Abstract
The transforming growth factor-β (TGF-β) signaling pathway serves a key role in the pathogenesis of liver cancer. To investigate the association between TGF-β1 and the following proteins: Proliferating cell nuclear antigen (PCNA), gankyrin, general vesicular transport factor p115 (p115), X-linked inhibitor of apoptosis protein (XIAP) and survivin, HepG2 liver cancer cells were transfected with small interfering RNA (siRNA) directed against TGF-β1, or were treated with exogenous TGF-β1. TGF-β1 protein expression levels were assessed at 72 and 96 h using western blotting, cell growth was evaluated using a Cell Counting kit-8 assay, and flow cytometry was used to examine cell cycle distribution and apoptosis. In addition, PCNA, gankyrin, p115, XIAP and survivin protein levels were evaluated using western blotting. TGF-β1 protein expression levels were decreased at 72 and 96 h following siRNA transfection, indicating that the siRNA against TGF-β1 was effective. In the TGF-β1-knockdown group, the HepG2 cells exhibited G1 or S-phase cell cycle arrest; therefore, the number of G2-phase cells was decreased, cell growth was inhibited and apoptotic peaks were observed. By contrast, no significant alteration in cell cycle distribution or apoptosis was observed in the cells treated with exogenous TGF-β1. In the exogenous TGF-β1 group, PCNA and XIAP protein expression levels were increased, whereas gankyrin, p115 and survivin protein expression was observed to be dependent on the duration of treatment. By contrast, PCNA, gankyrin, XIAP and survivin protein expression decreased following TGF-β1 knockdown; however, p115 protein expression increased. In conclusion, the TGF-β1 signaling pathway may affect cell growth, cell cycle distribution and apoptosis through the regulation of PCNA, gankyrin, p115, XIAP and survivin protein expression in liver cancer. The results of the present study may improve the current understanding of the role of the TGF-β signaling pathway during the pathogenesis of liver cancer.
Keywords: liver cancer, transforming growth factor-β 1, gankyrin, proliferating cell nuclear antigen, Golgi vesicular transport protein, X-linked inhibitor-of-apoptosis protein
Introduction
Liver cancer is one of the most common human malignancies. A number of signaling pathways, including the transforming growth factor β (TGF-β)/mothers against decapentaplegic homolog (1), proto-oncogene Wnt/β-catenin (2), rat sarcoma GTPase/mitogen-activated protein kinase (MAPK) (3), phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) (4), c-Jun N-terminal kinase (JNK)/signal transducer and activator of transcription (STAT) (5,6), hedgehog and tumor protein 53 transduction pathways, serve key roles in the pathogenesis of liver cancer. Among these signaling pathways, the TGF-β signaling pathway is one of the most important (1).
TGF-β1 is as a member of the TGF-β family able to produce tumor-inhibiting and promoting effects (7–9). TGF-β1 has been associated with immunosuppression, tumor angiogenesis, tumor cell migration, proliferation, differentiation, development, apoptosis and invasion, as well as other processes, in numerous types of cancer (10,11). For example, TGF-β1 expression is increased in liver cancer (12), intrahepatic cholangiocarcinoma (13), prostate cancer (14,15) and in head and neck squamous cell carcinoma (16), and leads to increased tumor growth. Conversely, TGF-β1 expression levels in patients with leukemia are significantly decreased compared with healthy subjects (17).
Cell growth, the cell cycle and apoptosis are closely associated with the genesis and development of liver cancer, and multiple factors are involved in their regulation, including proliferating cell nuclear antigen (PCNA), gankyrin, general vesicular transport factor p115 (p115), X-linked inhibitor of apoptosis protein (XIAP), survivin and caspase-3 (18–21). PCNA is a highly conserved protein; in addition to DNA replication, the functions of PCNA are associated with other vital cellular processes, including chromatin remodeling, DNA repair, sister-chromatid cohesion and cell cycle control (22). Recent studies have reported that tumor cells express increased levels of PCNA, identifying it as a potential target for cancer therapy (23,24). Gankyrin is a chaperone of the ubiquitin-proteasome and a novel oncogene, and has been demonstrated to be overexpressed in numerous types of cancer (25–27), including liver cancer (28–30), breast cancer (31,32), colorectal cancer (33), estrogen-driven endometrial carcinoma (26) and oral cancer (34). Gankyrin serves an essential role in tumor occurrence and development (25–34). p115 is a tether protein that has an important role in a number of signaling pathways required for cell proliferation, and has been extensively studied (35,36). A previous study demonstrated that p115 is a potential tumor biomarker and therapeutic target that is overexpressed in human gastric cancer cells (36). The inhibitor of apoptosis protein (IAP) family comprises internal apoptosis suppressors, and its members are able to bind to caspase and inhibit cell apoptosis. XIAP, an important member of the IAP family, possesses inhibitory activity and serves an important role in cell apoptosis (19,20). Survivin, a novel member of the IAP family with the lowest relative molecular weight, is the most potent suppressor of apoptosis that has been identified so far (21). However, there are currently few studies concerning the association between TGF-β1 and PCNA, gankyrin, p115, XIAP and survivin in liver cancer.
Following previous observations, TGF-β1 expression was knocked down in the present study using small interfering RNA (siRNA). Subsequently, cell growth, cell cycle distribution and apoptosis, as well as PCNA, gankyrin, p115, XIAP and survivin protein expression, was observed in HepG2 liver cancer cells. The present study aimed to further elucidate the association between TGF-β1 and the aforementioned factors, and the role of TGF-β1 during the genesis and development of liver cancer.
Materials and methods
Cell culture
HepG2 liver cancer cells were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and plated in culture flasks. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% v/v fresh fetal calf serum (FCS; TBD Biotechnology Corporation, Tianjin, China), at 37°C in a humidified atmosphere containing 5% CO2. To maintain the cell line, cells were replated following digestion for 1 min in 0.25% trypsin when they reached confluency.
Transient transfection
siRNA directed against TGF-β1 was designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The TGF-β1 siRNA sequences were as follows: Sense, 5′-GAC ACC AAC UAU UGC UUC ATT-3′ and antisense, 5′-UGA AGC AAU AGU UGG UGU CTT-3′. The scrambled negative control siRNA sequences were as follows: Sense 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and antisense, 5′-ACG UGA CAC GUU CGG AGA ATT-3′.
