Abstract
Although it is possible to use a tartrate-resistant acid phosphatase (TRAP) stain to assist in identifying osteoclasts, a separate method is needed to determine the bone resorption activity of osteoclasts. Since osteoclasts leave “pits” after bone matrix resorption (Charles et al., 2014), it is possible to stain pits as a method of measuring osteoclast bone resorption activity. The pit assay protocol enables researchers to stain bony slices that were co-cultured with osteoclasts with toluidine blue in order to allow the visualization, capture, and analysis of osteoclast resorptive activity based on the number, size and depth of pits (Zhou et al., 2015). The pit assay protocol is separated into three sequential stages: Preparation of bone slices (1); preparation of osteoclast precursors (Ross et al., 2006; Teitelbaum et al., 2000) (2), and bone resorption pit assay (3).
Materials and Reagents
Parafilm
Razor blade
Tissue culture tubes (standard 15 or 50 ml conical TC tubes)
5 ml syringe with 25 G needle
Cell strainer (70 μm) (Corning, catalog number: 352350)
Cell culture 96 well plates
Tissue culture petri dish
Whatman filter paper (Sigma-Aldrich, catalog number: WHA10347509)
6- to 10-week-old mice
Phosphate buffered saline (PBS), sterilized, pH 7.4 (Thermo Fisher Scientific, catalog number: 10010023)
Dulbecco’s modified Eagle medium, high glucose (Thermo Fisher Scientific, Gibco™, catalog number: 11965092)
Penicillin/streptomycin (10,000 U/ml) (Thermo Fisher Scientific, catalog number: 15140-122)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, Gibco™, catalog number: 16000044)
MEMα, nucleosides (Thermo Fisher Scientific, catalog number: 12571-063)
Macrophage-stimulating factor (M-CSF) (PeproTech, catalog number: 315-02) and receptor activated nuclear factor k-B ligand (RANKL) (PeproTech, catalog number: 315-11)
Bovine femurs (from meat market)
Ethanol
Glutaraldehyde (Sigma-Aldrich, catalog number: G5882)
Toluidine blue (Sigma-Aldrich, catalog Number: T3260-5G)
MilliQ water
Acid phosphatase, leukocyte (TRAP) Kit (Sigma-Aldrich, catalog number: 387A-1KT)
Sodium borate 10-hydrate (Fisher Scientific, catalog number: 02-003-999)
Trypan-blue (Thermo Fisher Scientific, catalog number: T6146-5G)
Ammonium chloride (NH4Cl) (Sigma-Aldrich, catalog number: A9434-500G)
Trizma® hydrochloride (Tris-HCl) (Sigma-Aldrich, catalog number: T5941-100G)
Red blood cell (RBC) lysis buffer (Sigma-Aldrich, catalog number: R7757) (see Recipes)
Osteoclast precursor culture media (see Recipes)
Osteoclast differentiation media (see Recipes)
1% toluidine blue (see Recipes)
Glutaraldehyde solution (see Recipes)
Equipment
Forceps scissors
Hacksaw
Scalpel
Water ultrasonicator
Isomet low speed saw (BUEHLER, catalog number: 111280) with wafering blade
Light microscope
Procedure
A. Preparation of bony slices
Note: If you have pre-made bone slices, please skip to step B.
The diaphysis of bovine femurs is cut transversely into 2–3 cm cylinders with a hacksaw as shown in Figure 1A.
Cut the cylinder into three segments and lift the marrow out as shown in Figure 1B. Any adherent muscle, marrow or periosteum is cleaned off with a scalpel.
The segments are then sonicated in warm water with a little detergent several times until the bone is clean. Repeat two to three times until the bone is clean. In the original experiment, segments were sonicated for 2 min at 50–60 Hz.
Bony segments are given a final clean of any adherent tissue with a scalpel, wrapped in parafilm and stored under frozen condition (−20 °C). Segments from the same cylinder are given the same label.
To make 4.4 × 4.4 × 0.2 mm bone slices, bony segment is transversely cut into 200 μm thick block with the Isomet low speed saw with the wafering blade. These 200 μm slices are used to produce the final 4.4 × 4.4 × 0.2 mm bone slices.
