Abstract
Many attempts have been made to characterize the activities of brassinosteroids (BRs), which are important plant hormones. The crosstalk between light perception and the BR signalling pathway has been extensively studied regarding its effects on photomorphogenesis, especially in elongating etiolated hypocotyls. In contrast, how and where the light induces BR biosynthesis remain uncharacterized. DWF4 is one of the main enzymes involved in the BR biosynthesis pathway in Arabidopsis thaliana. We established DWF4-GUS A. thaliana lines in a homozygous dwf4-102 genetic background, but functionally complemented with a genomic DWF4 sequence fused in-frame with a β-glucuronidase (GUS) marker gene. The DWF4-GUS plants enabled the visualization of the accumulation of DWF4 under different conditions. We investigated the effects of aboveground light on root and hypocotyl growth. We observed that root length increased when shoots were maintained under light irrespective of whether roots were exposed to light. We also determined that light perception in aerial tissues enhanced DWF4 accumulation in the root tips. Overall, our data indicate that BR biosynthesis is promoted in the root tip regions by an unknown mechanism in distantly located shoot tissues exposed to light, leading to increased root growth.
Introduction
Brassinosteroids (BRs) are phytohormones commonly found in higher plants. They induce diverse responses, including cell division, cell elongation, and tissue differentiation1–6. Components involved in BR biosynthesis, signal perception, and cytosolic signal cascades have been determined through studies of many loss-of-function mutants or mutants insensitive to applications of exogenous BRs. Such BR-defective mutants often exhibit severe dwarfism, with the production of small leaves and short flower stems under light conditions, and stunted hypocotyls even in darkness. Additionally, mutants defective in BR biosynthesis are easily distinguished by the suppression of the dwarf phenotype following the application of exogenous BRs (DWF1/CBB1 7–9, CPD/DWF3/CBB3 7, 10, 11, DWF4 12, 13, DWF5 14, DET2/DWF6 15–17, and DWF7/STE1 18). In contrast, mutants in which the application of exogenous BRs failed to suppress mutant phenotypes or those that exhibited decreased sensitivity to BR biosynthesis inhibitors have provided clues regarding the genes involved in the perception of BRs at the plasma membrane (BRI1/DWF2/CBB2 7, 19, 20). These mutants also provided information about downstream cytoplasmic phosphorylation cascades (BIN2 21–23, BSKs 24, BSU1 25, and BSS1 26) and transcription factors (BES1 27, 28 and BZR1 29, 30). In addition to the BR receptor complex, co-receptors and inhibitory regulators have been identified (BAK131–34 and BKI135–37).
Previous studies analysed the crosstalk between light and BR signalling pathways38, 39. Many of the findings of these BR-related studies focused on hypocotyl growth in darkness. In contrast, BR-related mutants exhibit stunted growth in darkness. Dominant positive mutations of transcription factor genes located downstream of the BR signalling pathway result in hypocotyl elongation even in the presence of BRZ, which is an inhibitor of BR biosynthesis, or in bri1 mutants27, 29. It was assumed that light conditions influence BR biosynthesis/accumulation in plants and that BRs promote, for instance, hypocotyl elongation depending on their abundance. Eventually, hypocotyl elongation in darkness became widely used as a marker of BR-induced responses10, 13, 15. It is assumed that during the elongation of hypocotyls, BRs are synthesized in hypocotyls and induce localized rapid cell elongation40, 41. The long-distance transport of BRs has not been considered.
The abovementioned analyses were conducted using aerial plant tissues. Relatively few studies have focused on the root tissues of plants growing in the shade or in darkness42–44. In this study, we investigated the effects of light conditions on root growth, in parallel with hypocotyl growth used as a control. We observed that roots grew more extensively when the shoots were maintained under light, regardless of whether roots were exposed to light or darkness.
We established DWF4-GUS A. thaliana lines in a dwf4-102 homozygous genetic background. These lines were functionally complemented with a genomic DWF4 sequence fused in-frame with a β-glucuronidase (GUS) marker gene. The DWF4-GUS plants enabled us to visualize the accumulation of DWF4 under various conditions. Our experiments involving DWF4-GUS plants revealed that exposure to light stimulates aerial tissues to induce the expression of DWF4, which encodes a BR biosynthesis enzyme, in root tips located distally to the illuminated tissues. Our findings strongly suggest that BR biosynthesis is promoted in roots by an unknown mechanism occurring in distantly located shoot tissues, and that the resulting BRs induce root growth.
