Abstract
MicroRNAs (miRNAs/miRs) have been identified as important post-transcriptional regulators in healthy liver physiology and liver diseases. However, the clinical significance of miR-638 in hepatocellular carcinoma (HCC) remains unclear. The aim of the present study was to investigate the status of miR-638 expression in HCC and to determine its clinical significance. The expression of miR-638 was evaluated in 60 HCC tissues samples and HCC SMMC-7721, HepG2 and Hep3B cell lines using reverse transcription-quantitative polymerase chain reaction. The association between the expression of miR-638 and the clinicopathological characteristics of patients with HCC was analyzed. The proportion of HCC patients with low miR-638 expression was identified as 68.3% (41/60). Furthermore, significantly lower miR-638 expression was identified in HCC tissue samples compared with the healthy control group (P=0.031). miR-638 expression was significantly lower in SMMC-7721 (P=0.021), HepG2 (P=0.005) and Hep3B (P=0.003) cells compared with the healthy human hepatic HL-7702 cell line. In addition, miR-638 expression was correlated with α-fetoprotein levels (P=0.042) and portal vein invasion (P=0.025). The area under curve was identified as 0.71 (95% confidence interval=0.63–0.79; P=0.001). The cut-off value for miR-638 was the median 2−Δ∆Cq=0.125. In conclusion, miR-638 may be involved in the progression of HCC and act as a potential biomarker for the prediction of HCC.
Keywords: miR-638, hepatocellular carcinoma, clinicopathological characteristics, formalin-fixed paraffin-embedded tissue, reverse transcription-quantitative polymerase chain reaction
Introduction
MicroRNAs (miRNAs or miRs) are small (19–25 nucleotides) endogenous, non-coding RNAs that regulate gene expression via inhibiting translation and/or promoting the degradation of target messenger RNA (mRNA) at the post-transcriptional level (1,2). Evidence highlights the significance of miRNAs as essential regulators of numerous biological processes, including cell proliferation, differentiation, apoptosis and metastasis (3). However, the precise mechanisms underlying the function of the majority of identified miRNAs remain unclear.
Dysregulation of miRNAs has been identified in numerous types of human tumors (4–6), suggesting that they serve essential roles in tumorigenesis and tumor development. Previous studies have reported that miR-638 expression is significantly downregulated, and may serve a role as a cancer-suppressor gene in human gastric cancer (7), breast cancer (8), basal cell carcinoma (9) and chronic lymphocytic leukemia (10). A previous study reported that the expression of miR-638 was markedly upregulated in hepatocellular liver cancer compared with healthy liver tissue (11). Notably, by using microarrays, miR-638 has been identified as one of the miRNAs that serve a role in the invasive-metastatic cascade in hepatocellular carcinoma (HCC) (12). In addition, the downregulation of miR-638 promotes the invasion and proliferation of human colorectal carcinoma (13) and non-small cell lung cancer (NSCLC) (14). miR-638 has been functionally associated with the hepatitis B virus (HBV) life cycle (15). However, the clinical significance of miR-638 in the treatment of patients with HCC remains unclear. In the present study, the expression of miR-638 in HCC was investigated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Furthermore, the association between the expression of miR-638 and the clinicopathological characteristics of patients with HCC was analyzed.
Materials and methods
Human tissue specimens
Formalin-fixed paraffin-embedded (FFPE) tissue samples, including 60 HCC and adjacent healthy liver tissue samples, were collected from patients who underwent curative hepatic resection for HCC between January 2008 and June 2010 at the Department of Pathology of the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China). None of the patients had received local or systemic therapy prior to surgery, and the tumor and matched adjacent healthy tissue samples were histologically confirmed. Written informed consent was obtained from all patients and the study was approved by the Institute Research Ethics Committee at the Cancer Center of Xi'an Jiaotong University (Xi'an, China). The relevant clinicopathological characteristics of the patients were collected from their clinical records.