The stack of 200 μm slices was then clamped and a strip 4.4 mm wide cut was made. The stack of bone strips is then clamped at 90 degrees to the clamp and the final bone slices are produced by cutting at 4.4 mm intervals. These cuttings of the bony segments finally led to produce the 4.4 × 4.4 × 0.2 mm bone slices.
Store bone slices in 70% ethanol until use.
Figure 1. Illustration of transverse and segmental cuts of bovine femur.
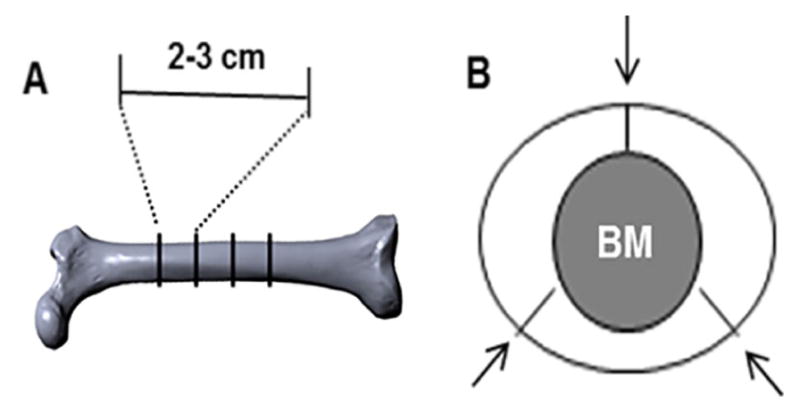
A. An image of bovine femur depicting diaphyseal portions that are transversely cut into 2–3 cm cylinders; B. Bony cylinders referred in (A) are further cut into three segments as indicated (arrows).
B. Preparation of bone marrow-derived osteoclast precursors
Dissect femur and tibia from 6- to 10-week-old mice under sterile condition and place them in cold PBS. Remove all soft tissues, such as muscle, fat, etc. with forceps scissors and razor blade.
Wash femurs and tibia with sterile PBS twice and cut both their ends.
Flush marrow cells into tissue culture tubes (standard 15 or 50 ml conical TC tubes) from both ends with 5 ml syringe (25 G needle) and DMEM (supplemented with P/S and 1% FBS). About 40 ml DMEM (supplemented with P/S and 1% FBS) is needed to flush marrow cells.
Pelletize marrow cells by centrifuging 637 × g for 5 min at room temperature and discard the supernatant; Re-suspend cells into 3 ml of red blood cell (RBC) lysis buffer at room temperature for 3–4 min to destroy red blood cells.
Add 7 ml αMEM media (with 10% FBS) to cells to neutralize the effect of RBC buffer; Pass the cells through a 70 μm cell strainer into a 50 ml conical tube.
Pelletize cells by centrifuging 637 × g for 5 min at room temperature and discard the supernatant; Wash cells once with 10 ml sterilized PBS and count cell numbers.
Re-suspend cells at 5 × 105 cells/ml into complete αMEM media supplemented with 20 ng/ml of M-CSF.
Culture cells at 5% CO2 and 37 °C overnight. Collect non-adherent cells and count their numbers and discard adherent cells (most of them are fibroblasts, mature macrophages and stromal cells). The expected yield of non-adherent cells is 5 × 106 cells/ml.
For pit assay, incubate the cells with the bone slice as described in step A6.
C. Bone resorption pit assay
Label bone slices and sterilize them in a sterile hood with UV light the day before the experiment.
On the day of the experiment, place bone slices in a 96 well (1 slice/well) labeled side down. Prepare bone marrow cells and plate at 50,000 cells/well or appropriate cell numbers and culture them under 200 μl of osteoclast differentiation media in the well.
Cells are cultured for 10–14 days with feeding every 2–3 days. Culture media must be replenished completely during each feeding.
Bone slices are then fixed with 2.5% glutaraldehyde in PBS for 30 min at room temperature.
-
TRAP (Sidqui et al., 1995) stain cells and examine microscopically to observe TRAP (+) osteoclasts.
Figure 2 shows an example of TRAP staining from tibia sections in 8-week old male mice. The three red arrows point to TRAP+ osteoclasts (which are stained red). A magnified image of an individual TRAP+ osteoclast is shown in the upper right hand corner.