A total of 3×105 cells were plated in 6-well plates in triplicate and grown to 30–50% confluency. For the transfection, 10 µl X-tremeGENE siRNA Transfection Reagent (Roche Diagnostics, Basel, Switzerland) and 2 µg siRNA were mixed in 200 µl DMEM for 15–20 min. The 210-µl mixture was added to the cells alongside 2 ml DMEM supplemented with 10% v/v FCS, and the plates were subsequently agitated. The cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. Cells were harvested at 72 and 96 h, and the proteins were isolated for further analysis. In addition, a negative control was created using the control siRNA and subsequently analyzed. Untreated cells were also analyzed. All experiments were performed in triplicate.
Detection of TGF-β1 protein expression using western blotting
Transfected cells were lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with phenylmethylsulfonyl fluoride (1:200) and phosphatase inhibitors (1:200) on ice for 15 min, at 72 and 96 h following transfection, prior to protein isolation. The total protein concentration was determined using a bicinchoninic acid kit (Beyotime Institute of Biotechnology). Protein samples (60 µg) were separated using 12% SDS-PAGE and then transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked by incubation in PBS with 5% skimmed milk at room temperature for 1 h. The membranes were subsequently incubated with primary antibodies against TGF-β1 (1:200; cat. no. sc-146; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and β-actin (1:200; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.) at 4°C overnight. The membranes were washed three times using PBS, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:5,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) at 37°C for 1 h. The membranes were washed three times using PBS and visualized using a BeyoECL Plus kit (cat. no. BYT-P0018; Beyotime Institute of Biotechnology). Images were captured using a fluorescence imager (Champgel 5500; Beijing Sage Creation Science and Technology Ltd., Beijing, China) and analyzed using Quantity One® software (version 4.5.2; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The quantitative results of grayscale analysis were used for the statistical analysis.
Detection of cell growth using the Cell Counting Kit-8 (CCK-8) assay
The HepG2 groups analyzed were as follows: i) the control group; ii) the negative siRNA control group; iii) the TGF-β1 siRNA transfected group. The following doses of exogenous TGF-β1 were added to HepG2 cells (the exogenous TGF-β1 group), 0, 5, 10, 20, 30, 40 and 50 µg/l.
A total of 1.5×104 transfected cells were plated in 96-well plates in triplicate and grown to 30–50% confluency, prior to transfection. A total of 0.8 µl X-tremeGENE siRNA Transfection Reagent and 0.15 µg siRNA were mixed in 30 µl DMEM for 15–20 min. The 30 µl mixture, or increasing doses of exogenous TGF-β1, were administered to the cells, in addition to 150 µl DMEM supplemented with 10% v/v FCS, and the plates were subsequently agitated. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. A total of 20 µl CCK-8 reagent (Beyotime Institute of Biotechnology) was added to the cell medium 24, 48, 72 and 96 h following transfection or treatment with exogenous TGF-β1. Following a 2-h incubation at 37°C, the absorbance was measured using a microplate reader (MK3; Thermo Labsystems, Inc., Beverly, MA, USA) at 450 nm in order to determine the number of viable cells. The control and negative control groups were also analyzed. All experiments were performed in triplicate. The data were normalized to their respective controls. The cell growth inhibition rates were calculated as follows: (the absorbance ratio of the control group-the absorbance ratio of the transfection group)/the absorbance ratio of the control group ×100. Following analysis, the optimum duration and dose of treatment was used in subsequent experiments.
Detection of cell cycle distribution using flow cytometry
The following groups were assessed: i) the control group; ii) the 24 h exogenous TGF-β1 group; iii) the 48 h exogenous TGF-β1 group; iv) the 72 h exogenous TGF-β1 group; v) the 72 h TGF-β1-knockdown group; vi) the 96 h TGF-β1-knockdown group. Cells in the exogenous TGF-β1 groups were treated with 25 µg/l TGF-β1.
At the respective time points the culture medium was removed, and the cells were washed in PBS, trypsinized, harvested, washed in PBS, centrifuged three times at 4°C and 1,000 × g for 5 min, added to 2 ml ice-cold 70% ethanol and preserved at 4°C. The cells were washed three times, and RNases and proteins were removed using a Cell Cycle Detection kit (BD Biosciences, Franklin Lakes, NJ, USA). The cells were subsequently incubated in 10 g/ml propidium iodide (PI) at 4°C for 10 min in the dark and the cell cycle distribution was analyzed using a FACScan flow cytometer (Sysmex Partec GmbH, Görlitz, Germany) and CyViewTM software version 6.0 (Sysmex Partec GmbH) within 2 h. All experiments were performed in triplicate.
Detection of apoptosis using flow cytometry
The cell groups described in the cell cycle distribution section were used. Cell apoptosis was detected using an Annexin V-fluorescein isothiocyanate (FITC)/PI Apoptosis Detection kit (BD Biosciences). At the respective time points, the cells were collected, centrifuged three times at 4°C and 1,000 × g for 5 min, and resuspended in 500 µl 1X binding buffer. The cell density was adjusted to 1×106 cells/ml. A total of 100 µl cells were incubated with 5 µl Annexin V-FITC and 5 µl PI for 15 min in the dark at room temperature. Cell apoptosis was evaluated using a FACScan flow cytometer. For each determination, a minimum of 50,000 cells was analyzed. All experiments were performed in triplicate.
Viable cells stained negative for PI and annexin V-FITC, early apoptotic cells stained positive for annexin V-FITC and negative for PI, and late apoptotic cells stained positive for annexin V-FITC and PI. Nonviable cells, which underwent necrosis, stained positive for PI but negative for annexin V-FITC.