Cell lines and culture conditions
Human SMMC-7721, HepG2 and Hep3B liver cancer cell lines, and the healthy human HL-7702 liver cell line were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). These cells were cultured in Dulbecco's modified Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and RPMI 1640 medium (GE Healthcare Life Sciences) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 100 U/ml penicillin and 100 U/ml streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C with 5% CO2.
RNA extraction
Total RNA was extracted from human FFPE tissue samples and all cell lines using an E.Z.N.A.® FFPE RNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA) and TriPure RNA Isolation Reagent (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturers' protocols. The RNA concentration and purity were determined and evaluated using the NanoDrop® ND-1000 (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). The absorbance (A)260:A280 ratio was used to estimate the purity of total RNA.
RT-qPCR
Complementary DNA (cDNA) was synthesized from 1 µg of RNA following the manufacturer's protocol (Takara Biotechnology Co., Ltd, Dalian, China). The 10-µl final reaction volume consisted of 1 µg of total RNA, 2 µl 5X PrimeScript® Buffer, 0.5 µl PrimeScript® RT Enzyme Mix and 1 µl RT primer (Takara Biotechnology Co., Ltd). The reaction was incubated for 15 min at 37°C followed by 5 sec at 85°C.
qPCR analyses were performed using Power SYBR® Green PCR Master Mix (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. qPCR reactions were performed using the Applied Biosystems® 7500 PCR System (Thermo Fisher Scientific, Inc.). The forward and reverse primers for miR-638 and U6 are presented in Table I. The following thermocycling conditions were performed: 95°C for 1 min; 40 cycles of 95°C for 10 sec; and 58°C for 40 sec. The 20-µl qPCR reaction volume consisted of 10 µl SYBR Prime Ex Taq™ II (2X) (Takara Biotechnology Co., Ltd), 1 µl forward primer (10 mM), 1 µl reverse primer (10 mM), 2 µl cDNA (<100 ng used per reaction) and 6 µl H2O. Results were normalized to the expression of U6 and the relative quantification of miRNA expression was calculated with the 2−ΔΔCq method, whereby 2−ΔΔCq = 2−[ΔCq (HCC) - ΔCq (control)] and ΔCq = Cq miR-638 - Cq U6 (16). A Cq value of 35 was assigned as the cut-off value for defining samples as non-detected. All reactions were performed in triplicate.
Table I.
RT, forward and reverse primers for RT-quantitative polymerase chain reaction analysis of miR-638 and U6.
| Primer | Sequence |
|---|---|
| miR-638 | |
| RT | 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCG |
| GCAATTGCACTGGATACGACAGGCCGC-3′ | |
| Forward | 5′-ATCCAGTGCGTG TCGTG-3′ |
| Reverse | 5′-TGCTAGGGATCGC GGGCGGGTG-3′ |
| U6 | |
| RT | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| Forward | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| Reverse | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
RT, reverse transcription; miR, miRNA.
miRNA target prediction
Established miRNA-target prediction tools were used to identify potential target genes of miR-638. The following eight prediction databases were used: DIANA TOOLS (http://diana.imis.athena-innovation.gr/), microRNA.org (http://www.microrna.org/microrna/home.do), miRDB (http://mirdb.org/miRDB/download.html), TargetMiner (http://www.isical.ac.in/~bioinfo_miu/mirnalist.html), TargetScan (http://targetscan.org/), RNA22-HSA (https://cm.jefferson.edu/rna22/), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) and RegRNA (http://regrna.mbc.nctu.edu.tw/html/tutorial.html). The top 100 target genes in the majority of databases were recorded and a comparison was made between them. Experimentally verified targets and only predicted target genes in mammals that appeared >4 times were noted in the current study.
