Abstract
The quantity of milk and milk fat and proteins are particularly important traits in dairy livestock. However, little is known about the regions of the genome that influence these traits in goats. We conducted a genome wide association study in French goats and identified 109 regions associated with dairy traits. For a major region on chromosome 14 closely associated with fat content, the Diacylglycerol O-Acyltransferase 1 (DGAT1) gene turned out to be a functional and positional candidate gene. The caprine reference sequence of this gene was completed and 29 polymorphisms were found in the gene sequence, including two novel exonic mutations: R251L and R396W, leading to substitutions in the protein sequence. The R251L mutation was found in the Saanen breed at a frequency of 3.5% and the R396W mutation both in the Saanen and Alpine breeds at a frequencies of 13% and 7% respectively. The R396W mutation explained 46% of the genetic variance of the trait, and the R251L mutation 6%. Both mutations were associated with a notable decrease in milk fat content. Their causality was then demonstrated by a functional test. These results provide new knowledge on the genetic basis of milk synthesis and will help improve the management of the French dairy goat breeding program.
Introduction
In Europe, goat farming mainly targets cheese production. The average milk production level of the animals differs between regions of the world, partly due to different farm management systems but also to different genetics (breeds, selection). Goat breeding programs are still rare. Some countries have created collective structures to control performance and to estimate breeding values. The French breeding scheme is unique in the number of animals it includes and the high AI rate (40%). Like in the case of dairy sheep and dairy cows, the objectives of selection are generally the quantity and composition of the milk. Dairy traits are fundamental in livestock production. The efficiency of the French breeding program has been responsible for an annual genetic gain of +13 kg per year for milk yield and of +0.1 g/kg per year for fat and protein contents for the past ten years. Today France is the first producer of goat milk in the European Union, it produces 27% of the total volume of milk from only 10% of the animals, and is the fifth largest producer in the world1.
The composition of goat milk differs from that of cattle. It contains more minerals and more calcium, particularly due to its specific casein composition, which results in larger micelles2,3. The fatty acid composition of goat milk also differs, with a higher proportion of short and medium fatty acid chains which are also grouped in smaller fat globules3,4. The proportion of caproic (C6 :0), caprylic (C8 :0), capric (C10 :0) and lauric acids (C12 :0) is considerably higher in goat milk3,5, and these are the fatty acids that give a typical flavor to goat milk and cheese6. The composition of goat milk makes it easier to digest than that of cows, it has a higher nutritional value and is healthier and less allergenic3,7,8.
The availability of genome sequencing data has opened up new fields of investigation in domestic ruminant species with sequenced genomes9–11. The development of high-density single nucleotide polymorphism (SNP) arrays and their application in genome-wide association studies has facilitated the identification of regions that control quantitative traits in dairy cattle (for example12–20). However, very little is known about the loci controlling milk traits in goats. Using a candidate gene approach, the effect of caseins, especially that of the αs1 casein (CSN1S1), on the composition of milk (mainly protein content) is well documented in goats21–23. Many variants (>15) have been found for the different caseins24–28. Among the CSN1S1 variants, the effect varies from +3.6 g/L to almost no casein synthesized by homozygous animals carrying the null allele.
However, no genome wide association study of milk production has been conducted in goats. The GoatSNP50 BeadChip was released in 201129 and made this type of analysis possible. A large family design was therefore implemented in France to provide more information about the genetic control of milk traits.
The aim of the present work was to perform linkage analyses (LA) and linkage disequilibrium (LD) analyses, based on GoatSNP50 BeadChip data, to identify the genomic regions responsible for the quantity and composition of goat milk.
Results
Discovery of QTLs for milk production traits
QTLs associated with milk yield (MY), protein and fat yield (PY and FY, respectively) as well as protein content (PC) and fat content (FC), were mapped by means of a genome scan (29 autosomes) using both haplotype-based linkage and association analyses in 1,941 dairy goats distributed in 20 half-sib families. All the goats and their 20 sires were genotyped with the 50 K GoatSNP50 Beadchip (Illumina, San Diego, CA). Analyses were first conducted independently in each breed and then combined in a joint analysis.
Haplotype-based linkage was used to detect 24 QTLs at a 1% chromosome-wide threshold of significance (Table 1). Among them, 11 hits in four regions of chromosomes CHI 1, 6, 14, and 21 exceeded the 5% genome-wide threshold of significance. Many more QTLs were detected using association mapping, giving a total of 85 hits, which exceeded the 5% genome-wide threshold of significance (Fig. 1A,B, Table S1, Fig. S1). Genome scans are shown in Table S2.
Table 1.
Genome scan for milk production traits in a daughter design of 1,941 dairy goats, based on haplotype-based linkage analyses.
| CH I | Trait | Breed | Significance level | LRT | Position (Mb) | 95% –CI (min max) | Substitution effect | Candidate genes |
|---|---|---|---|---|---|---|---|---|
| 1 | PC | Alpine | *** | 41.6 | 1.382 | 1.362–1.449 | 0.36 | PDE9A |
| 1 | PC | All | ** | 51.8 | 1.418 | 1.404–1.434 | 0.30 | |
| 2 | FY | All | ** | 51.8 | 0.239 | 0.237–0.241 | 0.33 | |
| 5 | PC | Saanen | ** | 31.5 | 0.205 | 0.196–0.224 | 0.32 | |
| 6 | FC | All | *** | 66.2 | 0.764 | 0.743–0.812 | 0.32 | |
| 6 | FC | Saanen | *** | 44.3 | 0.805 | 0.746–0.872 | 0.44 | Caseins cluster |
| 6 | PC | Alpine | *** | 62.5 | 0.814 | 0.786–0.848 | 0.37 | Caseins cluster |
| 6 | PC | All | *** | 161.3 | 0.824 | 0.792–0.842 | 0.50 | Caseins cluster |
| 6 | PC | Saanen | *** | 99.6 | 0.824 | 0.791–0.845 | 0.66 | Caseins cluster |
| 7 | FC | Alpine | ** | 36.8 | 0.037 | 0.013–0.042 | 0.30 | SLC27A1 |
| 11 | PC | Alpine | ** | 39.9 | 0.900 | 0.887–0.906 | 0.35 | PAEP |
| 14 | FY | Alpine | *** | 39.9 | 0.034 | 0.025–0.046 | 0.30 | |
| 14 | FY | All | ** | 53.8 | 0.037 | 0.025–0.048 | 0.28 | |
| 14 | FC | All | *** | 126.2 | 0.111 | 0.092–0.124 | 0.48 | DGAT1 |
| 14 | FC | Saanen | *** | 60.8 | 0.123 | 0.092–0.143 | 0.50 | |
| 14 | FC | Alpine | *** | 70.8 | 0.155 | 0.146–0.160 | 0.42 | |
| 19 | PY | All | ** | 52.3 | 0.285 | 0.248–0.290 | 0.27 | PLD2 GGT6 ALOX12, ALOX 12B, ALOX 15 |
| 21 | FC | All | ** | 52.0 | 0.554 | 0.526–0.576 | 0.27 | |
| 21 | FC | Alpine | ** | 37.2 | 0.571 | 0.538–0.588 | 0.27 | |
| 21 | PC | All | ** | 49.9 | 0.578 | 0.570–0.585 | 0.27 | |
| 21 | PY | All | *** | 58.0 | 0.634 | 0.627–0.640 | 0.31 | |
| 21 | MY | All | ** | 53.2 | 0.635 | 0.627–0.642 | 0.29 | |
| 25 | FC | Saanen | ** | 32.4 | 0.101 | 0.099–0.103 | 0.35 | |
| 28 | PC | Alpine | ** | 33.7 | 0.322 | 0.295–0.361 | 0.31 |
Significance level: ***: 5% genome-wide; ** 1% chromosome-wide. The 95% confidence intervals of the QTL locations were estimated by logarithm of odds drop-off.
Figure 1.
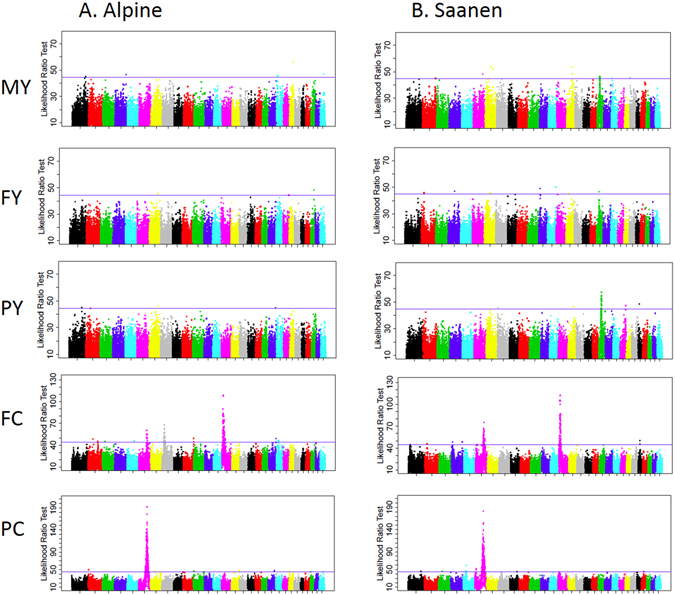
Manhattan plot of likelihood ratio test profiles for five milk production traits: milk yield (MY), fat yield (FY), protein yield (PY), fat content (FC) and protein content (PC) in Alpine (A) and Saanen goat breeds (B). The solid horizontal lines represent the 5% genome-wide thresholds (averaged over the 29 autosomes).