Evaluation of PCNA, gankyrin, p115, XIAP and survivin expression using western blotting
The cell groups described in the cell cycle distribution section were used. At the respective time points, total protein was extracted. The protein levels were evaluated using western blotting, following the aforementioned protocol used for the TGF-β1 protein. The following primary antibodies were used at 4°C overnight: Gankyrin (1:500; cat. no. GTX48519; GeneTex, Inc., Irvine, CA, USA), p115 (1:1,000; cat. no. GTX115115; GeneTex, Inc.), PCNA, XIAP (each 1:500; cat. no. BS1289 and BS1609, respectively; Bioworld Technology, Inc., St. Louis Park, MN, USA), and survivin (1:200; cat. no. sc-10811; Santa Cruz Biotechnology, Inc.). The horseradish peroxidase-conjugated secondary antibody was obtained from Santa Cruz Biotechnology, Inc. (1:5,000; cat. no. sc-2004) and incubated with the membrane for 1 h at 37°C.
Statistical analysis
Values are expressed as the mean ± standard deviation of triplicate data, and were compared using the Student's t-test and a one-way analysis of variance. Statistical analyses were conducted using GraphPad Prism (version 6; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
TGF-β1 protein expression
Following siRNA transfection, the TGF-β1 protein expression levels at 72 and 96 h were evaluated using western blotting, and were observed to be significantly decreased (P=0.0016 and P=0.0055, respectively; Fig. 1). No significant difference was observed in the TGF-β1 protein levels of the negative siRNA control group, as compared with the control group. These results indicate that the siRNA against TGF-β1 was effective.
Figure 1.
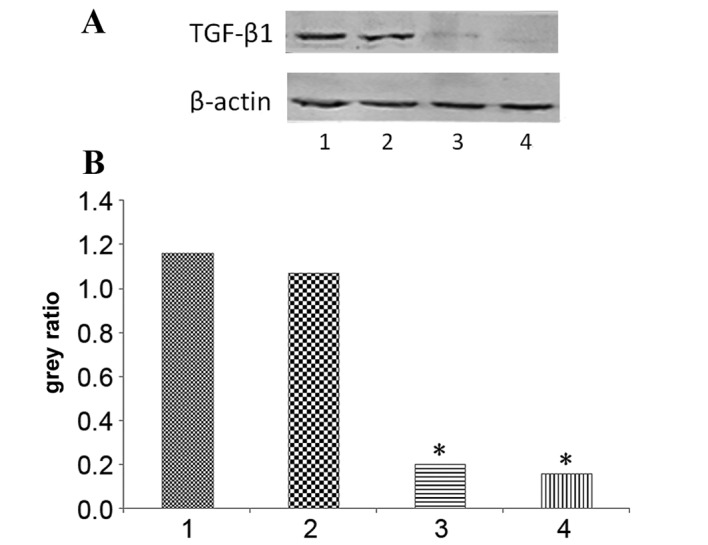
TGF-β1 protein expression levels decreased following TGF-β1 siRNA knockdown in HepG2 cells. (A) Representative protein bands from the western blotting. (B) Protein band quantification. 1, control group; 2, negative siRNA control group; 3, 72 h TGF-β1 siRNA transfection group; 4, 96 h TGF-β1 siRNA transfection group. Values were normalized to β-actin and expressed as the mean ± standard deviation of triplicate data. *P<0.05, vs. the control group. TGF-β1, transforming growth factor-β-1; siRNA, small interfering RNA.
Effect of TGF-β1 on HepG2 cell growth
Treatment with exogenous TGF-β1 inhibits HepG2 cell growth
The results of the CCK-8 assays are presented in Fig. 2. Treatment with 0–20 µg/l TGF-β1 inhibited HepG2 cell growth in a dose-dependent manner; however, at doses >30 µg/l, the inhibitory effect of treatment with TGF-β1 on HepG2 cell growth was decreased. Therefore, the optimum dose of TGF-β1 to inhibit HepG2 cell growth was between 20 and 30 µg/l (Fig. 2). Therefore, 25 µg/l TGF-β1 was used in subsequent experiments.
Figure 2.
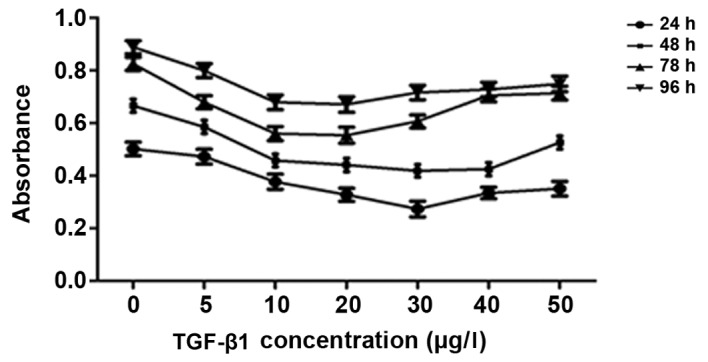
Treatment with exogenous TGF-β1 decreases HepG2 cell growth. TGF-β1, transforming growth factor-β-1.
The effect of silencing TGF-β1 on HepG2 cell growth
The results of the CCK-8 assays are presented in Fig. 3. No significant difference in cell viability was observed in the control group compared with the negative siRNA control group, whereas a significant decrease in the number of viable cells was observed in the TGF-β1 siRNA-transfected group (P=0.042; Fig. 3). The cell growth inhibition rates induced by the TGF-β1 siRNA were 12.9% at 24 h, 21.0% at 48 h, 34.3% at 72 h and 35.0% at 96 h. These results indicated that HepG2 cell growth was inhibited, or that cell death was increased, between 24 and 96 h following transfection, and that the optimum inhibition times were 72 and 96 h (Fig. 3). These results suggest that short-term inhibition of cell growth or promotion of cell death occurs following transient siRNA transfection.
Figure 3.
