Economic burden associated with multi-resistant Gram-negative organisms compared with that for methicillin-resistant Staphylococcus aureus in a university teaching hospital. J Hosp Infect. 2006;62:214–218. doi: 10.1016/j.jhin.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 3.The cost of antibiotic resistance: effect of resistance among Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudmonas aeruginosa on length of hospital stay. Infect Control Hosp Epidemiol. 2002;23:106–108. doi: 10.1086/502018. [DOI] [PubMed] [Google Scholar]
- 4.Giske CG, Monnet DL, Cars O, Carmeli Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother. 2008;52:813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans HL, Lefrak SN, Lyman J, et al. Cost of gram-negative resistance. Crit Care Med. 2007;35:89–95. doi: 10.1097/01.CCM.0000251496.61520.75. [DOI] [PubMed] [Google Scholar]
- 6.Safdar N, Maki DG. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann Intern Med. 2002;136:834–844. doi: 10.7326/0003-4819-136-11-200206040-00013. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg BM, Atmar RL, Stager CE, Greenberg SB. Bacteraemia in the elderly: predictors of outcome in an urban teaching hospital. J Infect. 2005;50:288–295. doi: 10.1016/j.jinf.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Mylotte JM, Tayara A, Goodnough S. Epidemiology of bloodstream infection in nursing home residents: evaluation in a large cohort from multiple homes. Clin Infect Dis. 2002;35:1484–1490. doi: 10.1086/344649. [DOI] [PubMed] [Google Scholar]
- 9.Pop-Vicas AE, D'Agata EM. The rising influx of multidrug-resistant gram-negative bacilli into a tertiary care hospital. Clin Infect Dis. 2005;40:1792–1798. doi: 10.1086/430314. [DOI] [PubMed] [Google Scholar]
- 10.Pop-Vicas A, Tacconelli E, Gravenstein S, Bing L, D'Agata E. Influx of multidrug-resistant, gram-negative bacteria in the hospital setting and the role of elderly patients with bacterial bloodstream infection. Infect Control Hosp Epidemiol. 2009;30:325–31. doi: 10.1086/596608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Fallon E, Pop-Vicas A, D'Agata E. The emerging threat of multidrug-resistant gram-negative organisms in long-term care facilities. J Gerontol A Biol Sci Med Sci. 2009;64:138–141. doi: 10.1093/gerona/gln020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiener J, Quinn JP, Bradford PA, et al. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA. 1999;281:517–523. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]
- 13.Smith PW, Bennett G, Bradley S, et al. SHEA/APIC guideline: infection prevention and control in the long-term care facility, 2008. Infect Control Hosp Epidemiol. 2008;29:785–814. doi: 10.1086/592416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley SE. Double, double, toil and trouble: infections still spreading in long-term-care facilities. Infect Control Hosp Epidemiol. 2005;26:227–230. doi: 10.1086/502531. [DOI] [PubMed] [Google Scholar]
- 15.Bradley SF. Issues in the management of resistant bacteria in long-term-care facilities. Infect Control Hosp Epidemiol. 1999;20:362–366. doi: 10.1086/501637. [DOI] [PubMed] [Google Scholar]
- 16.Bonomo RA. Multiple antibiotic-resistant bacteria in long-term-care facilities: an emerging problem in the practice of infectious diseases. Clin Infect Dis. 2000;31:1414–1422. doi: 10.1086/317489. [DOI] [PubMed] [Google Scholar]
- 17.Trick WE, Weinstein RA, DeMarais PL, et al. Colonization of skilled-care facility residents with antimicrobial-resistant pathogens. J Am Geriatr Soc. 2001;49:270–276. doi: 10.1046/j.1532-5415.2001.4930270.x. [DOI] [PubMed] [Google Scholar]
- 18.Pop-Vicas A, Mitchell SL, Kandel R, Schreiber R, D'Agata EM. Multi-drug-resistant gram-negative bacteria in a long-term care facility: prevalence and risk factors. J Am Geriatr Soc. 2008;56:1276–1280. doi: 10.1111/j.1532-5415.2008.01787.x. [DOI] [PubMed] [Google Scholar]
- 19.Siegel JD, Rhinehart E, Jackson M, Chiarello L. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control. 2007;35:S165–193. doi: 10.1016/j.ajic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 20.D'Agata E, Venkataraman L, DeGirolami P, Samore M. Molecular epidemiology of acquisition of ceftazidime-resistant gram-negative bacilli in a nonoutbreak setting. J Clin Microbiol. 1997;35:2602–2605. doi: 10.1128/jcm.35.10.2602-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wingard E, Shlaes JH, Mortimer EA, Shlaes DM. Colonization and cross-colonization of nursing home patients with trimethoprim-resistant gram-negative bacilli. Clin Infect Dis. 1993;16:75–81. doi: 10.1093/clinids/16.1.75. [DOI] [PubMed] [Google Scholar]