Statistical analysis
All statistical analyses were performed using SPSS software (version 16.0; SPSS, Inc., Chicago, IL, USA). Student's t test was performed to analyze the significance of differences between groups. To identify the association between clinicopathological characteristics of patients with HCC and the expression of miR-638, the c2 and Fisher's exact tests were performed. A receiver operating characteristic (ROC) curve was produced to evaluate the efficacy of miR-638 expression when distinguishing between the HCC and healthy liver tissue samples. All tests were two-tailed. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-638 expression is frequently decreased in human HCC tissue and cell lines compared with the healthy control groups
The expression of miR-638 was determined in 60 HCC and corresponding healthy tissue samples using RT-qPCR and normalized to the control U6. As illustrated in Fig. 1, miR-638 expression was significantly lower in HCC tissue compared with that in healthy tissue samples (P=0.031). According to the median tumor (T)/non-tumor (N) tissue ratio of miR-638 expression among the 60 HCC samples analyzed, 41 (T/N>1.0, 68.3%) cases demonstrated low expression of miR-638 in HCC tissue compared with healthy tissue samples. To further validate these results, the expression of miR-638 in cultured HCC cells was analyzed, and it was identified that the miR-638 expression was significantly lower in SMMC-7721 (P=0.021), HepG2 (P=0.005) and Hep3B (P=0.003) cells compared with that in HL-7702 cells (Fig. 2).
Figure 1.
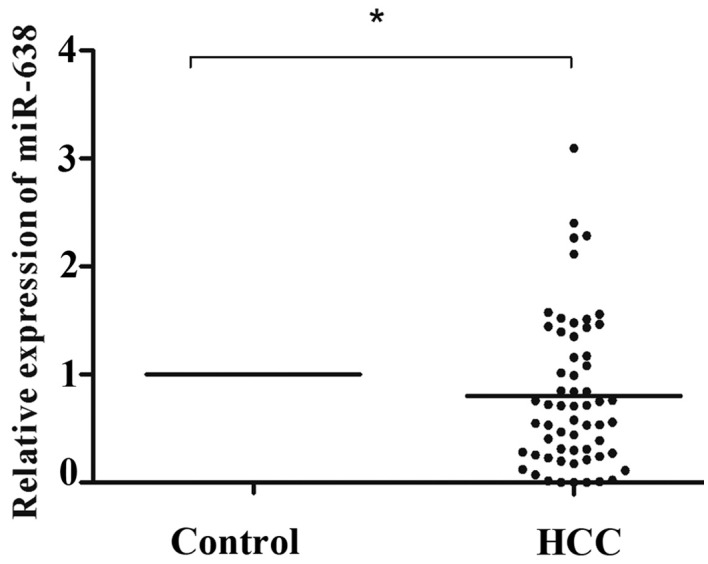
Low expression of miR-638 relative to U6 in 60 HCC tissue samples compared with healthy tissue samples (P=0.031). *P<0.05, Student's t test. miR, microRNA; HCC, hepatocellular carcinoma.
Figure 2.
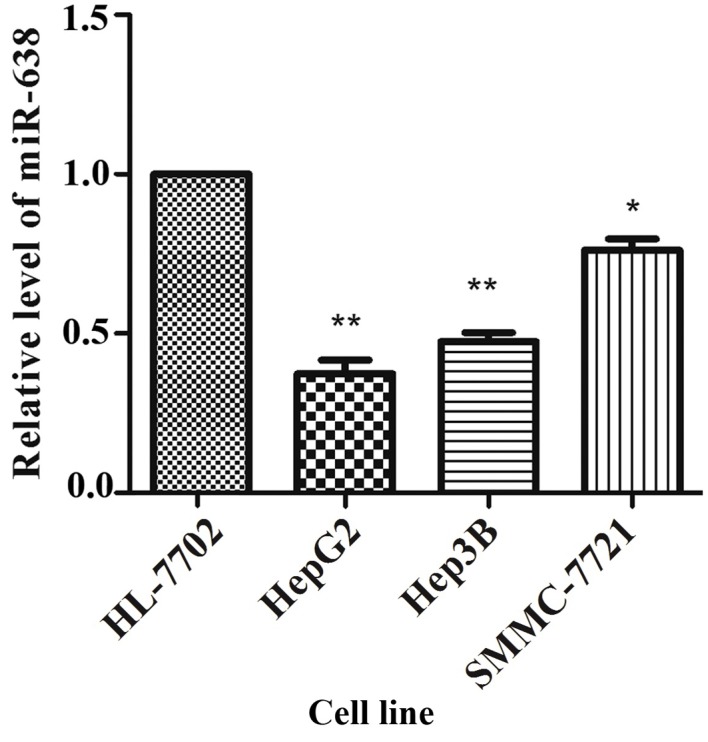
Low expression of miR-638 relative to U6 in the hepatocellular carcinoma cell lines SMMC-7721 (P=0.021), HepG2 (P=0.005) and Hep3B (P=0.003) compared with the healthy human hepatic HL-7702 cell line. **P<0.01, *P<0.05, Student's t test. miR, microRNA.