-
Remove cells from bone slice by sonicating 5–15 min in distilled water at 50–60 Hz to dislodge the cells from the bone slice.
Note: Sonicate bone slices individually in glass beaker in order to prevent misidentification of the bone slices if identification numbers come off.
Re-number the bone slices and completely dry bones on Whatman filter paper before staining.
Prepare 1% toluidine blue the day before experiment.
-
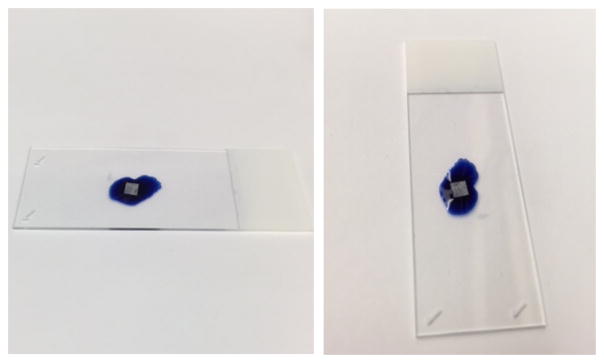
Stain bones 4 min total on 1% toluidine blue in 1% sodium borate. Stain by placing droplet (20 μl) of stain onto a Petri dish. Carefully place the bone slice onto the side of the droplet so that the slice is leaning onto the surface of the stain and not submerged under the surface of the drop. In other words, the bone slice should not break the surface tension of the droplet and get submerged by the droplet; once placed in a leaning position on the surface of the droplet, it should remain on the surface of the droplet.
Two images of the result of step C9 are shown below in Figure 3. In each image, the bone slice is leaning on the 1% toluidine blue in 1% sodium borate droplet. The bone slice in each picture is not submerged under the 1% toluidine blue in 1% sodium borate droplet.
-
Remove the bone slice from the droplet stain, and rinse by swirling bone slice in distilled water. Air dry and examine with light microscope.
Use Olympus scope and turn on light source from box on left so that the light source comes from the top of the scope.
-
Take pictures under microscope and evaluate the number and size of pit formation.
Note: Pits should be stained in the color of toluidine.
Figure 2. Image of osteoclasts positive for TRAP on the tibia section of an 8-week old male mouse.
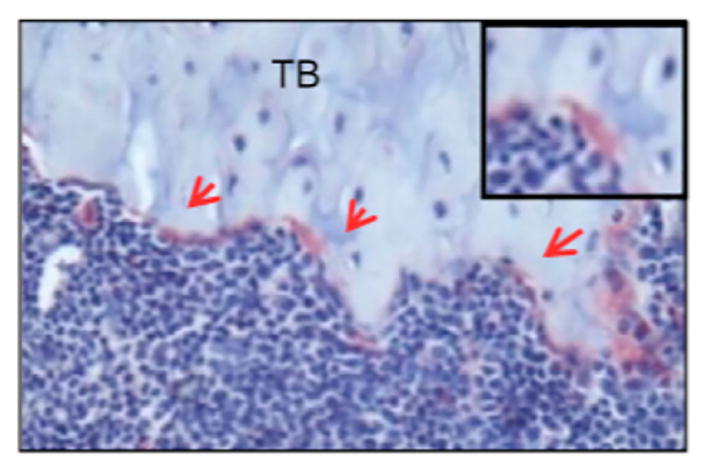
Tibia sections from 8-week old mice were stained with TRAP and counterstained with hematoxylin blue. Multinuclear osteoclasts appeared in red on trabecular bone surface (arrows). This representative example was taken from “SHP2 regulates osteoclastogenesis by promoting preosteoclasts fusion”, which is published in The FASEB Journal (Zhou et al., 2015).
Figure 3. Illustrative example of the proper position of bone slice on toluidine stain.
A droplet of toluidine stain was pipetted on a microscope slide. Using tweezers, the bone slice was placed onto the surface of the toluidine stain. As shown in Figure 3, the proper configuration of the bone slice is on the surface of the toluidine stain droplet.
Representative data
The following three representative examples have been taken from Zhou et al. (2015).