- 22.MacArthur RD, Lehman MH, Currie-McCumber CA, Shlaes DM. The epidemiology of gentamicin-resistant Pseudomonas aeruginosa on an intermediate care unit. Am J Epidemiol. 1988;128:821–827. doi: 10.1093/oxfordjournals.aje.a115035. [DOI] [PubMed] [Google Scholar]
- 23.Bradley SF, Terpenning MS, Ramsey MA, et al. Methicillin-resistant Staphylococcus aureus: colonization and infection in a long-term care facility. Ann Intern Med. 1991;115:417–422. doi: 10.7326/0003-4819-115-6-417. [DOI] [PubMed] [Google Scholar]
- 24.Hawes C, Morris JN, Phillips CD, Mor V, Fries BE, Nonemaker S. Reliability estimates for the Minimum Data Set for nursing home resident assessment and care screening (MDS) Gerontologist. 1995;35:172–178. doi: 10.1093/geront/35.2.172. [DOI] [PubMed] [Google Scholar]
- 25.Morris JN, Hawes C, Fries BE, et al. Designing the national resident assessment instrument for nursing homes. Gerontologist. 1990;30:293–307. doi: 10.1093/geront/30.3.293. [DOI] [PubMed] [Google Scholar]
- 26.Reisberg B, Ferris SH, de Leon MJ, Crook T. The Global Deterioration Scale for assessment of primary degenerative dementia. Am J Psychiatry. 1982;139:1136–1139. doi: 10.1176/ajp.139.9.1136. [DOI] [PubMed] [Google Scholar]
- 27.Katz S. Assessing self-maintenance: activities of daily living, mobility, and instrumental activities of daily living. J Am Geriatr Soc. 1983;31:721–727. doi: 10.1111/j.1532-5415.1983.tb03391.x. [DOI] [PubMed] [Google Scholar]
- 28.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 29.Facklam RR, Sahm DF. Enterococcus. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. 6th. Washington, DC: ASM Press; 1995. pp. 308–314. [Google Scholar]
- 30.Murray PR, Barron EJ, Pfaller MA. Manual of Clinical Microbiology. Washington, DC: ASM Press; 1998. [Google Scholar]
- 31.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemmen SW, Hafner H, Zolldann D, Stanzel S, Lutticken R. Distribution of multi-resistant gram-negative versus gram-positive bacteria in the hospital inanimate environment. J Hosp Infect. 2004;56:191–197. doi: 10.1016/j.jhin.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Scott E, Bloomfield SF. The survival and transfer of microbial contamination via cloths, hands and utensils. J Appl Bacteriol. 1990;68:271–278. doi: 10.1111/j.1365-2672.1990.tb02574.x. [DOI] [PubMed] [Google Scholar]
- 34.Bures S, Fishbain JT, Uyehara CF, Parker JM, Berg BW. Computer keyboards and faucet handles as reservoirs of nosocomial pathogens in the intensive care unit. Am J Infect Control. 2000;28:465–471. doi: 10.1067/mic.2000.107267. [DOI] [PubMed] [Google Scholar]
- 35.Catalano M, Quelle LS, Jeric PE, Di Martino A, Maimone SM. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J Hosp Infect. 1999;42:27–35. doi: 10.1053/jhin.1998.0535. [DOI] [PubMed] [Google Scholar]
- 36.Neely AN. A survey of gram-negative bacteria survival on hospital fabrics and plastics. J Burn Care Rehabil. 2000;21:523–527. doi: 10.1097/00004630-200021060-00009. [DOI] [PubMed] [Google Scholar]
- 37.D'Agata EM, Venkataraman L, DeGirolami P, Samore M. Molecular epidemiology of ceftazidime-resistant gram-negative bacilli on inanimate surfaces and their role in cross-transmission during nonoutbreak periods. J Clin Microbiol. 1999;37:3065–3067. doi: 10.1128/jcm.37.9.3065-3067.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drinka PJ, Stemper ME, Gauerke CD, Miller JM, Reed KD. The identification of genetically related bacterial isolates using pulsed field gel electrophoresis on nursing home units: a clinical experience. J Am Geriatr Soc. 2004;52:1373–1377. doi: 10.1111/j.1532-5415.2004.52371.x. [DOI] [PubMed] [Google Scholar]
- 39.Terpenning MS, Bradley SF, Wan JY, Chenoweth CE, Jorgensen KA, Kauffman CA. Colonization and infection with antibiotic-resistant bacteria in a long-term care facility. J Am Geriatr Soc. 1994;42:1062–1069. doi: 10.1111/j.1532-5415.1994.tb06210.x. [DOI] [PubMed] [Google Scholar]
- 40.Wendt C, Svoboda D, Schmidt C, Bock-Hensley O, von Baum H. Characteristics that promote transmission of Staphylococcus aureus nursing homes in German nursing homes. Infect Control Hosp Epidemiol. 2005;26:816–821. doi: 10.1086/502499. [DOI] [PubMed] [Google Scholar]