Results
Establishment of plants producing the DWF4 -GUS fusion protein
The BR biosynthesis pathway has been characterized in detail. Among the genes corresponding to the associated enzymes, we focused on DWF4. This gene encodes a C-22 hydroxylase, which is a key enzyme in the BR biosynthesis pathway13, 45–48. Among several alleles, dwf4-102 is the most frequently used for characterizing the dwf4 mutant49. It is a null allele that harbours a T-DNA insertion in the fifth exon (Fig. 1a). The dwf4-102 mutant plants exhibit severe dwarfism due to a BR-deficiency as is often observed for other BR-related mutants7–11, 14–23. The mutants produce very small leaves during the vegetative stage (Fig. 1b), and there is a lack of petal expansion, stamen elongation, and stem elongation in the flowers generated during the reproductive stage (Fig. 1c). Wild-type (WT) plants grown in darkness have elongated hypocotyls, while dwf4-102 mutant plants do not, which is common among BR mutants12, 13.
Figure 1.
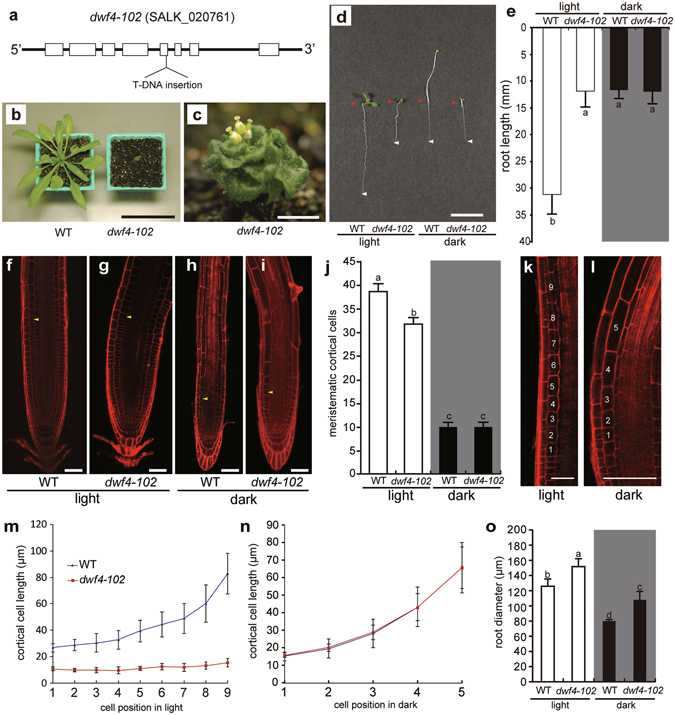
dwf4-102 mutants. (a) The dwf4-102 mutant plants contained a T-DNA insertion in the fifth exon of the DWF4 gene. These plants exhibited dwarfism during the vegetative stage (b) and produced defective flowers during the reproductive stage (c). The roots of dwf4-102 plants were relatively short under light and dark conditions (d,e) [wild-type (WT): n = 48; dwf4-102: n = 16]. Samples were stained with propidium iodine for analyses of the root apexes of WT (f,h) and dwf4-102 mutants (g,i) incubated under light (f,g) or in darkness (h,i). Arrowheads indicate the first cell of the elongation zone. Root diameters were measured in the region corresponding to the first cell of the elongation zone. (j) Cell division activity in the root apical meristems. The cortical cells were counted. There were no differences between WT and dwf4-102 plants incubated in darkness [WT: n = 42; dwf4-102: n = 24]. (k–n) Cell elongation activity in root tips. Under light conditions, cell elongation was observed in WT plants but not in dwf4-102 plants (k,m) [WT: n = 16; dwf4-102 n = 12]. On the other hand, the number of cells in the elongation zone was reduced, and there were no differences between WT and dwf4-102 plants incubated in darkness (l,n) [WT: n = 16; dwf4-102: n = 12]. (o) Root diameter. The roots of dwf4-102 plants were thicker than those of the WT plants under light and dark conditions [WT: n = 40; dwf4-102: n = 15]. Means denoted by the same letter are not significantly different (P < 0.05) according to the Tukey–Kramer method (j,o). Scale bars represent 50 mm (b), 10 mm (c,d), and 50 µm (f–i,k,l). Error bars represent standard deviations.
In addition to shorter stems, the roots of dwf4-102 plants exposed to light are considerably shorter than those of WT plants (Fig. 1d,e). In darkness, the roots of WT and dwf4-102 plants are similarly short. These results suggest that light positively affects root growth in WT plants, but not in dwf4-102 plants. To clarify the reasons for the differential root growth under light conditions, an examination of cell division and elongation is required.
We first analysed root cells using confocal laser scanning microscopy after staining samples with propidium iodine (PI) (Fig. 1f–i). We examined cell division activity in root apical meristems (RAMs) to determine whether it influences root length. We counted the number of cortical cells as described by Silva-Navas et al.44. There were fewer meristematic cells in the root meristems of WT plants grown in darkness than in plants exposed to light (Fig. 1j). This result suggests that cell division was less active in darkness. The meristems in darkness had about one-fourth as many cells as those grown under light. Exposure to light increased the number of meristematic cortical cells also in dwf4-102 plants (Fig. 1j).