Among the many QTLs detected, two highly significant (5% genome-wide threshold) regions were mapped to similar locations in the Saanen and Alpine breeds by both the association and linkage analyses (Table 1, Fig. 1A,B, Table S1, Fig. S1). The first QTL was on CHI 6, and was associated with PC in all analyses in the region of the casein genes, i.e. 82.5–82.8 Mb (Table S1, Fig. 2). This region was also associated with FC in all association analyses and in two of the three linkage analyses (in the Saanen breed and in the two breeds combined analyses). The second QTL was on CHI 14 associated with FC (Table 1, Fig. 3, Table S1) in the region of the DGAT1 gene, which codes for a key enzyme involved in the synthesis of milk triglycerides. Both regions exhibited the highest average substitution effects, from 0.37 to 0.66 standard deviation (Table 1). In addition, CHI 6 and CHI 14 haplotypes (association analysis) explained 39.1% and 6.8% of variance of the PC and FC traits in the analyses of the two breeds, respectively. A region of CHI 21, spanning 11.6 Mb (52.6–64.2), was associated with PC (Table 1, Table S1), as well as with FC, PY and MY (Table 1). The most significant breed specific QTLs were found for PC on CHI 1, 136.2–14.9 Mb (linkage analysis, Table 1) and for FC on CHI 8, 22.8–23.1 Mb (association analysis, Fig. 1A, Table S1) in Alpine goats. In the Saanen breed, a region of chromosome 19 was highly significantly associated with yield, including PY, FY, and MY (Fig. 1B, Table S1). In this breed, this region spanned a confidence interval of 4 Mb, from 22.0 to 26.0 Mb (Table S1).
Figure 2.
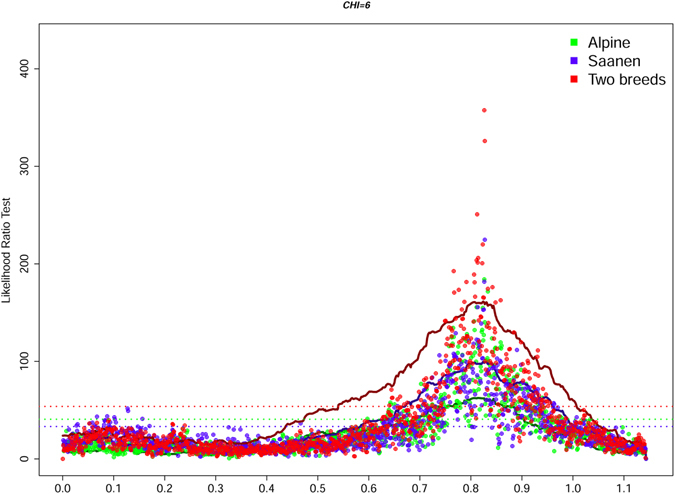
Global likelihood ratio test profiles for protein content on CHI 6 based on both linkage and haplotype-based linkage (solid lines) and association (dotted line) analyses. The dotted horizontal lines represent 5% genome-wide thresholds.
Figure 3.
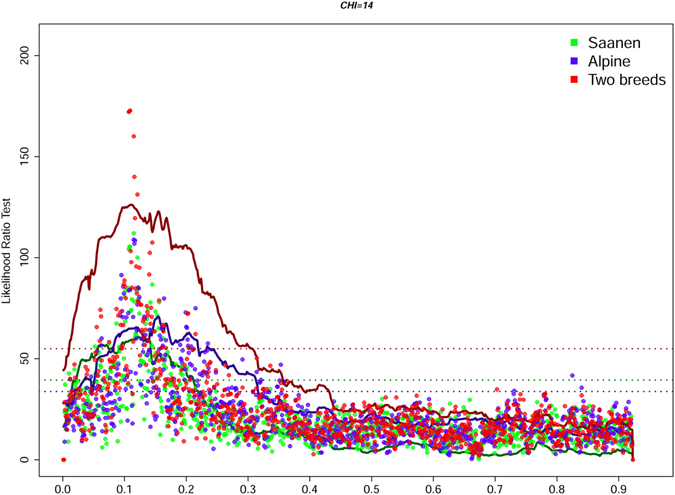
Global likelihood ratio test profiles for fat content on CHI 14 based on both linkage and haplotype-based linkage (solid lines) and association (dotted line) analyses. The dotted horizontal lines represent 5% genome-wide thresholds.
Fine-mapping identification of non-synonymous mutations in the DGAT1 gene
Discovery of candidate non-synonymous mutations in the DGAT1 gene for the CHI14 QTL.
Using the cattle DGAT1 mRNA sequence, we identified part of exon 1, exons 3–5 and 13–17. The combination of classic and long range PCR amplification followed by Sanger sequencing enabled us to construct an improved sequence by filling 7,725 out of the 8,728 undetermined nucleotides belonging to N blocks in the reference genome. This 37,251 bp sequence is available under accession number LT221856.
The SNP discovery in 2 Alpine and 2 Saanen animals with extreme phenotypes led to the identification of 29 polymorphisms (27 SNPs, a 1 bp and a 8-bp insertion) on the DGAT1 gene. The genotypes of the 20 bucks of the QTL design were determined for those 29 SNPs. SNPs were submitted to NCBI under ss numbers 1971466334–1971466359 and 1971466361–1971466363. Among these polymorphisms, 27 were intronic and two were located in exons 8 (NCBI_ss# 1971466363) and 15 (NCBI_ss# 1971466359), being responsible for the R251L and R396W substitutions in the DGAT1 protein sequence, respectively (Fig. 4).
Figure 4.
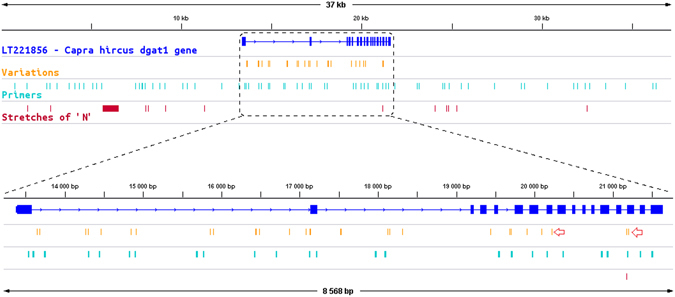
Determination of DGAT1 gene structure and polymorphism. The intron/exon structure of the LT221856 sequence is shown together with the SNPs detected and the primers used for sequencing. The location of the remaining N stretches is shown, together with their length (bp). A zoom on the coding region is also shown. The red arrows indicate the position of the R251L and R396W mutations.
Conservation of the DGAT1 protein sequence between ruminants
The goat DGAT1 protein sequence encompassed 489 amino acids, like in sheep and cattle. This sequence is extremely well conserved between these three species, as can be seen in Fig. S2. There was no difference between the goat and the sheep protein sequence, whereas four divergent amino acid sites were found in the cattle sequence.
The DGAT1 protein is still highly conserved at the scale of all ruminant species, with at least 88% of homology, as shown in Table S3. No variation across species was found at positions 251 and 396, where goat mutations were detected in the present study.
DGAT1 mutation frequency
The frequency of the genomic T mutation responsible for the R396W polymorphism was 12.0% in Saanen females and 8.1% in Alpine females,. The frequencies observed in AI males differed: 14.4% in the Saanen breed and 5.6% in the Alpine breed. No change in T frequency was observed in AI males between the year of birth 1998 and 2011 in either breed (Fig. S3, annual regression coefficient = 0.1).
At the R251L locus, the frequency of the T-allele was 4.4% in females and 2.6% in AI males of the Saanen breed. In the Alpine breed, no AI males were carriers of the T mutation and the frequency in females was 0.7%.
Association of the DGAT1 genotype with milk production traits and fatty acid composition of milk
Analysis of variance confirmed that the T-allele at codons 251 and 396 led to a dramatic decrease in milk fat content. The average FC (LS means of yield deviations) of females of each genotype at the R396W locus is shown in Fig. 5A. Concerning the effect of the R251L and R396W, significant differences between genotypes were also observed in FY with a difference of 3.05 kg (i.e 0.57 SD) between the two homozygous types (Fig. 5B). No significant effect of the R396W mutation was found on MY, PY or PC.
Figure 5.
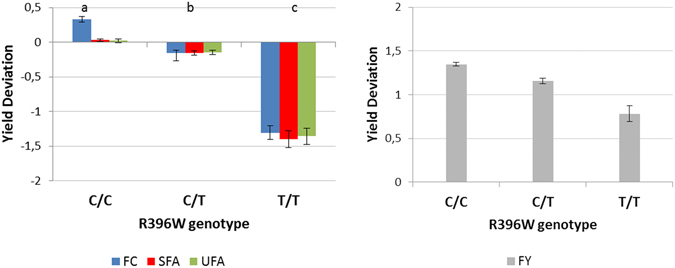
Effect of R396W genotype of DGAT1 gene on fat content (FC), quantity of saturated fatty acids (SFA) in milk, quantity of unsaturated fatty acids (UFA) in milk and fat yield (FY). The LS means have been estimated by using a mixed model including the genotype and the sire effect. Error bars indicate standard errors. Traits are expressed as the standard deviation of yield deviations. Lower case letters show significant differences in the trait between genotypes, as determined by a t-test at p < 0.05 for FY, and p < 0.005 for FC, SFA and UFA.