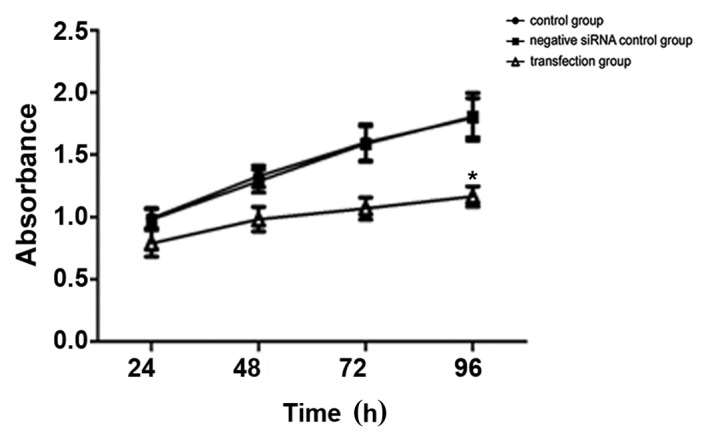
TGF-β1 knockdown decreases HepG2 cell growth. *P<0.05, vs. the control group. TGF-β1, transforming growth factor-β-1; siRNA, small interfering RNA.
Effect of altered TGF-β1 expression on HepG2 cell cycle distribution
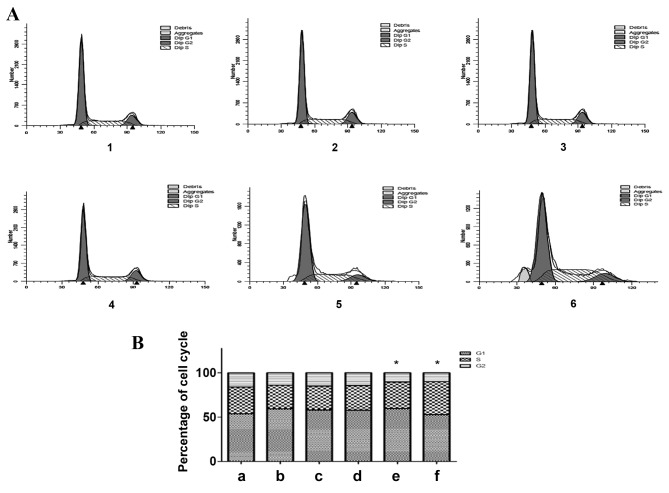
The flow cytometric results demonstrated that the percentage of G1-phase cells increased and the percentage of S-phase and G2-phase cells decreased, 24, 48 and 72 h following treatment with exogenous TGF-β1, as compared with the control group (Fig. 4). A total of 72 h following TGF-β1 knockdown, the percentage of G1-phase cells increased, the percentage of S-phase cells exhibited no significant alteration and the percentage of G2-phase cells decreased, as compared with the control group (P=0.0425). A total of 96 h following TGF-β1 knockdown, the percentage of G1-phase cells decreased, the percentage of S-phase cells increased and the percentage of G2-phase cells decreased, as compared with the control group (P=0.0326). In the exogenous TGF-β1 group, cells were arrested in the G1 phase, and the number of cells in the S and G2 phases decreased. In addition, cells in the TGF-β1-knockdown group were also arrested in the G1 phase, and the number of cells in the S phase remained unchanged and decreased in the G2 phase 72 h following transfection. By contrast, 96 h following knockdown, the cells were arrested in the S phase, the number of cells in the G2 phase decreased and the apoptosis peak was visible. The effect of TGF-β1 knockdown on cell cycle distribution was considered to be statistically significant. These results indicate that TGF-β1 knockdown inhibits cell cycle progression, therefore inhibiting cell growth.
Figure 4.
Effect of altered TGF-β1 expression on the HepG2 cell cycle distribution. (A) Flow cytometric analysis of HepG2 cell cycle distribution and (B) quantification of the flow cytometric data. 1, control group; 2, 24 h exogenous TGF-β1 group; 3, 48 h exogenous TGF-β1 group; 4, 72 h exogenous TGF-β1 group; 5, 72 h TGF-β1-knockdown group; 6, 96 h TGF-β1-knockdown group. *P<0.05, vs. the control group. TGF-β1, transforming growth factor-β-1.
Effect of altered TGF-β1 expression on HepG2 cell apoptosis
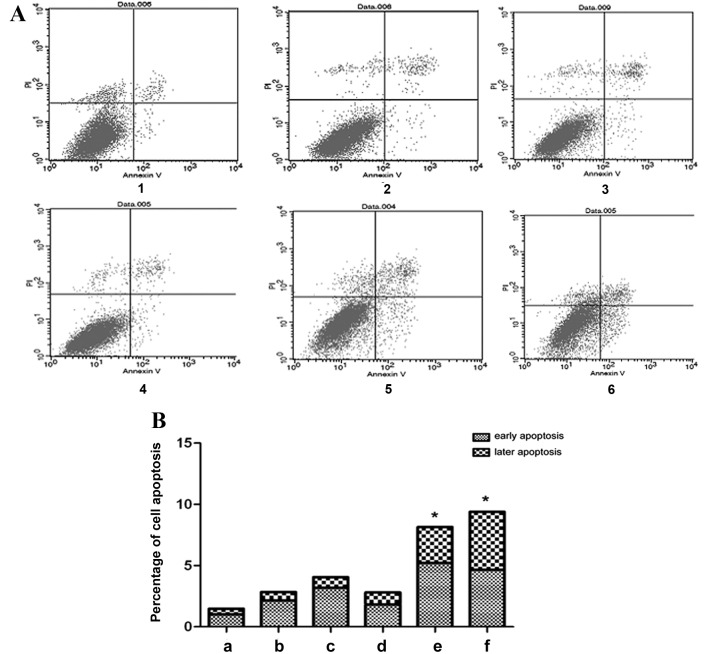
The flow cytometric results demonstrated that in the exogenous TGF-β1 group, the percentage of early apoptotic cells increased compared with the control group, particularly at 48 h (Fig. 5). However, the percentage of late apoptotic cells did not change significantly following treatment with exogenous TGF-β1 compared with the control group (Fig. 5). In the TGF-β1-knockdown group, early apoptosis significantly increased at 72 h, and early and late apoptosis significantly increased at 96 h, as compared with the control group (P=0.0461 and P=0.0433, respectively; Fig. 5). The results indicate that TGF-β1 knockdown increases cell apoptosis, therefore inhibiting cell growth.
Figure 5.