We then measured cell lengths of elongating cells in elongation zone (nine cells in light condition (Fig. 1k,m) and five cells in darkness (Fig. 1l,n)) to examine whether cell elongation activity influences root length. Under light condition, the elongation of cortical cells in dwf4-102 plants was remarkably reduced (Fig. 1m), however there were almost no difference between WT plants and dwf4-102 plants in darkness (Fig. 1n). Interestingly, the cortical cell elongation in darkness must have been promoted by BR independent pathway. Thus, reduced cell elongation also contributed to the development of short roots in darkness. These results suggest that light enhanced root growth in WT plants by promoting cell division and elongation. It is likely that this enhanced growth is related to BR biosynthesis because it was not observed in the dwf4-102 mutants. The roots of the dwf4-102 mutant plants were thicker than those of the WT plants, regardless of the presence of light (Fig. 1f–i,o).
We hypothesized that light enhances DWF4 expression and the subsequent root growth. To test the idea that light induces root-specific DWF4 expression, we attempted to establish complementation lines that express a chimeric gene consisting of the DWF4 genomic sequence (with its promoter and coding sequence) and the GUS gene sequence produced by translational fusion (Supplementary Fig. S1(a)). We selected three independent DWF4-GUS homozygous lines with a dwf4-102 homozygous background from the T2 generation (i.e., DWF4-GUS plants #1, #12, and #17). Plants from each line exhibited normal cotyledon development and root elongation during the juvenile phase (Supplementary Fig. S1(b)), and rosette leaf development during the vegetative phase (Supplementary Fig. S1(c)). Additionally, these plants underwent bolting and developed normal fertile flowers almost simultaneously with WT plants.
DWF4-GUS expression in shoots and roots under light and dark conditions
Using the established DWF4-GUS plants, we first analysed tissue-specific GUS production. In shoots exposed to light, strong GUS activity was observed in the shoot apical meristem and its periphery (Fig. 2a), as well as in the young trichomes and guard cells of the leaf epidermis (Fig. 2b). In roots, GUS activity was observed in the RAM as well as in the cell division and expansion zones (Fig. 2c) and the central cylinder (Fig. 2d). To examine the possible correlation between root growth and the DWF4-GUS activity level, we compared GUS activity between the roots of DWF4-GUS plants grown under light and in darkness. At 7 days after germination (DAG), the GUS activity level was much lower in DWF4-GUS plants grown in darkness (Fig. 2f) than in plants exposed to light (Fig. 2e). In darkness, GUS activity in the RAM was observed in the cell division zone, but not in the cell elongation zone (Fig. 2f).
Figure 2.
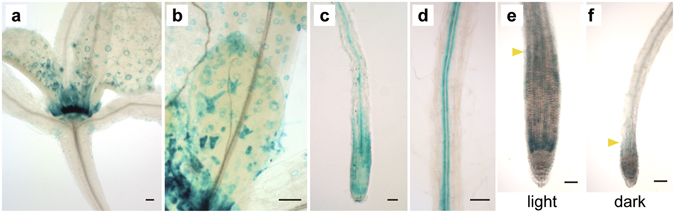
DWF4-GUS expression pattern. (a) High expression levels in the shoot apical meristem and surrounding region. (b) Expression levels in guard cells and developing trichomes. Expression levels in the root apex and young central cylinder (c) as well as the central cylinder (d) DWF4-GUS expression levels in roots under light (e) and in darkness (f) as determined by differential interference contrast microscopy. Arrowheads indicate the borders between the cell division and elongation zones. Scale bars represent 50 µm.
We monitored the growth of roots and hypocotyls from 1 to 7 DAG (i.e., during the juvenile stage) in plants incubated in darkness or under light (Fig. 3a–d). The roots of plants grown in darkness were slightly longer than the roots of plants exposed to light until 3 DAG (Fig. 3c). In contrast, hypocotyls grown in darkness were much longer than hypocotyls incubated under light (Fig. 3d). However, after 3 DAG, the roots of plants incubated under light grew faster than the roots maintained in darkness. To reveal the spatio-temporal changes to the DWF4-GUS accumulation pattern, we examined the roots of DWF4-GUS plants from 1 to 7 DAG. Analyses of the younger seedlings revealed that GUS activity in the root cell expansion region of dark-grown seedlings did not increase between 2 and 3 DAG (Fig. 3f,h). These results imply that light promotes DWF4 accumulation in root tissues (Fig. 3e,g).
Figure 3.