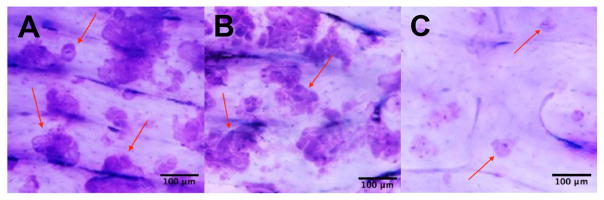
The “pits” created by the pit assay procedure are the blue areas in Figure 4A, Figure 4B, and Figure 4C. The red arrows in Figure 4A, 4B and 4C point to the pits created. The blue staining is due to the toluidine blue used in the procedure. The samples represented in Figure 4A and 4B both have a decent amount of pits. The sample represented in figure 4C has a relative absence of pits.
Figure 4. Illustrative examples of positive results of pit assays.
The pit assay experiments shown above were conducted on dentin slices co-cultured with osteoclasts for 10 days in vitro. The pit assay results in 4A and 4B were generated without the addition of an extraneous SHP2 inhibitor. In 4C, however, SHP2 was inhibited with NSC-87877 during the co-culture process to block mature osteoclast formation (Zhou et al., 2015). Hence, a lack of mature osteoclasts was induced, which caused a lack of resorption pits as shown in 4C. Scale bar, 100 μm.
The mice strain used in the original experiment is the C57BL/6 strain.
Recipes
-
RBC lysis buffer
0.747% NH4Cl
0.017% Tris-HCl
-
Osteoclast precursor culture media
αMEM supplemented with 1% penicillin/streptomycin (P/S), 10% FBS (heat-inactivated) and 20 ng/ml of M-CSF
-
Osteoclast differentiation media
Osteoclast precursor culture media supplemented with 100 ng/ml RANKL
-
1% toluidine blue
1% toluidine blue is prepared by combining 1 g sodium borate 10-hydrate in 100 ml dH2O.
Dissolve 1 g toluidine blue in this solution and allow it to stand overnight
Filter staining solution using Whatman filter paper and store at room temperature
-
Glutaraldehyde solution
Glutaraldehyde solution is 25% in H2O
Dilute to 2.5% with ddH2O for use
Acknowledgments
This protocol was adapted from the previously published paper Zhou et al. (2015). This work was supported in part by the U. S. National Institutes of Health (NIH) National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R21AR57156 (to W. Y.) and NIH National Institute of General Medical Sciences Grant P20 GM103468. This study was also aided by a grant from the Pediatric Orthopaedic Society of North America and the Orthopaedic Research and Education Foundation (to W. Y.), and the U. S. Department of Agriculture Research Service program Grant #5450-51000-046-00D (to J. C.).
References
- 1.Cao J, Wu Q, Qin YX, Reginato AM, Ehrlich MG, Yang W. SHP2 regulates osteoclastogenesis by promoting preosteoclast fusion. FASEB J. 2015;29(5):1635–1645. doi: 10.1096/fj.14-260844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charles JF, Aliprantis AO. Osteoclasts: more than ‘bone eaters’. Trends Mol Med. 2014;20(8):449–459. doi: 10.1016/j.molmed.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross FP. M-CSF, c-Fms, and signaling in osteoclasts and their precursors. Ann N Y Acad Sci. 2006;1068:110–116. doi: 10.1196/annals.1346.014. [DOI] [PubMed] [Google Scholar]
- 4.Sidqui M, Collin P, Vitte C, Forest N. Osteoblast adherence and resorption activity of isolated osteoclasts on calcium sulphate hemihydrate. Biomaterials. 1995;16(17):1327–1332. doi: 10.1016/0142-9612(95)91048-4. [DOI] [PubMed] [Google Scholar]
- 5.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289(5484):1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Mohan A, Moore DC, Lin L, Zhou FL, Cao J, Wu Q, Qin YX, Reginato AM, Ehrlich MG, Yang W. SHP2 regulates osteoclastogenesis by promoting preosteoclast fusion. FASEB J. 2015;29(5):1635–1645. doi: 10.1096/fj.14-260844. [DOI] [PMC free article] [PubMed] [Google Scholar]