Effect of altered TGF-β1 on HepG2 cell apoptosis. (A) Flow cytometric analysis of HepG2 cell apoptosis and (B) quantification of the flow cytometric data. 1, control group; 2, 24 h exogenous TGF-β1 group; 3, 48 h exogenous TGF-β1 group; 4, 72 h exogenous TGF-β1 group; 5, 72 h TGF-β1-knockdown group; 6, 96 h TGF-β1-knockdown group. In the scatter diagram, the upper left quadrant represents mechanically damaged cells, the upper right quadrant represents late apoptotic cells, the lower left quadrant represents normal cells and the lower right quadrant represents early apoptotic cells. *P<0.05, vs. the control group. TGF-β1, transforming growth factor-β-1; PI, propidium iodide.
Effect of altered TGF-β1 expression on PCNA, gankyrin, p115, XIAP and survivin protein expression
HepG2 cells were treated with exogenous TGF-β1 or TGF-β1 siRNA, and protein expression was evaluated using western blotting. In the exogenous TGF-β1 group, PCNA protein expression was significantly decreased at 24, 48 and 72 h following treatment compared with the control group (P=0.0016, P=0.0051 and P=0.0109, respectively); however, the most significant decrease was observed at 24 h (Fig. 6). Gankyrin expression was significantly decreased at 24 h (P=0.039), significantly increased at 72 h (P=0.028) and unchanged at 48 h compared with the control group (Fig. 6). p115 protein expression was significantly increased at 24 h (P=0.0382), but significantly decreased at 48 h (P=0.0289) and 72 h (P=0.0026) compared with the control group (Fig. 6). XIAP protein expression was significantly decreased compared with the control group (P24 h=0.0015, P48 h=0.0289, P72 h=0.0025; Fig. 7). Survivin protein expression at 24 h was significantly decreased compared with the control group (P=0.0041); however, no significant difference was observed at 48 and 72 h (P=0.1177 and P=0.5671, respectively; Fig. 7). In the TGF-β1-knockdown group, PCNA, gankyrin, XIAP and survivin protein expression was significantly decreased compared with the control group (PPCNA72 h=0.0004, PPCNA96 h=0.0106, Pgankyrin72 h=0.028, Pgankyrin96 h=0.023, PXIAP72 h=0.0026, PXIAP96 h=0.0227, Psurvivin72 h=0.0135, Psurvivin96 h=0.0203; Figs. 6 and 7). p115 expression was significantly increased at 96 h compared with the control group (P=0.0292; Fig. 6). PCNA, XIAP and survivin protein expression increased following TGF-β1 knockdown in a time-dependent manner compared with 72 h and 96 h following transfection, whereas gankyrin exhibited the opposite pattern (Figs. 6 and 7).
Figure 6.
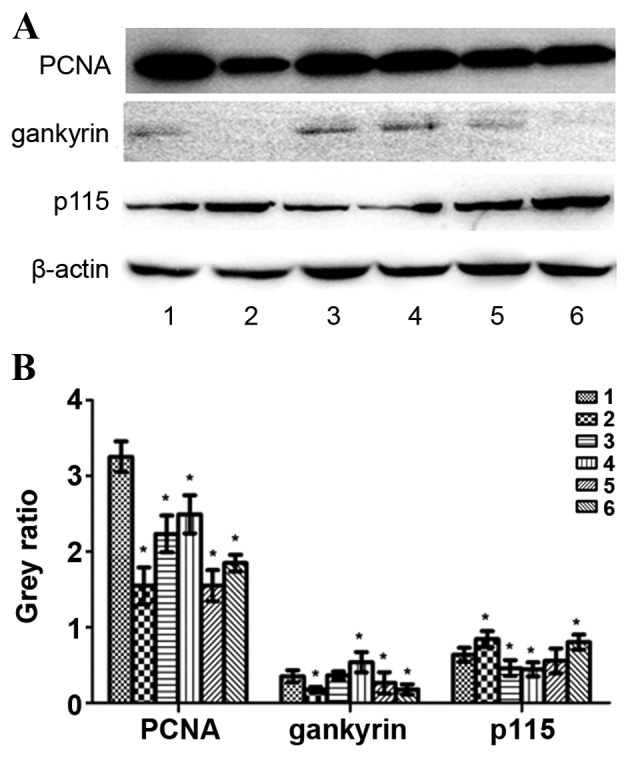
Protein expression levels of PCNA, gankyrin, p115 and β-actin following treatment with exogenous TGF-β1 or TGF-β1 siRNA in HepG2 cells. (A) Representative protein bands from the western blotting. (B) Protein band quantification. 1, control group; 2, 24 h exogenous TGF-β1 group; 3, 48 h exogenous TGF-β1 group; 4, 72 h exogenous TGF-β1 group; 5, 72 h TGF-β1-knockdown group; 6, 96 h TGF-β1-knockdown group. *P<0.05, vs. the control group. PCNA, proliferating cell nuclear antigen; p115, general vesicular transport factor p115; TGF-β1, transforming growth factor-β-1; siRNA, small interfering RNA.
Figure 7.
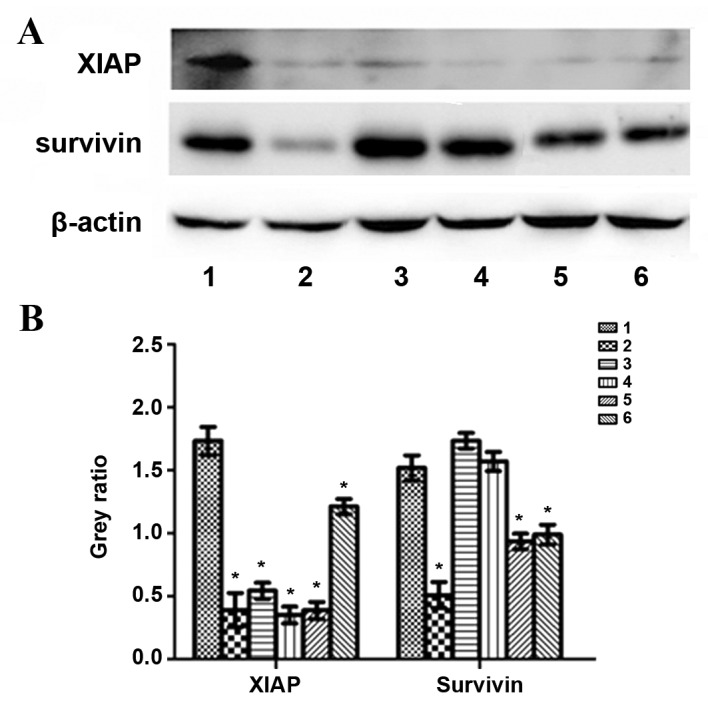
Protein levels of XIAP, survivin and β-actin following treatment with exogenous TGF-β1 or TGF-β1 siRNA in HepG2 cells. (A) Representative protein bands from the western blotting. (B) Protein band quantification. 1, control group; 2, 24 h exogenous TGF-β1 group; 3, 48 h exogenous TGF-β1 group; 4, 72 h exogenous TGF-β1 group; 5, 72 h TGF-β1-knockdown group; 6, 96 h TGF-β1-knockdown group. *P<0.05 vs. the control group. XIAP, X-linked inhibitor of apoptosis protein; TGF-β1, transforming growth factor-β-1; siRNA, small interfering RNA.