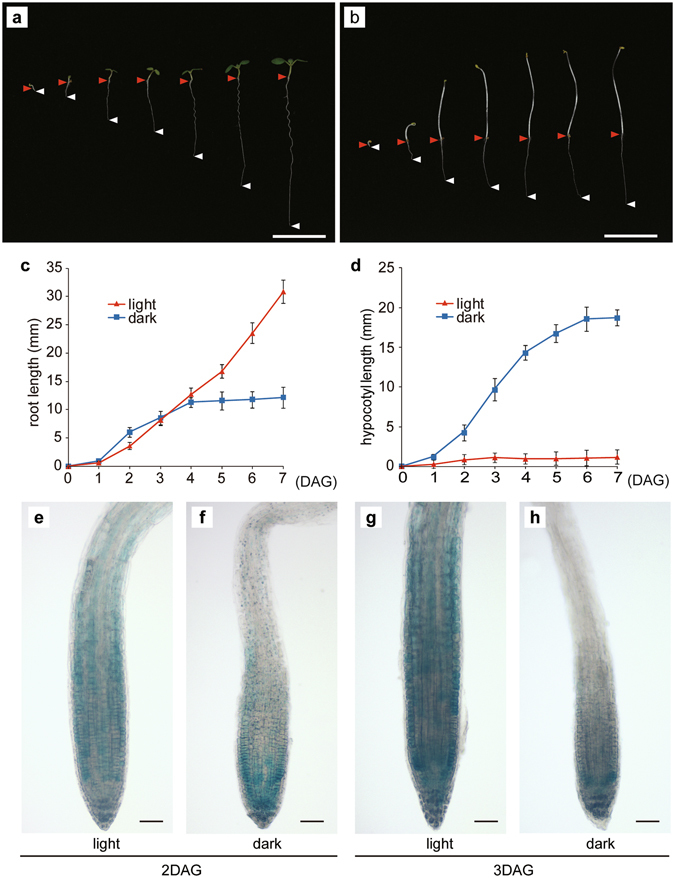
Root and hypocotyl growth and DWF4-GUS expression patterns. Plant growth under light (a) and in darkness (b) from 1 to 7 days after germination (DAG). Red arrowheads indicate the junctions between the hypocotyl and root, while white arrowheads indicate the root apex positions. Root (c) and hypocotyl (d) lengths from 1 to 7 DAG [n = 24]. After 3 DAG, the plants grown in darkness exhibited decreased root growth. (e–h) DWF4-GUS expression pattern in the root meristem. At 2 DAG, there were no differences in DWF4-GUS signals between the root meristems of plants incubated under light (e) or in darkness (f). At 3 DAG, the DWF4-GUS signal intensity in the roots incubated in darkness (h) started to decrease in the meristematic region and cell elongation zone. Scale bars represent 10 mm (a,b) and 50 µm (e–h). Error bars represent standard deviations.
Light perception in shoots affects root elongation and DWF4 expression in the root apical meristem
The results described in the previous section imply that light perception in shoots may induce DWF4 accumulation in the RAM through an unknown systemic mechanism. However, it is possible that the roots were able to perceive light under our experimental conditions. To eliminate this possibility, we developed a “bottom half covered growth (BCG) test”, in which the lower half of plates was covered with black polyethylene film. This resulted in only the shoots being exposed to light, while the roots remained in darkness (Supplementary Fig. S2). Seeds were aseptically placed on Murashige and Skoog (MS) agar medium in the darkened sides of plates, and were then covered with sterilized black film. The plates were positioned vertically and incubated under light until 7 DAG. The seeds were allowed to germinate in darkness under the film. When the elongated hypocotyls became approximately 5 mm long, the shoots were able to perceive light, and the hypocotyls stopped growing (Fig. 4a,b). In the BCG test, there were no significant root length differences between plants grown in darkness and those exposed to light until the shoots started to perceive light (i.e., when hypocotyls were about 5 mm long) as indicated in Fig. 3c,d. The roots began to grow after the plants started to perceive light. At 7 DAG, the seedlings used for the BCG test had much longer roots than the seedlings grown in darkness, and slightly shorter roots than the seedlings grown under light (Fig. 4a,c). To eliminate the possibility that contact effects of the film used to cover the seeds were influencing root growth, we used a clear film to cover control samples in the BCG test (Fig. 4a–c). There were no significant root length differences between the clear film-covered and black film-covered plants in the BCG test. These results suggest that light perception in shoot tissue promotes root growth, even though the roots themselves are maintained in darkness.
Figure 4.