miR-638 expression and clinicopathological characteristics
The associations between clinicopathological factors and miR-638 levels were analyzed using the chi-square test and Fisher's exact test, and the patients' clinicopathological characteristics are illustrated in Table II. No significant association was identified between low miR-638 expression and age (P=0.781), gender (P=0.089), HBV infection (P=0.114), tumor size (P=0.774), tumor node metastasis stage (P=0.146) or distant organ hepatic metastasis (P=0.083). However, the relative miR-638 expression levels were positively correlated with α-fetoprotein (AFP) levels and portal vein invasion (P=0.042, P=0.025, respectively).
Table II.
Association between the relative expression of miR-638 and the clinicopathological characteristics of patients with hepatocellular carcinoma.
| miR-638 expression | ||||
|---|---|---|---|---|
| Clinicopathological characteristics | No. of cases (n=60) | High (19) | Low (41) | P-value |
| Age, years | 0.781 | |||
| <50 | 26 | 9 | 17 | |
| ≥50 | 34 | 10 | 24 | |
| Gender | 0.089 | |||
| Female | 13 | 7 | 6 | |
| Male | 47 | 12 | 35 | |
| HBV infection status | 0.114 | |||
| + | 44 | 11 | 33 | |
| − | 16 | 8 | 8 | |
| Tumor size, cm | 0.774 | |||
| ≤5 | 22 | 6 | 16 | |
| >5 | 38 | 13 | 25 | |
| AFP level, µg/l | 0.042 | |||
| ≤20 | 20 | 10 | 10 | |
| >20 | 40 | 9 | 31 | |
| TNM stage | 0.146 | |||
| I+II | 20 | 9 | 11 | |
| III+IV | 40 | 10 | 30 | |
| Portal vein invasion status | 0.025 | |||
| Yes | 22 | 3 | 19 | |
| No | 38 | 16 | 22 | |
| Distant organ hepatic metastasis status | 0.083 | |||
| Yes | 19 | 3 | 16 | |
| No | 41 | 16 | 27 | |
miR, microRNA; HBV, hepatitis B virus; TNM, tumor node metastasis; AFP, α-fetoprotein.
ROC curve analysis for the diagnostic value of miRNA-638 expression in HCC tissue
ROC curve analysis was implemented to identify the predictive value of miRNA-638 level in HCC. As illustrated in Fig. 3, the area under the curve (AUC) was 0.71 (95% confidence interval=0.63–0.79; P=0.001). The cut-off value for miR-638 expression was the median 2−Δ∆Cq =0.125.
Figure 3.
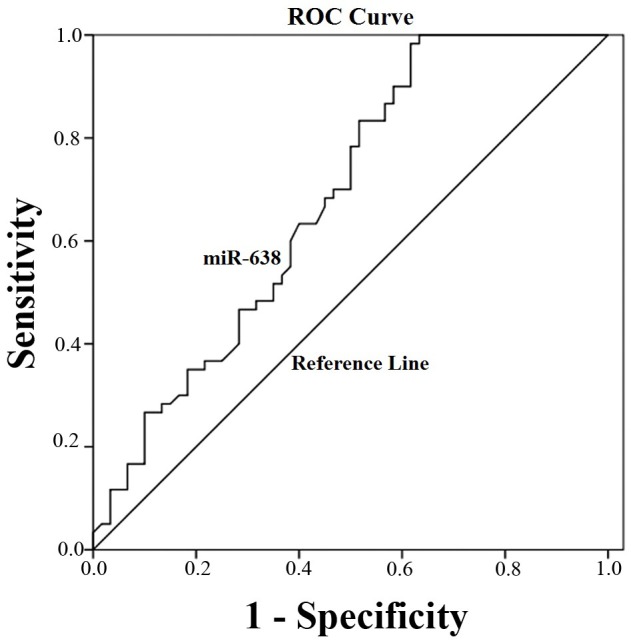
Receiver operating characteristic curve of miR-638 for hepatocellular carcinoma. The area under curve for miR-638 was 0.71 (95% confidence interval=0.63–0.79; P=0.001). miR, microRNA.