Concerning fatty acids, the R396W mutation consistently caused a significant decrease in the quantity of saturated and unsaturated fatty acids in milk (Fig. 5A). But the respective proportions of saturated and unsaturated fatty acids in the fat were not significantly affected. The R251L locus only affected fat content. Heterozygous carriers (G/T) had a significantly lower average fat content (−1.11 g/kg, i.e. −0.36 SD) than the homozygous wild type (G/G). The LS means of homozygous individuals (T/T)) were not estimated because too few females carried this genotype. The variance in fat content explained by the DGAT1 mutation differed significantly depending on the mutation: 46% for the R396W mutation and 6% for the R251L mutation, respectively.
Effect of the R251L and R396W mutations on the enzymatic activity of recombinant DGAT1
These two exonic mutations led to modifications in the amino acid sequence of the DGAT1 protein and were associated with changes in milk composition, e.g. a decrease in fat content. But even though these were the most likely causal mutations, an indirect effect due to linkage with the true genetic variant could not be excluded.
A functional test was consequently performed to prove their causality. First, using a baculovirus expression system in Sf21 insect cells (details in the Materials and Method section), one wild type and three different types of recombinant DGAT1 protein were produced: one with the R251L mutation only, one with the R396W mutation only and one carrying both mutations. Membrane fractions containing the proteins were prepared and the amount of neo-synthesized triglyceride from diglyceride produced by each membrane preparation under three different conditions was measured by gas chromatography – flame ionization detector (GCFID). The three conditions consisted in different amounts of incubated microsomes (10; 7.5 and 5 µL of protein solution) associated with an inversely proportional incubation time (2.5; 3.3 and 5 min), expected to yield identical amounts of product for a given protein, assuming enzymatic stability and lack of inhibition. The results are presented in Fig. 6. No activity was observed for the negative control membrane fraction. Analysis of these results with a generalized linear model showed that the condition effect was not significant (P = 0.10) thereby confirming that we were working in steady-state conditions, as required. In these conditions, the amount of synthesized triglycerides is indeed directly correlated with protein activity. In contrast, the construct effect was highly significant (P < 0.0001), indicating that the amount of triglyceride produced differed between the type of DGAT1 proteins included in the membrane fraction. DGAT1 constructions harboring the R251L and R396W mutation synthesized only 35% and 12% of the triglycerides, respectively, compared to the wild-type DGAT1 construction. The DGAT1 construction harboring both mutations showed a relative triglyceride synthesis of 10%.
Figure 6.
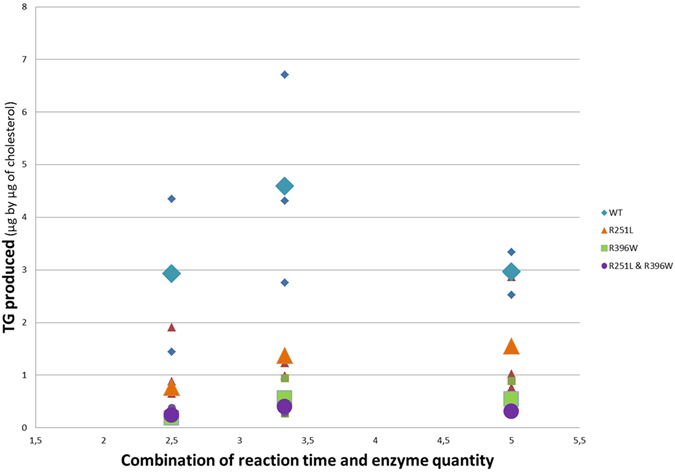
Quantity of triglyceride produced in each combination of reaction time and enzyme quantity for the four DGAT1 constructs: wild type, with the R251L mutation, with the R396W mutation, and with both mutations. This quantity of triglyceride has been corrected by an internal standard (TG19) and by the amount of measured cholesterol. The small symbols correspond to individual measurements and the large symbols are the mean of the corresponding group.
Discussion
In genome-wide association studies, we identified a large number of QTLs in 13 chromosomal regions, supporting the polygenic nature of dairy traits. Interestingly, a number of QTL regions included strong functional candidate genes or corresponded to orthologous regions associated with milk traits in cattle, as reported in the QTL database QTLdb (http://www.animalgenome.org/QTLdb).
A large number of QTLs with significant overlap across the cattle, sheep, and goat genome
Only some of the loci associated with production traits were common to the two breeds. Indeed, 13% to 19% of the QTLs for LA were detected in one breed only and 27% to 30% of the QTLs for LD were breed specific. These results suggest that the main genes responsible for milk composition - or allele frequencies - differ between the two goat breeds and that there is some level of genetic heterogeneity in the determinism of dairy traits in the Saanen and Alpine goat breeds.
The different QTLs found on CHI 220,30, CHI 531–33, CHI 1819,34,35, CHI 1932,34,36–38, CHI 2139,40, CHI 2441, CHI 2542 and even on CHI 8 detected only in LD (Table S1)43, all point to orthologous bovine QTL regions. However, the number of QTLs detected for milk traits in cattle is very high: QTLdb indeed accounts for more than 6,000 cattle data points, most of which concern milk traits44. Moreover, some of these bovine QTLs detected using low density marker panels (microsatellites), span large confidence intervals, together covering almost the whole genome, so it is not surprising that our QTLs overlap some of them. The QTL on CHI 28 was the only one of our significant signals with no corresponding bovine QTL. Under the conservative assumption that underlying causal genetic variants are the same (or are at least located on the same genes), the present study could help refine the confidence intervals of these bovine QTLs. We also found an orthologous ovine QTL for CHI 545, for CHI 19 but with a large interval (up to 30 Mb)46,47 and for CHI 2148.
Four strong candidate genes for protein and fat QTLs
A QTL for percentage protein was found at the orthologous bovine position of our QTL on the CHI 1 in Chinese Holstein cows49. Later, 11 more SNPs were identified on the phosphodiesterase 9A (PDE9A) gene, located in the QTL confidence interval, associated with the different milk production traits50. This gene is involved in the activation of the cGMP dependent pathway and is therefore a strong candidate for our QTL for PC on CHI1. In return, our data could help find the causal SNP - or exclude false positive SNPs - for the cattle QTL.
The QTL found for PC on the CHI 5 in the association analyses is located at a distance of one Mb from the lactalbumin alpha (LALBA) gene, which is a strong functional candidate gene. This gene codes for a major whey protein of the milk that is involved in the lactose synthase binary complex51. Polymorphisms in this gene influence milk traits in mice52, cattle42,53 and sheep51.
Concerning the QTL for fat content on CHI 7, the long chain fatty acid transport protein 1 gene (SLC27A1) has been proposed as a possible candidate gene for a QTL in the orthologous cattle genomic region54. Indeed, in an association study of 48 Chinese Holstein, Lv et al.54 identified a synonymous SNP in exon 3 of the gene associated with milk yield. This gene, which is involved in lipid metabolism, is a strong functional candidate gene. However, its localization on the goat chromosome 7 at 5.2 Mb is outside the confidence interval of our QTL on CHI 7 [1.3–4.2 Mb].
Finally, for the QTL for PC on CHI 11, the confidence interval includes the progestagen-associated endometrial protein gene (PAEP) which codes for the β lactoglobulin precursor and is known for its effect on milk protein content in cattle55. The β-lactoglobulin, which is absent from human milk, is one of the major whey proteins in ruminants and, moreover is considered to be a dominant milk allergen56. Implementing selection on this locus could be one way to reduce β lactoglobulin, thereby reducing the allergenic properties of goat milk.
In the Alpine breed, the QTL for FC on CHI 8 [22.8–23.1] spans the region of a single gene, the myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 gene (MLLT3) with no straightforward functional role in lipid metabolism. In contrast, the confidence interval for the QTL on CHI 19 (22–27.6 Mb), included 192 annotated genes. Among them, a few are related to fatty acid and lipid metabolism pathways: phospholipase D2 (PLD2), gamma-glutamyltransferase 6 (GGT6) and arachidonate lipoxygenase (ALOX) 12, −12B, −15.
The major region detected on CHI 6 corresponds to the casein cluster
The effect of caseins, especially CSN1S1, on the PC of milk and on other milk traits is well known in goats. A large number of variants has been found for the different caseins21–28. Nine of the existing variants (A, B1, B2, B3, B4, C, H, L and M are described as having a marked effect on the quantity of casein produced, two (E and I) have intermediate effects, three (D, F and G) have a weak effect and three others (O1, O2, N) are depicted as null alleles57,58. Alleles A, B, C, E and F were found in the French dairy goat population, with a preponderance of alleles E and F in the Saanen and Alpine breeds before 200059. Some alleles, including the E allele, contain insertions that cannot be typed with the 50 K chip60.