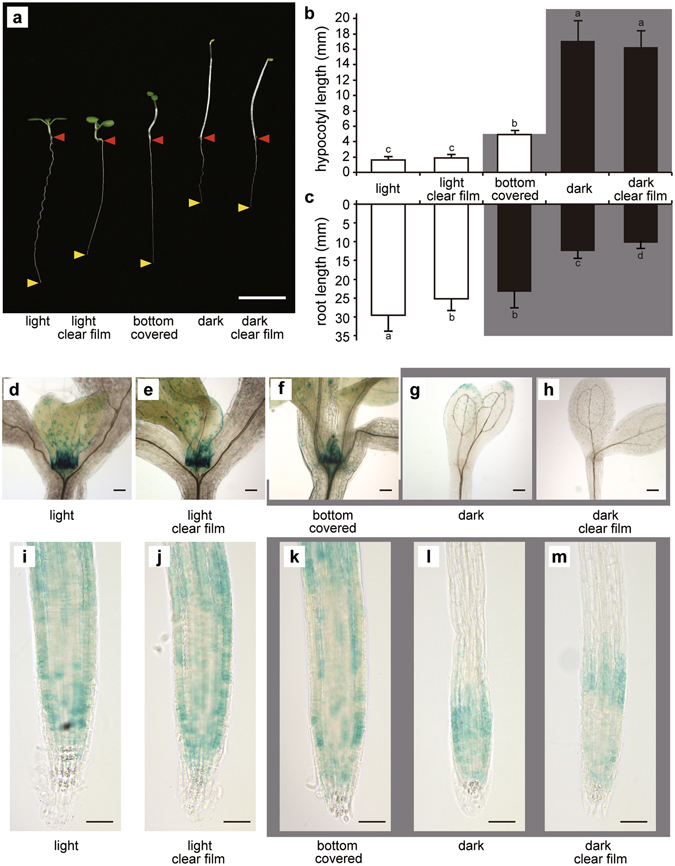
Root elongation and DWF4-GUS expression under light and in darkness according to the bottom half covered growth (BCG) test. The results of the BCG test revealed that light perception in shoots promotes DWF4 expression in the root apex. (a) Root elongation. Red arrowheads indicate the junctions between the hypocotyl and root, while yellow arrowheads indicate the root apex positions. Hypocotyl (b) and root (c) lengths according to the BCG test. The grey background indicates which part was darkened (n = 32). Means denoted by the same letter are not significantly different (P < 0.05) according to the Tukey–Kramer method. DWF4-GUS expression around the shoot apical meristem (d–h) and in the root apex (i–m). Light conditions with no film (d,i) or with a clear film (e,j). Shoots under light and roots in darkness covered with a black film (f,k). Although the root apex was shaded, the GUS signals in the root meristem were stronger than those of plants maintained in darkness. Dark conditions with no film (g,l) or with a clear film (h,m). Bars represent 10 mm (a) and 50 µm (d–m). Error bars represent standard deviations.
We subsequently analysed DWF4-GUS accumulation in plants used for the BCG test (BCG plants). The roots and shoots of BCG plants exhibited high GUS activity levels similar to plants grown under light, but in contrast to plants incubated in darkness (Fig. 4d–m). These results suggest that light perception in shoots promotes DWF4 accumulation in the root apex. Additionally, high DWF4 activity levels in roots may enhance root growth through the activation of cell division and elongation.
To confirm that the light perception in shoots enhances root growth, we devised a “top half covered growth (TCG) test”, in which the shoots are covered with a black film, while the roots are exposed to light (Fig. 5). To avoid phototactic shoot movements, TCG plates were incubated in complete darkness until 3 DAG to promote hypocotyl growth. The plates were then exposed to light. The roots of plants used for the TCG test (TCG roots) were shorter than those of plants covered with clear film, but were almost the same length as the roots of plants grown in darkness (Fig. 5a–c). These results indicate that light perception in roots does not promote root growth. We also analysed DWF4-GUS accumulation in the TCG test (Fig. 5d–m). The GUS activity levels in the root apex of TCG plants (Fig. 5k) were as weak as those of roots incubated in darkness (Fig. 5l,m), suggesting that the perception of light in roots does not promote DWF4 accumulation in the root apex. We conclude that light perception in shoots, but not in roots, promotes BR biosynthesis in roots, which may enhance root growth.
Figure 5.