Target prediction of miR-638
Following the target prediction analysis using eight databases, 6 validated target genes and 10 qualified target genes were identified in ≥3 different established miRNA-target prediction programs. They were as follows: Tetraspanin 1 (TSPAN1), cyclin-dependent kinase 2 (CDK2), Sp2 transcription factor (Sp2), tumor protein p53 inducible nuclear protein 2 (TP53INP2), SRY-box 2 (SOX2), breast cancer 1 (BRCA1), dishevelled binding antagonist of beta catenin 3 (DACT3), StAR-related lipid transfer (START), StAR-related lipid transfer domain containing 10 (STARD10), protein O-linked mannose N-acetylglucosaminyltransferase 1 (β 1,2-) (POMGNT1), transcription elongation regulator 1 like (TCERG1L), zinc finger protein 281 (ZNF281), vascular endothelial growth factor A (VEGFA), hepatic leukemia factor (HLF), basic, immunoglobulin-like variable motif-containing (BIVM), neuronal PAS domain protein 4 (NPAS4) and muskelin 1 (MKLN1) (Table III).
Table III.
Target prediction of microRNA-638 following searching on eight different databases.
| Database | ||||||||
|---|---|---|---|---|---|---|---|---|
| Target gene | RegRNA | miRDB | TargetScan | RNA22-HSA | Target Miner | DIANA TOOLS | microRNA. org | PITA |
| TSPAN1 | + | + | + | |||||
| CDK2 | + | + | ||||||
| Sp2 | + | + | + | + | ||||
| TP53INP2 | + | + | + | |||||
| SOX2 | + | + | + | + | + | + | ||
| BRCA1 | + | + | + | |||||
| DACT3 | + | + | + | + | ||||
| STARD10 | + | + | + | + | ||||
| POMGNT1 | + | + | + | + | + | |||
| TCERG1L | + | + | + | + | + | + | ||
| ZNF281 | + | + | + | + | ||||
| VEGF | + | + | + | + | ||||
| HLF | + | + | + | + | + | |||
| BIVM | + | + | + | + | + | |||
| NPAS4 | + | + | + | + | + | + | ||
| MKLN1 | + | + | + | + | + | |||
+, the gene appears in the corresponding database. TSPAN1, tetraspanin 1; CDK2, cyclin-dependent kinase 2; Sp2, Sp2 transcription factor; TP53INP2, tumor protein p53 inducible nuclear protein 2; SOX2, SRY-box 2; BRCA1, breast cancer 1; DACT3, dishevelled binding antagonist of beta catenin 3; START, StAR-related lipid transfer; STARD10, StAR-related lipid transfer domain containing 10; POMGNT1, protein O-linked mannose N-acetylglucosaminyltransferase 1 (β 1,2-); TCERG1L, transcription elongation regulator 1 like; ZNF281, zinc finger protein 281; VEGFA, vascular endothelial growth factor A; HLF, hepatic leukemia factor; BIVM, basic, immunoglobulin-like variable motif-containing; NPAS4, neuronal PAS domain protein 4; MKLN1, muskelin 1.