The frequencies of the CSN1S1 genotype in French goats were recently estimated28. The most frequent CSN1S1 genotypes in the French dairy goat population are genotypes AA and AE carried by more than 50% of all goats. More than 25% of the progeny-tested males carried the AA genotype. After estimating genetic parameters, Carillier-Jacquin et al.28 found that the polymorphism of CSN1S1 explained 24.4% of PC in the Saanen breed and 38.2% of PC in the Alpine breed.
QTL mapping to the goat casein cluster in the present study explained 39.1% of the variance in PC, suggesting that the GoatSNP50 BeadChip-based haplotypes mainly capture genetic variability due to the CSN1S1gene. Other polymorphisms probably play a role within the casein cluster (CSN1S2, CSN2, CSN3genes) and will be investigated in further comprehensive analyses of this region. QTLs for traits related to milk composition have been found in orthologous regions in other ruminant species14,19,20,61–64. Only a few polymorphisms in casein genes have been reported in cattle65–68. The large number of polymorphisms in CSN1S1 gene and their marked effect on milk composition is specific to the goat.
The DGAT1 protein is conserved between species but with a large number of existing variants
The DGAT1 gene is known to influence milk composition. This gene codes for a microsomal enzyme that catalyzes the last and limiting step of triglyceride synthesis, i.e. the transformation from a diacylglycerol to a triacylglycerol69,70. This enzyme, which was first known for its action in the formation of adipose tissue, has been shown to play a key role in lactation i.e DGAT1 knock-out mice were indeed unable to synthesize milk71.
Ours is the first study to report on the effect of the DGAT1 gene polymorphisms in the goat species and its influence on milk fat content. However, a bovine QTL for milk composition has been already associated with the DGAT1 gene in a genome scan15,72–75. The authors identified a non-synonymous mutation, K232A, which had an effect on milk fat content76–78 The results obtained by these authors strongly suggested that DGAT1 alone accounts for the QTL effect in the genomic region they had identified. and they showed that the enzymatic activity of the K232A recombinant DGAT1 protein is characterized by a lower Vmax. In sheep, a corresponding QTL has also been detected for dairy traits at the proximal end of ovine chromosome 9 (OAR9), the sheep homolog of BTA1461. Scata et al.79 completed the sheep DGAT1 sequence and found five polymorphisms, including two associated with an effect on milk composition.
Further, DGAT1 catalyzes the last step of triglyceride synthesis and its high protein sequence conservation between ruminant species supports the hypothesis that this protein plays a major role in biological functions. However, some variations have been reported within species. As DGAT1 was suspected to act in cattle and sheep, several authors already searched for polymorphisms in the goat DGAT1 gene by partially sequencing it, but most of the polymorphisms mapped to intronic and promoter regions80–84. The bovine mutation K232A was also the subject of unsuccessful searches85.
The DGAT1 protein is located on the membrane of the endoplasmic reticulum. Little is known about its three-dimensional structure, as no crystal structure of it or any closely homologous protein has yet been determined, but different hypotheses or predictions of the 2D or 3D structure have been proposed. Topology models predict eight86 or six87 transmembrane domains, whereas in vitro constructs predict only three and a N terminus oriented toward the cytosol86. If the active site was originally assumed to be on the cytosolic side of the membrane88,89, a more recent study suggests that a putative catalytic histidine is involved in the luminal side of the membrane. The histidine located at position 416, is not far from our major mutation R396W which, according to the present study, located in a very active part of the protein86. Based on their model, the R251L mutation would map near a transmembrane domain.
Importance of the DGAT1 mutations in goat
Like other DGAT1 mutations known to be associated with milk traits in ruminants, the major effect of the R251L and R396W mutations is on fat content76–79,82. The R396W mutation explains as much as 46% of the variance of the trait, which is similar to the variant with the strongest effect found in cattle76. We found no significant effect of the mutations on milk yield or protein content traits in contrast to what was found for the bovine K232A mutation76,78.
The frequency of the two mutations described here is lower than the frequency usually found for the alanine residue of the K232A mutation in different bovine populations78. The frequency of the bovine mutation is highly variable and differs among breeds and geographical areas. The positive effect of the mutation on MYand PC may have been selected in some breeding strategies, depending on the relative weight given to the different milk traits. Caprine mutations, although associated with lower fat contents, have not been eliminated, and their frequencies have remained stable in the male population in recent years.
The functional test revealed significant effects of the mutations on triglyceride production and strongly supports the hypothesis that these are causal mutations. We used a test that is quite similar to the functional test carried out in cattle: we measured the production of triglycerides using partially purified recombinant proteins. However, to enable more accurate estimations, we used the amount of cholesterol as a normalization factor of the membrane quantities used in the enzymatic reaction.
Another interesting point is that the R396W mutation has a stronger effect than the R251L, as shown both by the results of the in vitro functional test and of the in vivo milk analysis. However, the effect of the mutation estimated from these two experiments is not comparable.
The main possible future application of this work is the use of DGAT1 in the breeding scheme. We propose a molecular tool to test two novel mutations in the DGAT1 gene that can be genotyped on a routine and large scale basis. One short term application in goat breeding schemes would be to include information on the DGAT1 genotype in the estimation of breeding values for fat content. Recently, Carillier-Jacquin et al.28 investigated the benefits of including CSN1S1 major gene effect in the genetic evaluation of French dairy goats. Their results showed an improvement in predictive ability (from 6% to 27%) for the estimated breeding value, both in a genetic and genomic evaluation model (even if only males are genotyped). The potential gain of including the DGAT1 gene for the accuracy of the prediction could even be higher than the gain estimated for the multi-allelic CSN1S1 as they explain the same range of phenotypic variance in their respective traits and the DGAT1 mutations are only bi-allelic. The difficulty of predicting genotypes of ungenotyped animals is indeed the main limitation to improving the accuracy of the estimated breeding values and the prediction is easier for bi-allelic mutations. On the other hand, the DGAT1 genotyping result may help breeders chose among half-sib candidate bucks which one to bring to the breeding center for further selection and progeny testing.
Conclusions
In this study, we identified a large number of QTLs for dairy traits in goats. Further, we identified two mutations in the DGAT1 gene associated with a reduction in fat content and proved their causality with a functional test. These results advance our understanding of the genetic architecture of caprine milk composition and will be useful for breeding programs. Our results could also help develop a naturally low fat milk market, and are possibly transferable to other breeds by introgression or new genetic engeneering such as CRISPR-cas996, if and when this technique receives social acceoptance for the genetic improvement of animals.
Materials and Methods
Resource population
The data for QTL detection came from a total of 2,209 commercial French dairy goats sampled in 2010 as part of the national “Genomcap” and the EU “3SR” (www.3srbreeding.eu) projects. The 2,209 animals were distributed in 20 half-sib families sired by 9 Saanen and 11 Alpine artificial insemination (AI) bucks. Family size averaged 109 (±16) daughters per buck and ranged from to 73 to 126. AI bucks were chosen to be both representative of the genetic diversity of the whole population and to maximize the genetic diversity between families. They were chosen among the widely used AI bucks with large numbers of daughters in commercial farms in 2009 and 2010.
Ethics Statement
DNA samples for this study are stored at the Laboratoire d’Analyses Génétiques pour les Espèces Animales (LABOGENA, Jouy en Josas, France; www.labogena.fr). Neither sperm collection nor blood sampling was performed specifically for this study. Sperm was collected from bucks by the goat breeding organization Capgenes (Mignaloux Beauvoir, France; http://www.capgenes.com/), with the authorization of the DGAL (Direction Générale de l’ALimentation) FR CC 860. Sperm was collected at artificial insemination stations, and we used extra doses from this collection. Blood samples were taken at commercial farms. The animals were not part of any experimental design. They were sampled by veterinarians and/or under veterinarian supervision as part of routine management practices. Extra samples were requested when blood sampling took place.
Genome-wide SNP genotyping
All 2,209 animals were genotyped using the Illumina GoatSNP50 BeadChip (53,347 SNPs). DNA was extracted from blood samples and genotyping was performed at LABOGENA, following the manufacturer’s instructions.
Data were filtered using an in-house pipeline. Briefly, any individual with a call rate <95% (N = 16) or showing pedigree inconsistency (N = 228, i.e. 10%) was discarded. SNP quality control included the following inclusion criteria: call rate >99%, minor allele frequency >1% and Hardy-Weinberg P-value > 10–6. After filtering, a total of 44,612 SNPs distributed on goat autosomes were retained for further analyses. Marker order and positions were based on the caprine Assembly CHIR_1.0 downloaded (December 9, 2014) from the following link: http://bioinformatics.tecnoparco.org/SNPchimp/index.php/download/download-goat-data.
Phenotypic measurements
The traits considered were MY, PY, FY, PC and FC. Traits were standardized to 250 days of lactation. For QTL detection, yield deviations (YD) for all five traits were provided by the French national genetic evaluation computing centre97. For all traits, YD were raw data corrected for the fixed effects included in the genetic evaluation model: flock, age at kidding, month of kidding, dry period length, year by region combination and the random permanent environment. Next, YD were averaged over all the lactations of one animal (maximum 3 lactations per animal). A total of 1,941 phenotyped and genotyped animals were used for QTL detection. Family size averaged 97 (±16) daughters per buck (range: 60 to 115).