Discussion
RNA interference has been successfully used to study gene function, and has assisted in determining associations between upstream and downstream factors in various signaling pathways (37). In the present study, TGF-β1 protein expression was observed to be decreased 72 and 96 h following siRNA transfection, indicating that the siRNA against TGF-β1 was effective.
In the present study, treatment with exogenous TGF-β1 inhibited HepG2 cell growth; the degree of inhibition following treatment with concentrations between 20 and 30 µg/l was the most significant. The effect of treatment with exogenous TGF-β1 on cell growth may be due to TGF-β1 having a role as a tumor suppressor in the early stage of tumor development, and is involved in two-way regulation during the genesis and development of liver cancer (7–9). In addition, an siRNA directed against TGF-β1 inhibited HepG2 cell growth, potentially as TGF-β1 is overexpressed in liver cancer (12). However, it remains unclear why treatment with exogenous TGF-β1 and siRNA against TGF-β1 inhibited HepG2 cell growth, and the underlying molecular mechanisms require further study.
Previously, TGF-β1 has been revealed to induce G1-phase cell cycle arrest or prolong the time of the G1-S phase transition in mesothelioma and breast cancer (38), which is consistent with the results of the present study. In the exogenous TGF-β1 group, cells were arrested in the G1 phase, and the percentage of cells in the S and G2 phases decreased. The TGF-β1-knockdown cells were also arrested in the G1 and S phases 72 and 96 h following transfection, respectively. These results are consistent with the results of cell growth.
In the present study, treatment with exogenous TGF-β1 or siRNA against TGF-β1 inhibited HepG2 cell growth, cell cycle progression and apoptosis; however, the effect of TGF-β1 knockdown was more significant. This is potentially as exogenous TGF-β1 has inconsistent effects on the expression of related factors, including PCNA, gankyrin, p115, XIAP and survivin. As the effect was more significant in the TGF-β1-knockdown group, the changes due to TGF-β1 knockdown will be discussed. The effect of treatment with exogenous TGF-β1 requires further study.
PCNA serves an important role in the priming of cell proliferation and is therefore an indicator of cell proliferation. (18) For example, antisense TGF-β1 oligonucleotides may lead to significantly decreased expression levels of PCNA and inhibit cell growth in oral squamous cell carcinoma (39). Gankyrin, a novel oncogene, regulates the cell cycle and apoptosis (28). LBH589 inhibits the proliferation and metastasis of hepatocellular carcinoma through the inhibition of the gankyrin/STAT3/Akt signaling pathway (28). However, there are few studies on gankyrin, and the association between TGF-β1 and gankyrin remains unclear. In the present study, treatment with exogenous TGF-β1 resulted in decreased protein expression levels of PCNA and gankyrin compared with the control group; therefore, treatment with exogenous TGF-β1 may inhibit cell growth and enhance apoptosis. p115 is a potential tumor biomarker and therapeutic target in human gastric cancer (36). However, in the present study, TGF-β1 knockdown resulted in increased p115 protein expression levels as compared with the control group. The underlying molecular mechanism remains to be completely elucidated.
XIAP and survivin are considered to be IAPs and their decreased expression causes caspase-3 to become phosphorylated, therefore increasing cellular apoptosis. TGF-β may upregulate certain anti-apoptotic genes, including B-cell lymphoma-2 like 2 and XIAP (10), and XIAP knockdown abolishes the TGF-β1-induced proliferation of malignant meningioma cells (11). TGF-β signaling pathway antagonists similarly activate the survivin promoter, rendering cells refractory to further promoter activation by insulin-like growth factor-1 (40). Similarly, in the present study, TGF-β1 knockdown resulted in decreased XIAP and survivin protein expression levels as compared with the control group, therefore enhancing cellular apoptosis.
In conclusion, the TGF-β signaling pathway affects cell growth, the cell cycle and apoptosis by regulating the protein expression of PCNA, gankyrin, p115, XIAP and survivin. The results of the present study may improve current understanding of liver cancer pathogenesis and the respective role of the TGF-β signaling pathway. By understanding these processes in detail, it may be possible to treat tumors by modulating TGF-β signal transduction cascades within cells, and to specifically control cell growth, differentiation and apoptosis.