Discussion
miRNA alterations have been identified in numerous human cancer types (4), which can act as oncogenes and/or tumor suppressors (2). Each miRNA has hundreds or thousands of mRNA targets and target genetic regions, such as 3′-untranslated regions (UTRs), 5′-UTRs and coding regions at the transcriptional level, subsequently affecting protein expression (17,18). It has been demonstrated that miRNAs are well preserved in FFPE tissue due to their short length, thus underscoring the suitability of FFPE tissue specimens as appropriate resources for miRNA expression analyses (19–21). Currently, multiple methods are used to identify and quantify miRNAs in tumor samples, including microarray (22), RT-qPCR (23), RNA sequencing (24) and in situ hybridization (25).
In the present study, RT-qPCR was performed to assess the expression of miR-638 in FFPE tissue samples from patients with HCC. miRNAs that have been validated and possess the potential to affect cellular function are available at the miRNA database miRBase (www.mirbase.org/) (26). The primers for miR-638 were designed using the above miRNA database in the present study. Previous studies have demonstrated that miR-638 is downregulated in certain human tumor types, such as gastric cancer (7), breast cancer (8), NSCLC (14) and colorectal carcinoma (13). A previous study revealed that the downregulation of miR-638 was present in 68% (41/60) of primary human NSCLC tissue samples compared with that in the paired healthy samples (14).
Lin et al (11) detected the expression of miR-638 in high-density multiple organ tumor and healthy tissue microarrays using in situ hybridization. This study reported that the expression of miR-638 was markedly upregulated in hepatocellular liver cancer tissue (n=20) compared with healthy liver tissue (n=5) samples. However, in the present study, it was suggested that miR-638 serves a role as tumor suppressor in HCC. The proportion of low expression miR-638 was 68.3% (41/60) among the 60 patients with HCC and the relative expression of miR-638 in HCC tissue samples was significantly lower compared with the expression in the healthy control group. The conflicting data on miR-638 expression in HCC may be explained by various factors, such as tissue specificity, different populations and small sample sizes. Notably, miR-638 expression in HCC was detected in a small cohort and the clinicopathological characteristics were not evaluated in the previous study (11).
In the current study, miR-638 expression was detected in a relatively larger sample size, which minimized the effect of individual differences, and a full-panel analysis was performed between miR-638 expression and the clinicopathological characteristics of patients with HCC. In addition, a lower expression of miR-638 was identified in several HCC cell lines (HepG2, SMMC-7721 and Hep3B) compared with the healthy human hepatic HL-7702 cell line. Furthermore, the results of the present study demonstrated that serum miR-638 expression was significantly lower in patients with HCC compared with the healthy control group (P<0.001; data not shown).
To the best of our knowledge, the present study is the first to investigate the association between miR-638 expression and the clinicopathological characteristics of patients with HCC. The results of the current study suggest that low miR-638 expression correlates with AFP levels and portal vein invasion status. Although statistically significant, the correlation between AFP and low miR-638 expression was weak. AFP was predicted as one of the potential target genes of miR-638 following the use of established miRNA-target prediction tools. Further studies into whether AFP expression is regulated by miR-638 are warranted. The results of ROC analysis indicate that miR-638 possesses a moderate diagnostic value in HCC, with an AUC of 0.71. Several studies have demonstrated the potential of miRNAs as predictors of therapeutic response and overall survival rate in patients with cancer. A study performed by Parasramka et al (27) identified miR-638 as one of the garcinol-specific miRNA biomarkers that sensitize human pancreatic adenocarcinoma cells to the combination treatment of garcinol and gemcitabine. Furthermore, miR-638 was identified as a potential predictor of early virological response to interferon treatment in patients with chronic hepatitis B (15). In addition, miR-638 was one of the four miRNAs identified through genome-wide serum miRNA profiling that predict the survival rate of patients with nasopharyngeal carcinoma (28). A previous study demonstrated that the downregulation of miR-638 in colorectal cancer predicts poor survival (13).