Association mapping
For QTL detection, both linkage analyses (LA) and linkage disequilibrium (LD) using interval mapping were applied to the data using the QTLMap software (ref.98; http://dga7.jouy.inra.fr/qtlmap/). For LA, interval mapping99 was performed with the likelihood ratio test (LRT) using within-sire linear regression100. The QTL effect (average substitution effect) was expressed in deviation units (SD). Linkage disequilibrium was based on a regression analysis of the phenotypes onto founder haplotypes101. Analyses were performed for each haplotype of four consecutive SNPs along the chromosome. The computations of phase and transmission probabilities were optimized to be rapid and as exact as possible.
Chromosome-wide significance levels were calculated with QTLMap, using the current family structure and the MY phenotypes. For LA, the empirical 5% and 1% chromosome-wide significance levels of the test statistics were estimated from 1,000 within-family permutations102 for each chromosome. For LD, the empirical chromosome-wide significance level of the test statistics was estimated from 1,000 simulations for each chromosome, assuming a trait of heritability equal to 0.35. The 5% genome-wise thresholds were obtained by applying the Bonferroni correction Pgenome-wise = 1 - (1 - Pchromosome-wise)n, where n is the number of autosomes analyzed, i.e. 29.
The 95% confidence intervals of the QTL locations were estimated with the logarithm of odds drop-off99 implemented in QTLMap software. In practice, the bounds of the interval were the two locations where the likelihood was equal to the maximum likelihood minus 3.84 [].
Sequencing and SNP calling for the 20 AI buck
Because evidence already existed for high conservation between cattle and goat DGAT1 sequences80, the 1785 bp Bos taurus mRNA sequence (NM_174693.2) was used to identify the orthologous DGAT1 region in the Capra hircus CHIR_1.0 genome and the exonic regions, through sim4 program103. Using this information, N blocks (>9 consecutive undetermined nucleotides) were identified in the reference sequence in the DGAT1 orthologous region extended to 15 kb upward and forward. PCR amplifications were performed to produce DNA fragments containing one or several N blocks, using the Long PCR Enzyme Mix provided by Fermentas (http://www.fermentas.de). Either PCR or internal primers were used for the Sanger sequencing reaction with the BigDye Terminator v3.1 Cycle Sequencing Kit (http://www.appliedbiosystems.com), after ExoSAP treatment104, and amplicons were analyzed on a ABI3730 DNA analyzer (ThermoFisher Scientific).
Sequences were aligned using DNAbaser software (http://www.dnabaser.com/) to generate a consensus sequence from two Alpine and two Saanen animals with extreme phenotypes for fat content.
SNP discovery was carried out by Sanger sequencing on the same animals. The genotypes of the discovered SNPs were determined using the Sanger sequencing method described above for the relevant PCR products of the remaining bucks of the QTL design.
The primers used in this study are listed in Table S4.
Comparison of protein sequences among ruminants
The sequence of the goat DGAT1 gene was then translated and the corresponding protein sequence was compared to those of other ruminants, The sheep and bovine sequences were extracted from Uniprot (http://www.uniprot.org/ entries A8VJM4 and Q8MK44 respectively). The weblogo software (http://weblogo.threeplusone.com/) was used to obtain a graphical representation of the conservation of the protein between three domestic ruminant (cattle, sheep and goats). Predicted protein sequences of other ruminant species (Ceratotherium simum; Bulbalus bubalis; Camelus dromedaries; Camelus bactrianus and Vicugna_pacos) were extracted from NCBI (http://www.ncbi.nlm.nih.gov/gene/?term=dgat1). A percent identity matrix was then created between all the species retained using the Clustal omega software (http://www.ebi.ac.uk/Tools/msa/clustalo/).
DGAT1 genotyping
SNP ss# 1971466363 (corresponding to R251L mutation) and SNP ss# 1971466359 (corresponding to R396W mutation) were genotyped using a PCR-RFLP test, using BstUI and MspI (New England Biolabs) enzymes respectively. Briefly, 50 ng of the appropriate PCR product were digested with 10 U of enzyme according to the manufacturer’s instructions. After migration on a 3% TBE agarose gel, the genotypes were assessed independently by two different people. All the females from the resource population and 752 AI males were genotyped in this way.
Association of DGAT1 genotype with milk production traits and milk fatty acid composition
A single SNP test of association was performed for each of the two mutations by performing an analysis of variance (ANOVA), using the mixed procedure in the statistical analysis system (SAS 9.1). The dependent variables were yield deviations (YD). For milk production traits (MY, PY, FY, PC, FC), YD were calculated in the context of the national genetic evaluation, similar to those used for the QTL detection. For milk fatty acid composition, YD were estimated for two traits, saturated and unsaturated fatty acids, predicted from mid-infrared spectrometry105. Test-day fatty acid compositions from females during their first and second lactations were corrected for six fixed effects: herd-test-day, day in milk, parity, month of kidding, season of measurement, time of day (morning or evening) of milking, intra lactation stage, and spectrometers (n = 6 machines in three laboratories, MilkoScan FT6000 and MilkoScan FT+; Foss Electric, Hillerod, Denmark).
Fatty acid composition was expressed in two different measurement units: g per 100 g of milk and g per 100 g of fat. For each of the two mutations, the three possible genotypes were fitted as a fixed explanatory variable and a significance threshold of p < 0.01 was selected. The varcomp procedure of SAS was used to estimate the proportion of variance explained by the genotype. The sire was included as a random effect in both mixed and varcomp models. For the R396W polymorphism, analyses were performed on the two breeds pooled. For the R251L polymorphism, only the Saanen breed was analyzed because too few females were carriers of the mutation in the Alpine breed.
Traits are expressed as the standard deviation of YD. The values of standard deviations were 5.39 kg for FY, 3.12 g/kg for FC, 0.50 g per 100 g of milk for SFA, 0.17 g per 100 g of milk for UFA.
Expression of recombinant DGAT1 in Sf21 cells
DGAT1 cDNAs were obtained by RT-PCR from total RNA extracted from blood samples of two R396W heterozygous Alpine goats. The DGAT1 coding sequence was then amplified using primers 1 & 2 and cloned into TOPO TA vector (Invitrogen). The coding sequence was then reamplified from the TOPO plasmid for cloning into the pFastBac1 vector using primers 3 and 4. PCR amplifications were conducted on an ABI 9700 thermocycler (Applied Biosystems) with the following program: 1 min initial denaturation at 98 °C, 65 cycles of 30 s at 98 °C, 30 s extension at 68 °C and 75 s at 72 °C, followed by 10 min final extension at 72 °C. The 25 µL amplification mixture contained 100 ng of DNA, 1 unit Q5 high fidelity DNA polymerase (New England Biolabs), 5 µL Q5 PCR buffer, 5 µL Q5 enhancer, dNTP 200 µM and 12.5 pmol of each primer.
The PCR products were purified using QIAquick PCR purification kit (QIAgen) and double digested, as were the vectors, with BamHI-HF and XhoI (New England Biolabs). The digested inserts were then ligated into the pFastBac1 linearized vector. The different alleles (wild type, mutated R251L, mutated both R251L and R396W) were obtained from the mutated R396W clones using the QuickChange II Site-Directed Mutagenesis Kit (Agilent) to obtain the R251L mutation and to generate the wild-type codon at position 396. At each step, inserts of the clones were completely sequenced to check the integrity of the insert sequence. The primers used are listed in Table S4.
Transposition of the coding sequence into DH10Bac and multiplication and titration of recombinant virus using Sf21 cells were performed using a Bac-To-Bac Baculovirus Expression System, (Invitrogen) according to the manufacturer’s recommendations.
The harvested Sf21 cells obtained after infection were resuspended in 5 mL of a buffer containing 50 mM Tris HCl and 250 mM NaCl at pH 8. 200 µL of 25X Protease inhibitor was added and the solution was disrupted by sonication, 4 times x 10 pulses at 40%. Nuclei and large cellular debris were pelleted by centrifuging at 10,000 × g for 10 min. The supernatant was then ultracentrifuged for 1 h at 100,000 × g at 4 °C and the resulting pellet was resuspended in 250 µL of the same buffer as above.
DGAT1 Activity Assay
DGAT1 activity was tested using the wild type DGAT1, the three recombinant DGAT1 (mutated R251L, mutated R396W and mutated both R251L and R396W) and a membrane fraction of Sf21 transfected with DDX3X used as negative control. DDX3X was a 662 amino-acid protein with no known implication in lipid metabolism.