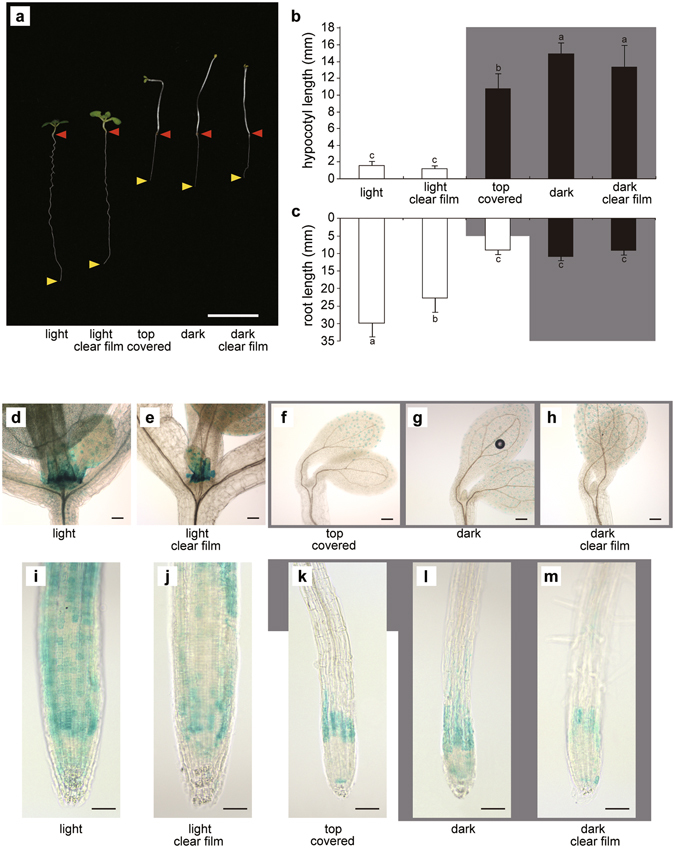
Top half covered growth (TCG) test results revealed that light perception in roots did not promote DWF4 expression in the root apex. (a) Root elongation. Red arrowheads indicate the junctions between the hypocotyl and root, while yellow arrowheads indicate the root apex positions. Hypocotyl (b) and root (c) lengths according to the TCG test. The grey background indicates which part was darkened [n = 32]. Means denoted by the same letter are not significantly different (P < 0.05) according to the Tukey–Kramer method. DWF4-GUS expression around the shoot apical meristem (d–h) and in the root apex (i–m). Light conditions with no film (d,i) or with a clear film (e,j). Shoots in darkness covered with a black film and roots under light (f,k). Although the root apex was exposed to light, the GUS signal in the root meristem was weaker than that of plants that were incubated entirely under light. Dark conditions with no film (g,l) or with a clear film (h,m). Bars represent 10 mm (a) and 50 µm (d–m). Error bars represent standard deviations.
Discussion
The BR signalling pathway has been well described, from the effects of light to plant growth (e.g., hypocotyl elongation), but there is relatively little information available regarding the sites where BR is actually synthesized. Our study results indicate that light perception in the aerial tissues affects root growth, possibly through the induction of BR biosynthesis in the root tip regions. Among the enzymes in the BR biosynthesis pathway, DET2, CPD, and DWF4 have been the most extensively studied. The DWF4 expression level was observed to be lower than that of CPD 40, 50–53. Consistent with this observation, DWF4-GUS plants exhibited localized accumulation patterns in the roots and leaves, which differed from the expression pattern of proCPD-GUS reporter plants11. Based on these studies, DWF4 is considered the rate-limiting enzyme in the BR biosynthesis pathway54, 55. Accordingly, we assumed that the DWF4 accumulation level and pattern significantly affect BR levels during plant growth and development.
We performed qRT-PCR analysis to compare DWF4 mRNA levels between plants tested in this study (Supplementary Fig. S3). The result showed that root tips under light condition expressed DWF4 transcript more than those in darkness. But the difference is much smaller than that we observed in DWF4-GUS staining which reflects DWF4 protein accumulation. It is suggested that light might regulate DWF4 protein stability as well.
Wild-type plants grown in darkness produced short roots similar to those of the dwf4 mutants grown under light or in darkness. This observation suggests that light perception promotes BR biosynthesis in roots, resulting in root elongation. We conducted two experiments to test this hypothesis. First, we completed BCG and TCG tests to reveal the tissue where light is perceived. Our observations strongly indicate that light perception in shoots, but not in roots, is a determining factor for the subsequent root elongation in WT plants, but not in dwf4 mutants. In general, BR is thought to function as a signal molecule that does not undergo long-distance transport40, 41. Second, we established DWF4-GUS transgenic lines in a dwf4-102 background to visualize DWF4 accumulation in tissues and estimate BR biosynthesis in roots. We observed a tight correlation between light perception in shoots and the DWF4 (DWF4-GUS) accumulation level in root tips. These results imply that some signal, conveyed from the shoots to the roots, induces BR biosynthesis in the root tips, and that the resulting BRs promote root growth via cell division and elongation.