As the function and role of miR-638 in HCC remain unclear, the present study aimed to identify the potential target genes of miR-638. Following searching in eight different established miRNA-target prediction programs, the following 16 genes were identified: TSPAN1, CDK2, Sp2, TP53INP2, SOX2, BRCA1, DACT3, STARD10, POMGNT1, TCERG1L, ZNF281, VEGF, HLF, BIVM, NPAS4 and MKLN1. Among these possible target genes, CDK2, Sp2, TP53INP2, SOX2, TSPAN1 and BRCA1 are verified target genes of miR-638. Previous studies have demonstrated that miR-638 inhibits cell proliferation by targeting Sp2 in gastric cancer (7); inhibits cell proliferation and invasion; and regulates cell cycle by targeting TSPAN1 in human colorectal carcinoma (13). The results of a previous study revealed that the downregulation of miR-638 promotes cell proliferation and invasion, and induces mesenchymal-like transition in NSCLC by directly targeting SOX2, whereas the upregulation of miR-638 can reverse the effect (14). These results suggested that miR-638 may serve as a novel tumor suppressor. However, the studies mentioned above were performed in vitro or on animal models, and there are no in vivo studies on miR-638 use as an anticancer therapy at present. In addition to the verified targets of miR-638, the potential target genes of miR-638, including DACT3, STARD10, TSPAN1, POMGNT1, TCERG1L, ZNF281 and VEGF, have been demonstrated to serve important roles in certain types of cancer. Previous studies have revealed that DACT3 serves a role as an epigenetic regulator in colorectal cancer (29), and the loss of STARD10 expression identifies a group of breast cancer patients with poor prognosis, independently of erb-b2 receptor tyrosine kinase 2 and triple negative expression status (30). Furthermore, previous studies have suggested that POMGNT1 serves as a prognostic factor for glioma patient survival (31) and that TCERG1L is a risk marker for colon cancer in patients with ulcerative colitis (32). ZNF281 has been identified to be involved in epithelial-mesenchymal transition and cancer (33). Previous studies have demonstrated that VEGF has various effects on several types of cancer, including the promotion angiogenesis, invasion and migration (34,35). Presently, to the best of our knowledge, there are no data that associate cancer with BIVM, NPAS4 or MKLN1. The predicted and experimentally verified targets of miR-638 may serve an essential role in tumorigenesis and progression. A limitation of the present study is the relatively small sample size. Thus, a larger cohort study is required to establish the diagnostic value of miR-638 in HCC. Furthermore, in vitro and in vivo experiments are required to define the role and underlying mechanism of miR-638 involvement in the development and progression of HCC.
In conclusion, the results of the present study have demonstrated for the first time that the expression of miR-638 is frequently decreased in HCC, and is correlated with AFP levels and portal vein invasion status. This suggests that miR-638 serves a significant role in the development and progression of HCC. The findings of the present study and the predicted target genes identified suggest that miR-638 acts as an oncomiR in HCC tumorigenesis and progression. Furthermore, it has been suggested that miR-638 has a moderate diagnostic value in HCC. Therefore, miR-638 may be considered as a potential novel predictor of HCC and a target for promising alternatives of specific therapeutic treatment for patients with HCC.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Berindan-Neagoe I, Pdel Monroig C, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: The key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014:896206. doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Zhao LY, Yao Y, Han J, Yang J, Wang XF, Tong DD, Song TS, Huang C, Shao Y. miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig Dis Sci. 2014;59:1743–1753. doi: 10.1007/s10620-014-3087-5. [DOI] [PubMed] [Google Scholar]