The activity of each sample was tested in a final volume of 160 µL containing 250 mM sucrose, 1 mM EDTA, 20 mM MgCl2, 100 mM Tris HCl (pH 7.5), 20 µg free fatty acid BSA, 20 µg of diacylglycerol 10:10 and 10 µg of 14-acylCoA. The reaction was triggered by adding to this assay mixture 10, 7.5 or 5 µL of protein solution at 37 °C. The time of incubation was inversely proportional to the amount of protein added: 2.5, 3.3 or 5 min. The reaction was stopped on ice. Lipids were then extracted using the Bligh and Dyer method106 in presence of an internal standard (glyceryl trinonadecanoate (Tg19) 8 mg) and solid phase extraction (SPE) was carried out. Briefly a SiOH glass cartridge (200 mg, Macherey Nagel) was equilibrated with 2 ml of dichloromethane, and the extract was then put down in 20 µl of 10% methanol in dichloromethane, and neutral lipids were eluted with 2 ml of the same mixture. The final extract was concentrated, dissolved in 20 µl of ethyl acetate and analyzed by gas chromatography with flame ionization detection using the FOCUS Thermo Electron system equiped with an Zebron-1 Phenomenex fused silica capillary columns (5 m × 0.32 mm i.d, 0.50 µm film thickness). Oven temperature was programmed from 200 °C to 350 °C at a rate of 5 °C per min and the carrier gas was hydrogen (0.5 bar). The injector and the detector were programmed at 315 °C and 345 °C respectively107.
The amount of measured triglyceride 10:10:14 was then corrected by an internal standard (TG19) and by the amount of measured cholesterol. As the cholesterol reflects the membrane concentration, it was used to normalize the amount of membrane between the samples. Each condition was repeated three times.
Avaliability of data and materials
SNPs were submitted to NCBI (dbSNP) under ss numbers 1971466334-1971466359 and 1971466361-1971466363. All other relevant data are within the article and its supplementary information files.
Electronic supplementary material
Acknowledgements
The authors thank Pierre Martin and all the Capgenes breeding organizations for providing the data and their helpful contributions. This work was supported by grants from French organizations through “PhénoFinlait” (a research programm including INRA, APIS-GENE, ALLICE (formerly UNCEIA), CAPGENES and FCEL) and EC (FP7/2007–2013), grant no. 245140, “3SR”, Sustainable Solutions for Small Ruminants (http://www.3srbreeding.eu/). The authors thank all the partners involved. The authors also thank Cécile Albenne for her precious help in enzymology, Stéphane Fabre, Marc Teisser and Julie Demars for their involvement and help with softwares and molecular part, and Aurélie Tircazes for her help with genotype calling. Lipidomic analyses were performed on the Toulouse MetaToul-Lipidomique Core Facility (I2MC, Inserm 1048, Toulouse, France), MetaboHUB-ANR-11-INBS-0010.
Author Contributions
R.R., I.P. and G.T.-K. conceived and designed th experiments. P.M., C.M., K.C.-T., J.S., F.W., I.R., H.B. and J.B.-M. performed the experiments. P.M., R.R., I.P., C.M. and P.B. analysed the data. P.M., R.R., G.T.-K. and I.P. wrote the article. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Rachel Rupp and Gwenola Tosser-Klopp contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-22118-x.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02052-0
Accession codes: The 37,251 bp goat DGAT1 sequence is publically available under accession number LT221856 (submitted to European Nucleotide Archive).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Maigret, C. Chiffres clés 2016, Productions Caprines lait et viande, Institut de l’Elevage et Confédération Nationale de l’Elevage http://idele.fr/?eID=cmis_download&oID=workspace://SpacesStore/f97384dd-f5cb-46bb-aaf5-d84f0326d422, 2016, (Date of access:04/01/2017).
- 2.Remeuf F. Influence du polymorphisme génétique de la caséine αs1 caprine sur les caractéristiques physico-chimiques et technologiques du lait. Lait. 1993;73:549–557. doi: 10.1051/lait:19935-652. [DOI] [Google Scholar]
- 3.Park YW, Juárez M, Ramos M, Haenlein GFW. Physico-chemical characteristics of goat and sheep milk. Small Rumin. Res. 2007;68:88–113. doi: 10.1016/j.smallrumres.2006.09.013. [DOI] [Google Scholar]
- 4.Boichard, D. et al. Déterminisme génétique de la composition en acides gras et protéines du lait des ruminants et potentialités de sélection. INRA Prod. Anim. 27, 283–298 (2014).
- 5.Alonso L, Fontecha J, Lozada L, Fraga MJ, Juárez M. Fatty acid composition of caprine milk: major, branched-chain, and trans fatty acids. J. Dairy Sci. 1999;82:878–884. doi: 10.3168/jds.S0022-0302(99)75306-3. [DOI] [PubMed] [Google Scholar]
- 6.Skjevdal T. Flavour of goat’s milk: A review of studies on the sources of its variations. Livest. Prod. Sci. 1979;6:397–405. doi: 10.1016/0301-6226(79)90007-1. [DOI] [Google Scholar]
- 7.Park YW. Hypo-allergenic and therapeutic significance of goat milk. Small Rumin. Res. 1994;14:151–159. doi: 10.1016/0921-4488(94)90105-8. [DOI] [Google Scholar]
- 8.Haenlein GFW. Goat milk in human nutrition. Small Rumin. Res. 2004;51:155–163. doi: 10.1016/j.smallrumres.2003.08.010. [DOI] [Google Scholar]
- 9.Bovine Genome Sequencing and Analysis Consortium et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science324, 522–528 (2009). [DOI] [PMC free article] [PubMed]
- 10.International Sheep Genomics Consortium et al. The sheep genome reference sequence: a work in progress. Anim. Genet. 41, 449–453 (2010). [DOI] [PubMed]
- 11.Dong Y, et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) Nat. Biotechnol. 2013;31:135–141. doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- 12.Ashwell MS, et al. Detection of putative loci affecting milk production and composition, health, and type traits in a United States Holstein population. J. Dairy Sci. 1998;81:3309–3314. doi: 10.3168/jds.S0022-0302(98)75896-5. [DOI] [PubMed] [Google Scholar]
- 13.Ashwell MS, Rexroad CE, Jr, Miller RH, VanRaden PM, Da Y. Detection of loci affecting milk production and health traits in an elite US Holstein population using microsatellite markers. Anim. Genet. 1997;28:216–222. doi: 10.1111/j.1365-2052.1997.00115.x. [DOI] [Google Scholar]
- 14.Georges M, et al. Mapping Quantitative Trait Loci Controlling Milk Production in Dairy Cattle by Exploiting Progeny Testing. Genetics. 1995;139:907–920. doi: 10.1093/genetics/139.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyen DW, et al. A genome scan for QTL influencing milk production and health traits in dairy cattle. Physiol. Genomics. 1999;1:165–175. doi: 10.1152/physiolgenomics.1999.1.3.165. [DOI] [PubMed] [Google Scholar]
- 16.Boichard D, et al. Detection of genes influencing economic traits in three French dairy cattle breeds. Genet. Sel. Evol. 2003;35:77–101. doi: 10.1186/1297-9686-35-1-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolbehdari D, et al. A whole genome scan to map QTL for milk production traits and somatic cell score in Canadian Holstein bulls. J. Anim. Breed. Genet. Z. Für Tierz. Zücht. 2009;126:216–227. doi: 10.1111/j.1439-0388.2008.00793.x. [DOI] [PubMed] [Google Scholar]
- 18.Meredith BK, et al. Genome-wide associations for milk production and somatic cell score in Holstein-Friesian cattle in Ireland. BMC Genet. 2012;13:21. doi: 10.1186/1471-2156-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen HG, et al. A genome scan for quantitative trait loci affecting milk production in Norwegian dairy cattle. J. Dairy Sci. 2002;85:3124–3130. doi: 10.3168/jds.S0022-0302(02)74400-7. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, et al. Mapping quantitative trait loci for milk production and health of dairy cattle in a large outbred pedigree. Genetics. 1998;149:1959–1973. doi: 10.1093/genetics/149.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grosclaude F, Mahé M-F, Brignon G, Di Stasio L, Jeunet R. A Mendelian polymorphism underlying quantitative variations of goat αs1-casein. Génétique Sélection Évolution. 1987;19:399–412. doi: 10.1186/1297-9686-19-4-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leroux C, Mazure N, Martin P. Mutations away from splice site recognition sequences might cis-modulate alternative splicing of goat αS1-casein transcripts. Structural organization of the relevant gene. J Biol Chem. 1992;267:47–57. [PubMed] [Google Scholar]
- 23.Boulanger A, Grosclaude F, Mahé M-F. Polymorphisme des caséines αs1 et αs2 de la chèvre (Capra hircus) Génétique Sélection Évolution. 1984;16:157–176. doi: 10.1051/gse:19840203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin P, Ollivier-Bousquet M, Grosclaude F. Genetic polymorphism of caseins: a tool to investigate casein micelle organization. Int. Dairy J. 1999;9:163–171. doi: 10.1016/S0958-6946(99)00055-2. [DOI] [Google Scholar]