Glossary
Abbreviations
- TGF-β
transforming growth factor-β
- MAPK
mitogen-activated protein kinase
- PI3K
phosphoinositide 3-kinase
- JNK
c-Jun N-terminal kinase
- STAT
signal transducer and activator of transcription
- PCNA
proliferating cell nuclear antigen
- p115
general vesicular transport factor p115
- XIAP
X-linked inhibitor of apoptosis protein
- siRNA
small interfering RNA
- DMEM
Dulbecco's modified Eagle's medium
- FCS
fetal calf serum
- CCK-8
Cell Counting kit-8
- PI
propidium iodide
- FITC
fluorescein isothiocyanate
References
- 1.Balzarini P, Benetti A, Invernici G, Cristini S, Zicari S, Caruso A, Gatta LB, Berenzi A, Imberti L, Zanotti C, et al. Transforming growth factor-beta1 induces microvascular abnormalities through a down-modulation of neural cell adhesion molecule in human hepatocellular carcinoma. Lab Invest. 2012;92:1297–1309. doi: 10.1038/labinvest.2012.94. [DOI] [PubMed] [Google Scholar]
- 2.Monga SP. β-catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology. 2015;148:1294–1310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM, Hu YW, Lin L, Chen J, Zheng L, Wang Q. HBx-related long non-coding RNA DBH-AS1 promotes cell proliferation and survival by activating MAPK signaling in hepatocellular carcinoma. Oncotarget. 2015;6:33791–33804. doi: 10.18632/oncotarget.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang H, Li RP, Liang P, Zhou YL, Wang GW. miR-125a inhibits the migration and invasion of liver cancer cells via suppression of the PI3K/AKT/mTOR signaling pathway. Oncol Lett. 2015;10:681–686. doi: 10.3892/ol.2015.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Guo Y, Li S, Han R, Ying J, Zhu H, Wang Y, Yin L, Han Y, Sun L, et al. A novel anti-cancer agent Icaritin suppresses hepatocellular carcinoma initiation and malignant growth through the IL-6/Jak2/Stat3 pathway. Oncotarget. 2015;6:31927–31943. doi: 10.18632/oncotarget.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang XH, Liu BR, Qu B, Xing H, Gao SL, Yin JM, Wang XF, Cheng YQ. Silencing STAT3 may inhibit cell growth through regulating signaling pathway, telomerase, cell, cycle, apoptosis and angiogenesis in hepatocellular carcinoma: Potential uses for gene therapy. Neoplasma. 2011;58:158–171. doi: 10.4149/neo_2011_02_158. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, Guo L, Dong L, Guo L, Li S, Zhang J, Sun M. TGF-beta1 signal pathway may contribute to rhabdomyosarcoma development by inhibiting differentiation. Cancer Sci. 2010;101:1108–1116. doi: 10.1111/j.1349-7006.2010.01512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sjölund J, Boström AK, Lindgren D, Manna S, Moustakas A, Ljungberg B, Johansson M, Fredlund E, Axelson H. The notch and TGF-β signaling pathways contribute to the aggressiveness of clear cell renal cell carcinoma. PLoS One. 2011;6:e23057. doi: 10.1371/journal.pone.0023057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gore AJ, Deitz SL, Palam LR, Craven KE, Korc M. Pancreatic cancer-associated retinoblastoma 1 dysfunction enables TGF-β to promote proliferation. J Clin Invest. 2014;124:338–352. doi: 10.1172/JCI71526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caja L, Bertran E, Campbell J, Fausto N, Fabregat I. The transforming growth factor-beta (TGF-β) mediates acquisition of a mesenchymal stem cell-like phenotype in human liver cells. J Cell Physiol. 2011;226:1214–1223. doi: 10.1002/jcp.22439. [DOI] [PubMed] [Google Scholar]
- 11.Gogineni VR, Gupta R, Nalla AK, Velpula KK, Rao JS. uPAR and cathepsin B shRNA impedes TGF-β1-driven proliferation and invasion of meningioma cells in a XIAP-dependent pathway. Cell Death Dis. 2012;3:e439. doi: 10.1038/cddis.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee D, Chung YH, Kim JA, Lee YS, Lee D, Jang MK, Kim KM, Lim YS, Lee HC, Lee YS. Transforming growth factor beta 1 overexpression is closely related to invasiveness of hepatocellular carcinoma. Oncology. 2012;82:11–18. doi: 10.1159/000335605. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Chen JR, Hsu CH, Li YH, Chen YM, Lin CY, Huang SJ, Chang ZK, Chen YC, Lin CH, et al. A zebrafish model of intrahepatic cholangiocarcinoma by dual expression of hepatitis B virus X and hepatitis C virus core protein in liver. Hepatology. 2012;56:2268–2276. doi: 10.1002/hep.25914. [DOI] [PubMed] [Google Scholar]
- 14.Darrington E, Zhong M, Vo BH, Khan SA. Vascular endothelial growth factor A, secreted in response to transforming growth factor-β1 under hypoxic conditions, induces autocrine effects on migration of prostate cancer cells. Asian J Androl. 2012;14:745–751. doi: 10.1038/aja.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuzio P, Ditonno P, Rutigliano M, Battaglia M, Bettocchi C, Loverre A, Grandaliano G, Perlino E. Regulation of TGF-β1 expression by androgen deprivation therapy of prostate cancer. Cancer Lett. 2012;318:135–144. doi: 10.1016/j.canlet.2011.08.034. [DOI] [PubMed] [Google Scholar]
- 16.Freudlsperger C, Bian Y, Wise Contag S, Burnett J, Coupar J, Yang X, Chen Z, Van Waes C. TGF-β and NF-κB signal pathway cross-talk is mediated through TAK1 and SMAD7 in a subset of head and neck cancers. Oncogene. 2013;32:1549–1559. doi: 10.1038/onc.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Chen P, Huang HF, Huang MJ, Chen YZ. Reduction of transforming growth factor-β1 expression in leukemia and its possible role in leukemia development. Leuk Lymphoma. 2012;53:145–151. doi: 10.3109/10428194.2011.603446. [DOI] [PubMed] [Google Scholar]