- 8.Tan X, Peng J, Fu Y, An S, Rezaei K, Tabbara S, Teal CB, Man YG, Brem RF, Fu SW. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014;16:435. doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, Altmeyer P, Bechara FG. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167:847–855. doi: 10.1111/j.1365-2133.2012.11022.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang YH, Liu P, Hong M, Miao KR, Liu P, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33:1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 11.Lin Y, Zeng Y, Zhang F, Xue L, Huang Z, Li W, Guo M. Characterization of microRNA expression profiles and the discovery of novel microRNAs involved in cancer during human embryonic development. PLoS One. 2013;8:e69230. doi: 10.1371/journal.pone.0069230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, Wang L, Zhou J, Ren ZG, Li YX, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Fei B, Wang Q, Song M, Yin Y, Zhang B, Ni S, Guo W, Bian Z, Quan C, et al. MicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinoma. Oncotarget. 2014;5:12083–12096. doi: 10.18632/oncotarget.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Y, Wu Y, Liu B, Wang P, Chen Y. Downregulation of miR-638 promotes invasion and proliferation by regulating SOX2 and induces EMT in NSCLC. FEBS Lett. 2014;588:2238–2245. doi: 10.1016/j.febslet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 18.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′UTR as in the 3′UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiró-Chova L, Peña-Chilet M, López-Guerrero JA, García-Giménez JL, Alonso-Yuste E, Burgues O, Lluch A, Ferrer-Lozano J, Ribas G. High stability of microRNAs in tissue samples of compromised quality. Virchows Arch. 2013;463:765–774. doi: 10.1007/s00428-013-1485-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10:513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siebolts U, Varnholt H, Drebber U, Dienes HP, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62:84–88. doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 23.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 24.Jin W, Grant J, Stothard P, Moore SS, Guan LL. Characterization of bovine miRNAs by sequencing and bioinformatics analysis. BMC Mol Biol. 2009;10:90. doi: 10.1186/1471-2199-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sempere LF, Korc M. A method for conducting highly sensitive microRNA in situ hybridization and immunohistochemical analysis in pancreatic cancer. Methods Mol Biol. 2013;980:43–59. doi: 10.1007/978-1-62703-287-2_4. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, Van Dongen S, Enright AJ. Mirbase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parasramka MA, Ali S, Banerjee S, Deryavoush T, Sarkar FH, Gupta S. Garcinol sensitizes human pancreatic adenocarcinoma cells to gemcitabine in association with microRNA signatures. Mol Nutr Food Res. 2013;57:235–248. doi: 10.1002/mnfr.201200297. [DOI] [PubMed] [Google Scholar]
- 28.Liu N, Cui RX, Sun Y, Guo R, Mao YP, Tang LL, Jiang W, Liu X, Cheng YK, He QM, et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int J Cancer. 2014;134:1359–1368. doi: 10.1002/ijc.28468. [DOI] [PubMed] [Google Scholar]
- 29.Jiang X, Tan J, Li J, Kivimäe S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–541. doi: 10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy NC, Biankin AV, Millar EK, McNeil CM, O'Toole SA, Segara D, Crea P, Olayioye MA, Lee CS, Fox SB, et al. Loss of STARD10 expression identifies a group of poor prognosis breast cancers independent of HER2/Neu and triple negative status. Int J Cancer. 2010;126:1445–1453. doi: 10.1002/ijc.24826. [DOI] [PubMed] [Google Scholar]
- 31.Lan J, Guo P, Lin Y, Mao Q, Guo L, Ge J, Li X, Jiang J, Lin X, Qiu Y. Role of glycosyltransferase PomGnT1 in glioblastoma progression. Neuro Oncol. 2015;17:211–222. doi: 10.1093/neuonc/nou151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TO, Park J, Kang MJ, Lee SH, Jee SR, Ryu DY, Yang K, Yi JM. DNA hypermethylation of a selective gene panel as a risk marker for colon cancer in patients with ulcerative colitis. Int J Mol Med. 2013;31:1255–1261. doi: 10.3892/ijmm.2013.1317. [DOI] [PubMed] [Google Scholar]
- 33.Hahn S, Hermeking H. ZNF281/ZBP-99: A new player in epithelial-mesenchymal transition, stemness, and cancer. J Mol Med (Berl) 2014;92:571–581. doi: 10.1007/s00109-014-1160-3. [DOI] [PubMed] [Google Scholar]
- 34.Fernández M, Semela D, Bruix J, Colle I, Pinzani M, Bosch J. Angiogenesis in liver disease. J Hepatol. 2009;50:604–620. doi: 10.1016/j.jhep.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Sampat KR, O'Neil B. Antiangiogenic therapies for advanced hepatocellular carcinoma. Oncologist. 2013;18:430–438. doi: 10.1634/theoncologist.2012-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]