- 25.Neveu C, Riaublanc A, Miranda G, Chich J-F, Martin P. Is the apocrine milk secretion process observed in the goat species rooted in the perturbation of the intracellular transport mechanism induced by defective alleles at the alpha(s1)-Cn locus? Reprod. Nutr. Dev. 2002;42:163–172. doi: 10.1051/rnd:2002015. [DOI] [PubMed] [Google Scholar]
- 26.Chessa S, et al. Short communication: the beta-casein (CSN2) silent allele C1 is highly spread in goat breeds. J. Dairy Sci. 2008;91:4433–4436. doi: 10.3168/jds.2008-1228. [DOI] [PubMed] [Google Scholar]
- 27.Chiatti F, et al. Effect of kappa-casein polymorphism on milk composition in the Orobica goat. J. Dairy Sci. 2007;90:1962–1966. doi: 10.3168/jds.2006-508. [DOI] [PubMed] [Google Scholar]
- 28.Carillier-Jacquin C, Larroque H, Robert-Granie C. Including as1 casein gene information in genomic evaluations of French dairy goats. Genet. Sel. Evol. 2016;48:54. doi: 10.1186/s12711-016-0233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosser-Klopp G, et al. Design and Characterization of a 52K SNP Chip for Goats. PLoS ONE. 2014;9:e86227. doi: 10.1371/journal.pone.0086227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashwell MS, et al. Detection of quantitative trait loci affecting milk production, health, and reproductive traits in Holstein cattle. J. Dairy Sci. 2004;87:468–475. doi: 10.3168/jds.S0022-0302(04)73186-0. [DOI] [PubMed] [Google Scholar]
- 31.Chamberlain AJ, et al. Validation of single nucleotide polymorphisms associated with milk production traits in dairy cattle. J. Dairy Sci. 2012;95:864–875. doi: 10.3168/jds.2010-3786. [DOI] [PubMed] [Google Scholar]
- 32.Bennewitz J, et al. Combined analysis of data from two granddaughter designs: A simple strategy for QTL confirmation and increasing experimental power in dairy cattle. Genet. Sel. Evol. 2003;35:319–338. doi: 10.1186/1297-9686-35-3-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrooten C, Bink MCaM, Bovenhuis H. Whole genome scan to detect chromosomal regions affecting multiple traits in dairy cattle. J. Dairy Sci. 2004;87:3550–3560. doi: 10.3168/jds.S0022-0302(04)73492-X. [DOI] [PubMed] [Google Scholar]
- 34.Bennewitz J, et al. Multiple quantitative trait loci mapping with cofactors and application of alternative variants of the false discovery rate in an enlarged granddaughter design. Genetics. 2004;168:1019–1027. doi: 10.1534/genetics.104.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutiérrez-Gil B, Wiener P, Richardson RI, Wood JD, Williams JL. Identification of QTL with effects on fatty acid composition of meat in a Charolais x Holstein cross population. Meat Sci. 2010;85:721–729. doi: 10.1016/j.meatsci.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 36.Morris CA, et al. Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2007;18:64–74. doi: 10.1007/s00335-006-0102-y. [DOI] [PubMed] [Google Scholar]
- 37.Mosig MO, et al. A whole genome scan for quantitative trait loci affecting milk protein percentage in Israeli-Holstein cattle, by means of selective milk DNA pooling in a daughter design, using an adjusted false discovery rate criterion. Genetics. 2001;157:1683–1698. doi: 10.1093/genetics/157.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bagnato A, et al. Quantitative trait loci affecting milk yield and protein percentage in a three-country Brown Swiss population. J. Dairy Sci. 2008;91:767–783. doi: 10.3168/jds.2007-0507. [DOI] [PubMed] [Google Scholar]
- 39.Bouwman AC, Visker MHPW, van Arendonk JAM, Bovenhuis H. Genomic regions associated with bovine milk fatty acids in both summer and winter milk samples. BMC Genet. 2012;13:93. doi: 10.1186/1471-2156-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Zas SL, Southey BR, Heyen DW, Lewin HA. Detection of quantitative trait loci influencing dairy traits using a model for longitudinal data. J. Dairy Sci. 2002;85:2681–2691. doi: 10.3168/jds.S0022-0302(02)74354-3. [DOI] [PubMed] [Google Scholar]
- 41.Li X, et al. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J. Dairy Sci. 2015;98:8152–8163. doi: 10.3168/jds.2015-9383. [DOI] [PubMed] [Google Scholar]
- 42.Schopen GCB, et al. Whole-genome association study for milk protein composition in dairy cattle. J. Dairy Sci. 2011;94:3148–3158. doi: 10.3168/jds.2010-4030. [DOI] [PubMed] [Google Scholar]
- 43.Bouwman AC, Bovenhuis H, Visker MHPW, van Arendonk JAM. Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011;12:43. doi: 10.1186/1471-2156-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z-L, Park CA, Wu X-L, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41:D871–D879. doi: 10.1093/nar/gks1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Gámez E, Gutiérrez-Gil B, Suarez-Vega A, de la Fuente LF, Arranz JJ. Identification of quantitative trait loci underlying milk traits in Spanish dairy sheep using linkage plus combined linkage disequilibrium and linkage analysis approaches. J. Dairy Sci. 2013;96:6059–6069. doi: 10.3168/jds.2013-6824. [DOI] [PubMed] [Google Scholar]
- 46.García-Fernández M, Gutiérrez-Gil B, García-Gámez E, Sánchez JP, Arranz JJ. The identification of QTL that affect the fatty acid composition of milk on sheep chromosome 11. Anim. Genet. 2010;41:324–328. doi: 10.1111/j.1365-2052.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- 47.Jonas E, et al. Mapping quantitative trait loci (QTL) in sheep. IV. Analysis of lactation persistency and extended lactation traits in sheep. Genet. Sel. Evol. GSE. 2011;43:22. doi: 10.1186/1297-9686-43-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crisà A, et al. Exploring polymorphisms and effects of candidate genes on milk fat quality in dairy sheep. J. Dairy Sci. 2010;93:3834–3845. doi: 10.3168/jds.2009-3014. [DOI] [PubMed] [Google Scholar]
- 49.Russo V, et al. A whole genome scan for QTL affecting milk protein percentage in Italian Holstein cattle, applying selective milk DNA pooling and multiple marker mapping in a daughter design. Anim. Genet. 2012;43(Suppl 1):72–86. doi: 10.1111/j.1365-2052.2012.02353.x. [DOI] [PubMed] [Google Scholar]
- 50.Yang S-H, et al. Validation of PDE9A Gene Identified in GWAS Showing Strong Association with Milk Production Traits in Chinese Holstein. Int. J. Mol. Sci. 2015;16:26530–26542. doi: 10.3390/ijms161125976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García-Gámez E, et al. GWA analysis for milk production traits in dairy sheep and genetic support for a QTN influencing milk protein percentage in the LALBA gene. PloS One. 2012;7:e47782. doi: 10.1371/journal.pone.0047782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stinnakre MG, Vilotte JL, Soulier S, Mercier JC. Creation and phenotypic analysis of alpha-lactalbumin-deficient mice. Proc. Natl. Acad. Sci. USA. 1994;91:6544–6548. doi: 10.1073/pnas.91.14.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lundén, A. & Lindersson. α-Lactalbumin polymorphism in relation to milk lactose. In Proceedings of the 6th World Congress on Genetics Applied to Livestock Production25 (1998).
- 54.Lv Y, et al. Association between polymorphisms in the SLC27A1 gene and milk production traits in Chinese Holstein cattle. Anim. Biotechnol. 2011;22:1–6. doi: 10.1080/10495398.2011.527567. [DOI] [PubMed] [Google Scholar]
- 55.Huang W, et al. Association between milk protein gene variants and protein composition traits in dairy cattle. J. Dairy Sci. 2012;95:440–449. doi: 10.3168/jds.2011-4757. [DOI] [PubMed] [Google Scholar]
- 56.Cui C, et al. Gene targeting by TALEN-induced homologous recombination in goats directs production of β-lactoglobulin-free, high-human lactoferrin milk. Sci. Rep. 2015;5:10482. doi: 10.1038/srep10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leroux C, et al. Real-time RT-PCR and cDNA macroarray to study the impact of the genetic polymorphism at the α s1 -casein locus on the expression of genes in the goat mammary gland during lactation. Reprod. Nutr. Dev. 2003;43:459–469. doi: 10.1051/rnd:2003032. [DOI] [PubMed] [Google Scholar]
- 58.Selvaggi M, Laudadio V, Dario C, Tufarelli V. Major proteins in goat milk: an updated overview on genetic variability. Mol. Biol. Rep. 2014;41:1035–1048. doi: 10.1007/s11033-013-2949-9. [DOI] [PubMed] [Google Scholar]
- 59.Martin, P. & Leroux, C. Le gène caprin spécifiant la caséine αs1: un suspect tout désigné aux effets aussi multiples qu’inattendus. INRA Productions Animales 125–132 (2000).
- 60.Grosclaude, F. et al. From gene to cheese: The caprine αs1-casein polymorphism, its effects and its evolution. INRA Productions animales 3–19 (1994).