- 18.Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL, Zhang J, Yang Y, Liu H, Wang F, et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349:87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 19.Bao W, Zhu F, Duan Y, Yang Y, Cai H. HtrA1 resensitizes multidrug-resistant hepatocellular carcinoma cells by targeting XIAP. Biomed Pharmacother. 2015;70:97–102. doi: 10.1016/j.biopha.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 20.Wu WY, Kim H, Zhang CL, Meng XL, Wu ZS. Clinical significance of autophagic protein LC3 levels and its correlation with XIAP expression in hepatocellular carcinoma. Med Oncol. 2014;31:108. doi: 10.1007/s12032-014-0108-3. [DOI] [PubMed] [Google Scholar]
- 21.Vogl TJ, Oppermann E, Qian J, Imlau U, Tran A, Hamidavi Y, Korkusuz H, Bechstein WO, Nour-Eldin NE, Gruber-Rouh T, et al. Transarterial chemoembolization of hepatocellular carcinoma in a rat model: The effect of additional injection of survivin siRNA to the treatment protocol. BMC Cancer. 2016;16:325. doi: 10.1186/s12885-016-2357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann Bot. 2011;107:1127–1140. doi: 10.1093/aob/mcq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dillehay KL, Lu S, Dong Z. Antitumor effects of a novel small molecule targeting PCNA chromatin association in prostate cancer. Mol Cancer Ther. 2014;13:2817–2826. doi: 10.1158/1535-7163.MCT-14-0522. [DOI] [PubMed] [Google Scholar]
- 24.Dillehay KL, Seibel WL, Zhao D, Lu S, Dong Z. Target validation and structure-activity analysis of a series of novel PCNA inhibitors. Pharmacol Res Perspect. 2015;3:e00115. doi: 10.1002/prp2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng T, Hong X, Wang J, Pei T, Liang Y, Yin D, Song R, Song X, Lu Z, Qi S, et al. Gankyrin promotes tumor growth and metastasis through activation of IL-6/STAT3 signaling in human cholangiocarcinoma. Hepatology. 2014;59:935–946. doi: 10.1002/hep.26705. [DOI] [PubMed] [Google Scholar]
- 26.Bai Z, Tai Y, Li W, Zhen C, Gu W, Jian Z, Wang Q, Lin JE, Zhao Q, Gong W, et al. Gankyrin activates IL-8 to promote hepatic metastasis of colorectal cancer. Cancer Res. 2013;73:4548–4558. doi: 10.1158/0008-5472.CAN-12-4586. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Yang Y, Zhang Z, He Y, Liu Z, Yu Y, Wu S, Cai B, Feng Y. Gankyrin plays an essential role in estrogen-driven and GPR30-mediated endometrial carcinoma cell proliferation via the PTEN/PI3K/AKT signaling pathway. Cancer Lett. 2013;339:279–287. doi: 10.1016/j.canlet.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 28.Song X, Wang J, Zheng T, Song R, Liang Y, Bhatta N, Yin D, Pan S, Liu J, Jiang H, Liu L. LBH589 Inhibits proliferation and metastasis of hepatocellular carcinoma via inhibition of gankyrin/STAT3/Akt pathway. Mol Cancer. 2013;12:114. doi: 10.1186/1476-4598-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Ding J, Wu K, Ning BF, Wen W, Sun HY, Han T, Huang L, Dong LW, Yang W, et al. Gankyrin-mediated dedifferentiation facilitates the tumorigenicity of rat hepatocytes and hepatoma cells. Hepatology. 2011;54:1259–1272. doi: 10.1002/hep.24530. [DOI] [PubMed] [Google Scholar]
- 30.Qian YW, Chen Y, Yang W, Fu J, Cao J, Ren YB, Zhu JJ, Su B, Luo T, Zhao XF, et al. 28(GANK) prevents degradation of Oct4 and promotes expansion of tumor-initiating cells in hepatocarcinogenesis. Gastroenterology. 2012;142:1547–1558. doi: 10.1053/j.gastro.2012.02.042. [DOI] [PubMed] [Google Scholar]
- 31.Gao L, Xie H, Dong L, Zou J, Fu J, Gao X, Ou L, Xiang S, Song H. Gankyrin is essential for hypoxia enhanced metastatic potential in breast cancer cells. Mol Med Rep. 2014;9:1032–1036. doi: 10.3892/mmr.2013.1860. [DOI] [PubMed] [Google Scholar]
- 32.Zhen C, Chen L, Zhao Q, Liang B, Gu YX, Bai ZF, Wang K, Xu X, Han QY, Fang DF, et al. Gankyrin promotes breast cancer cell metastasis by regulating Rac1 activity. Oncogene. 2013;32:3452–3460. doi: 10.1038/onc.2012.356. [DOI] [PubMed] [Google Scholar]
- 33.Mine H, Sakurai T, Kashida H, Matsui S, Nishida N, Nagai T, Hagiwara S, Watanabe T, Kudo M. Association of gankyrin and stemness factor expression in human colorectal cancer. Dig Dis Sci. 2013;58:2337–2344. doi: 10.1007/s10620-013-2627-8. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Knobloch TJ, Kresty LA, Zhang Z, Lang JC, Schuller DE, Weghorst CM. Gankyrin, a biomarker for epithelial carcinogenesis, is overexpressed in human oral cancer. Anticancer Res. 2011;31:2683–2692. [PubMed] [Google Scholar]
- 35.Millarte V, Boncompain G, Tillmann K, Perez F, Sztul E, Farhan H. Phospholipase C γ1 regulates early secretory trafficking and cell migration via interaction with p115. Mol Biol Cell. 2015;26:2263–2278. doi: 10.1091/mbc.E15-03-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XJ, Luo Y, Yi YF. P115 promotes growth of gastric cancer through interaction with macrophage migration inhibitory factor. World J Gastroenterol. 2013;19:8619–8629. doi: 10.3748/wjg.v19.i46.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Sun G, Sun X. RNA interference-mediated silencing of speckle-type POZ protein promotes apoptosis of renal cell cancer cells. Onco Targets Ther. 2016;9:2393–2402. doi: 10.2147/OTT.S91097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Wen G, Zhao Y, Tong J, Hei TK. The role of TGFBI in mesothelioma and breast cancer: Association with tumor suppression. BMC Cancer. 2012;12:239. doi: 10.1186/1471-2407-12-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SG, Song JY. Therapeutic targeting of oncogenic transforming growth factor-β1 signaling by antisense oligonucleotides in oral squamous cell carcinoma. Oncol Rep. 2012;28:539–544. doi: 10.3892/or.2012.1811. [DOI] [PubMed] [Google Scholar]
- 40.Song K, Shankar E, Yang J, Bane KL, Wahdan-Alaswad R, Danielpour D. Critical role of a survivin/TGF-β/mTORC1 axis in IGF-I-mediated growth of prostate epithelial cells. PLoS One. 2013;8:e61896. doi: 10.1371/journal.pone.0061896. [DOI] [PMC free article] [PubMed] [Google Scholar]