We analysed DWF4-GUS accumulation patterns under various conditions, with a particular focus on the meristematic region. We initially confirmed the difference in DWF4-GUS accumulation between light and dark conditions. The GUS signal in the shoot apical meristem and RAM was considerably lower in darkness than under light. We focused on the RAM because the dwf4-102 mutants exhibited retarded root growth under light and dark conditions. Recent studies have started to investigate BR signalling in A. thaliana roots56, 57. In the RAM of plants grown in darkness, the DWF4-GUS signal was weak and limited to the cell division zone. These results were consistent with the number of meristematic cells and the production of relatively short roots. In terms of the relationship between BRs and root development, the BRs are reportedly indispensable for cell-cycle progression and cell elongation in the RAM58–62. Our observations of DWF4-GUS plants complement the available tissue-specific information regarding BR biosynthesis in roots developing in darkness. Additionally, our daily observations of DWF4-GUS plants for 1 week revealed a decrease in the DWF4-GUS signal in the root apex under dark conditions after 3 DAG (Fig. 3f,h). The root meristem then started to atrophy. We thought that a stable BR supply and DWF4 activity level in the RAM would maintain root meristem activities and the resulting root elongation, especially after 3 DAG. However, BRs promote hypocotyl elongation in darkness. These results suggest that DWF4 expression is differentially regulated between the hypocotyl and RAM. Therefore, when seeds germinate in darkness, the seedlings may detect their underground position and prioritize hypocotyl elongation over root elongation to perceive light as soon as possible. This would likely increase the survival rate of seedlings during the plant germination stage. We believe these regulatory activities have been established not only for the BR signalling pathway, but also for BR biosynthesis. At this moment, we could not find out what kind of signal(s) carried from above ground tissue to RAM to turn on DWF4 expression, accumulation and consequent BR biosynthesis.
We introduced a new system, BCG and TCG system, to see plants with shoots and roots under light or dark condition. Xu et al. reported an agar-plate method to study root growth42. In that method aerial tissues are placed outside the container, where the physical conditions for shoots and roots would be different. Silva-Navas et al. reported another system, D-Root, to cultivate plants with roots under different light conditions43, 44. The strength of light illumination is more intense than that we used in this analysis. On top of that, our observation was performed with young seedling of less than 7 DAG, while other works were conducted using older plants than ours. When we discuss root growth, we should consider experimental differences and what we would like to compare with natural growth condition.
Methods
Plant materials and growth conditions
We used the A. thaliana Col-0 line as the WT control along with several homozygous and heterozygous dwf4-102 mutant lines (SALK_020761). Seeds were surface-sterilized with 70% (v/v) ethanol for 2 min, rinsed twice with distilled water, and incubated in 0.05% (v/v) plant preservative mixture (Plant Cell Technology; http://www.plantcelltechnology.com/) for 2 days at 4 °C in darkness. Sterilized seeds were germinated in plates containing half-strength MS basal salt medium supplemented with B5 vitamins, 1.0% (w/v) sucrose, 2 mM 2-morpholinoethanesulfonic acid monohydrate (pH 5.8), and 1.5% (w/v) agarose or in a soil mixture of vermiculite soil and Sakata Supermix-A (Sakata Seed Corporation, http://www.sakataseed.co.jp/). Plants were incubated at 22–23 °C under white light (FHF32EX-N-H, 25 µmol m−2 s−1; Panasonic, http://panasonic.jp/light). Hypocotyl and root lengths were measured using ImageJ software.
Plasmid construction
We constructed the pWAT2-proDWF4-DWF4-GUS vector to generate DWF4-GUS–complemented dwf4-102 mutant plants. We amplified the following DNA fragments using specific primer sets (Supplementary Table 1): DWF4 promoter region and the open reading frame (4,936 bp) from A. thaliana Col-0 genomic DNA, DWF4 terminator region (561 bp) from A. thaliana Col-0 genomic DNA, and the GUS gene (1,812 bp) from the pWAT208-MASS-GUS plasmid63. All amplified fragments were introduced into the pWAT2 plasmid63 to construct the pWAT2-pro DWF4-DWF4-GUS vector. The primers used to construct this vector are listed in Supplementary Table 1.
Plant transformation
We introduced pWAT2-proDWF4-DWF4-GUS into Agrobacterium tumefaciens strain GV3101 cells carrying the pSOUP plasmid63. These cells were then used to transform dwf4-102 heterozygotes using the floral dip method. The A. thaliana inflorescences were dipped in half-strength MS basal salt medium containing 1.0% (w/v) sucrose, 0.05% (v/v) Silwet L77 solution (pH 5.8), and transgene-carrying A. tumefaciens cells. The selected plants were then subjected to genotyping using the following primers: Lba1, SALK_020761-RP, and SmaI-DWF4 Term.-rev (Supplementary Fig. S1a, Supplementary Table S1).