- 61.Barillet F, Arranz J-J, Carta A. Mapping quantitative trait loci for milk production and genetic polymorphisms of milk proteins in dairy sheep. Genet. Sel. Evol. GSE. 2005;37:S109–S123. doi: 10.1186/1297-9686-37-S1-S109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kühn C, et al. Isolation and application of chromosome 6 specific microsatellite markers for detection of QTL for milk-production traits in cattle. J. Anim. Breed. Genet. 1996;113:355–362. doi: 10.1111/j.1439-0388.1996.tb00626.x. [DOI] [Google Scholar]
- 63.Spelman RJ, Coppieters W, Karim L, van Arendonk JA, Bovenhuis H. Quantitative trait loci analysis for five milk production traits on chromosome six in the Dutch Holstein-Friesian population. Genetics. 1996;144:1799–1808. doi: 10.1093/genetics/144.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bovenhuis H, Weller JI. Mapping and analysis of dairy cattle quantitative trait loci by maximum likelihood methodology using milk protein genes as genetic markers. Genetics. 1994;137:267–280. doi: 10.1093/genetics/137.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamiński S, Cieslińska A, Kostyra E. Polymorphism of bovine beta-casein and its potential effect on human health. J. Appl. Genet. 2007;48:189–198. doi: 10.1007/BF03195213. [DOI] [PubMed] [Google Scholar]
- 66.Prinzenberg EM, Krause I, Erhardt G. SSCP analysis at the bovine CSN3 locus discriminates six alleles corresponding to known protein variants (A, B, C, E, F, G) and three new DNA polymorphisms (H, I, A1) Anim. Biotechnol. 1999;10:49–62. doi: 10.1080/10495399909525921. [DOI] [PubMed] [Google Scholar]
- 67.Grosclaude, F. e polymorphisme génétique des principales lactoprotéines bovines. Relations avec la quantité, la composotop, et les aptitudes fromagères du lait. INRA Productions animales 5–7 (1988).
- 68.Rando A, et al. Characterization of the CSN1AG Allele of the Bovine αs1-Casein Locus by the Insertion of a Relict of a Long Interspersed Element1. J. Dairy Sci. 1998;81:1735–1742. doi: 10.3168/jds.S0022-0302(98)75741-8. [DOI] [PubMed] [Google Scholar]
- 69.Bell RM, Coleman RA. Enzymes of glycerolipid synthesis in eukaryotes. Annu. Rev. Biochem. 1980;49:459–487. doi: 10.1146/annurev.bi.49.070180.002331. [DOI] [PubMed] [Google Scholar]
- 70.Mayorek N, Grinstein I, Bar-Tana J. Triacylglycerol synthesis in cultured rat hepatocytes. The rate-limiting role of diacylglycerol acyltransferase. Eur. J. Biochem. FEBS. 1989;182:395–400. doi: 10.1111/j.1432-1033.1989.tb14844.x. [DOI] [PubMed] [Google Scholar]
- 71.Smith SJ, et al. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- 72.Riquet J, et al. Fine-mapping of quantitative trait loci by identity by descent in outbred populations: application to milk production in dairy cattle. Proc. Natl. Acad. Sci. USA. 1999;96:9252–9257. doi: 10.1073/pnas.96.16.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Looft C, et al. A mammary gland EST showing linkage disequilibrium to a milk production QTL on bovine Chromosome 14. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2001;12:646–650. doi: 10.1007/s003350020003. [DOI] [PubMed] [Google Scholar]
- 74.Farnir F, et al. Simultaneous mining of linkage and linkage disequilibrium to fine map quantitative trait loci in outbred half-sib pedigrees: revisiting the location of a quantitative trait locus with major effect on milk production on bovine chromosome 14. Genetics. 2002;161:275–287. doi: 10.1093/genetics/161.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coppieters W, et al. A QTL with major effect on milk yield and composition maps to bovine Chromosome 14. Mamm. Genome. 1998;9:540–544. doi: 10.1007/s003359900815. [DOI] [PubMed] [Google Scholar]
- 76.Grisart B, et al. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc. Natl. Acad. Sci. USA. 2004;101:2398–2403. doi: 10.1073/pnas.0308518100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winter A, et al. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA. 2002;99:9300–9305. doi: 10.1073/pnas.142293799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spelman RJ, Ford CA, McElhinney P, Gregory GC, Snell RG. Characterization of the DGAT1 Gene in the New Zealand Dairy Population. J. Dairy Sci. 2002;85:3514–3517. doi: 10.3168/jds.S0022-0302(02)74440-8. [DOI] [PubMed] [Google Scholar]
- 79.Scatà MC, et al. Ovine acyl CoA:diacylglycerol acyltransferase 1– molecular characterization, polymorphisms and association with milk traits. Anim. Genet. 2009;40:737–742. doi: 10.1111/j.1365-2052.2009.01909.x. [DOI] [PubMed] [Google Scholar]
- 80.Angiolillo A, et al. Identification of a single nucleotide polymorphism at intron 16 of the caprine acyl-coenzyme A: diacylglycerol acyltransferase 1 (DGAT1) gene. J. Dairy Res. 2007;74:47–51. doi: 10.1017/S0022029906002196. [DOI] [PubMed] [Google Scholar]
- 81.Milanesi, E. et al. Preliminary association studies between EBV and SNPs in 5 candidate genes for milk fat in goats. in (2010).
- 82.An XP, et al. Polymorphism identification in goat DGAT1 and STAT5A genes and association with milk production traits. Czech J. Anim. Sci. 2013;58:321–327. doi: 10.17221/6862-CJAS. [DOI] [Google Scholar]
- 83.Ozmen, O. & Kul, S. Polymorphism of goat DGAT1 gene and their association with milk production traits. Indian J. Anim. Sci. 84 (2014). [PMC free article] [PubMed]
- 84.Wang G, et al. Genetic polymorphism of DGAT1 gene association with lactation traits in Laoshan Dairy Goat. Sci. Agric. Sin. 2010;43:4717–4724. [Google Scholar]
- 85.Tăbăran, A. et al. Identification of polymorphism in Goat and Sheep DGAT1 gene associated with milk production traits. 71, 283–286 (2014).
- 86.McFie PJ, Stone SL, Banman SL, Stone SJ. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the N terminus in dimer/tetramer formation. J. Biol. Chem. 2010;285:37377–37387. doi: 10.1074/jbc.M110.163691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yen C-LE, Stone SJ, Koliwad S, Harris C, Farese RV. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coleman R, Bell RM. Evidence that biosynthesis of phosphatidylethanolamine, phosphatidylcholine, and triacylglycerol occurs on the cytoplasmic side of microsomal vesicles. J. Cell Biol. 1978;76:245–253. doi: 10.1083/jcb.76.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coleman R, Bell RM. Triacylglycerol synthesis in isolated fat cells. Studies on the microsomal diacylglycerol acyltransferase activity using ethanol-dispersed diacylglycerols. J. Biol. Chem. 1976;251:4537–4543. [PubMed] [Google Scholar]
- 90.Cases S, et al. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Talhari DT, et al. Interaction of a C-terminal peptide of Bos taurus diacylglycerol acyltransferase 1 with model membranes. Biochim. Biophys. Acta. 2009;1788:2320–2325. doi: 10.1016/j.bbamem.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 92.Lopes JLS, et al. Deconstructing the DGAT1 enzyme: Binding sites and substrate interactions. Biochim. Biophys. Acta. 2014;1838:3145–3152. doi: 10.1016/j.bbamem.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 93.Colón-González F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 94.Lopes, J. L. S., Beltramini, L. M., Wallace, B. A. & Araujo, A. P. U. Deconstructing the DGAT1 Enzyme: Membrane Interactions at Substrate Binding Sites. PLoS ONE10 (2015). [DOI] [PMC free article] [PubMed]
- 95.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 2000;25:111–112. doi: 10.1016/S0968-0004(99)01539-X. [DOI] [PubMed] [Google Scholar]
- 96.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clément, V., Boichard, D., Piacère, A., Barbat, A. & Manfredi, E. Genetic evaluation of French goats for dairy and type traits. In Proc. 7th Congress on Genetic Applied to Livestock Production 119–122 (2002).
- 98.Elsen J-M, Mangin B, Goffinet B, Boichard D, Le Roy P. Alternative models for QTL detection in livestock. I. General introduction. Genet. Sel. Evol. GSE. 1999;31:213–224. doi: 10.1186/1297-9686-31-3-213. [DOI] [Google Scholar]
- 99.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Knott SA, Elsen JM, Haley CS. Methods for multiple-marker mapping of quantitative trait loci in half-sib populations. Theor. Appl. Genet. 1996;93:71–80. doi: 10.1007/BF00225729. [DOI] [PubMed] [Google Scholar]
- 101.Legarra A, Fernando RL. Linear models for joint association and linkage QTL mapping. Genet. Sel. Evol. 2009;41:1–17. doi: 10.1186/1297-9686-41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bell J. A simple way to treat PCR products prior to sequencing using ExoSAP-IT. BioTechniques. 2008;44:834. doi: 10.2144/000112890. [DOI] [PubMed] [Google Scholar]
- 105.Ferrand-Calmels M, et al. Prediction of fatty acid profiles in cow, ewe, and goat milk by mid-infrared spectrometry. J. Dairy Sci. 2014;97:17–35. doi: 10.3168/jds.2013-6648. [DOI] [PubMed] [Google Scholar]
- 106.Bligh EG, Dyer WJ. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 107.Barrans A, et al. Hepatic lipase induces the formation of pre-beta 1 high density lipoprotein (HDL) from triacylglycerol-rich HDL2. A study comparing liver perfusion to in vitro incubation with lipases. J. Biol. Chem. 1994;269:11572–11577. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