Histochemical staining of DWF4-GUS plants
To analyse DWF4-GUS expression in transgenic plants, samples were fixed in 90% acetone for 15 min on ice and washed twice with 100 mM sodium phosphate buffer (pH 7.0). The samples were then incubated overnight at 37 °C in 100 mM sodium phosphate buffer (pH 7.0), 10 mM sodium EDTA, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 0.1% (v/v) Triton X-100, and 0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid. The stained samples were rinsed twice with 100 mM sodium phosphate buffer (pH 7.0) and then treated with 70% (v/v) ethanol to deplete chlorophyll. The bleached samples were then hydrated in an ethanol series (70, 30, and 15%). To enhance the size of RAM cells, samples were cleared with chloral hydrate.
Fluorescence and confocal microscopy
An IX70 epifluorescence microscope (Olympus, http://www.olympus-global.com/en/) equipped with a C7780-20 CCD digital camera (Hamamatsu Photonics, http://www.hamamatsu.com/jp/en/index.html) was used to observe GUS staining. A C2+ confocal microscope (Nikon, http://www.nikon.com/index.htm) was used to observe PI staining. Roots harvested from plants at 7 DAG were stained with 2.5 µg/ml PI for 2–3 min and then visualized after an excitation by a Sapphire 561-nm laser. The PI signal was detected with a 552–617-nm bandpass filter. Cell lengths in PI-stained samples were measured using ImageJ software.
RAM activity analysis
To investigate the activity of cell division and consequent cell elongation in root tips, we followed the method described by Silva-Navas et al.44. In this study, we assigned the border between meristem and elongating cells, where an adjacent cell had a size longer than by more than 50%.
Statistical analysis
All statistical analyses were completed using Microsoft Excel 2016 software. Significant differences in multiple comparisons were determined with the Tukey–Kramer method.
Electronic supplementary material
Acknowledgements
We are also grateful to the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutant used in this work. This work was supported by JSPS KAKENHI Grant numbers 25120707 and 15H04628 (to Y.W.).
Author Contributions
J.S. and Y.W. conceived and designed the experiments. J.S. conducted the experiments and analysed the results. J.S. and Y.W. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01872-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong H, Chu C. Brassinosteroid signaling and application in rice. J Genet. Genomics. 2012;39:3–9. doi: 10.1016/j.jgg.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J-Y, Sae-Seaw J, Wang Z-Y. Brassinosteroid signalling. Development. 2013;140:1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejonghe W, Mishev K, Russinova E. The brassinosteroid chemical toolbox. Current Opinion in Plant Biol. 2014;22:48–55. doi: 10.1016/j.pbi.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Fàbregas N, Caño-Delgado AI. Turning on the microscope turret: A new view for the study of brassinosteroid signaling in plant development. Physiologia Plantarum. 2014;151:172–183. doi: 10.1111/ppl.12130. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Bai M-Y, Chong K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014;33:683–696. doi: 10.1007/s00299-014-1578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauschmann A, et al. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. doi: 10.1046/j.1365-313X.1996.9050701.x. [DOI] [Google Scholar]
- 8.Klahre U, et al. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe S, et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 11.Mathur J, et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313X.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- 12.Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe S, et al. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choe S, et al. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- 15.Li J, et al. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 16.Fujioka S, et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noguchi T, et al. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999;120:833–839. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe S, et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- 19.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, et al. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choe S, et al. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol. 2002;130:1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mora-García S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada S, et al. Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell. 2015;27:375–390. doi: 10.1105/tpc.114.131508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z-Y, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/S1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 30.He J-X, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 32.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/S0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 33.Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlova R, et al. The Arabidopsis Somatic Embryogenesis Receptor-Like Kinase1 protein complex includes Brassinosteroid-Insensitive1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, et al. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014;24:1328–1341. doi: 10.1038/cr.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, et al. The intrinsically disordered protein BKI1 is essential for inhibiting BRI1 signaling in plants. Mol. Plant. 2015;8:1675–1678. doi: 10.1016/j.molp.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 38.Luo XM, et al. Integration of light- and brassinosteroid signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L, et al. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant. 2009;2:755–772. doi: 10.1093/mp/ssp039. [DOI] [PubMed] [Google Scholar]
- 40.Shimada Y, et al. Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 2003;131:287–297. doi: 10.1104/pp.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Symons GM, Reid JB. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 2004;135:2196–2206. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W, et al. An improved agar-plate method for studying root growth and response of Arabidopsis thaliana. Sci. Rep. 2013;3:1273. doi: 10.1038/srep01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva-Navas J, et al. D-Root: a system for cultivating plants with the roots in darkness or under different light conditions. Plant J. 2015;84:244–55. doi: 10.1111/tpj.12998. [DOI] [PubMed] [Google Scholar]
- 44.Silva-Navas J, et al. Flavonols Mediate Root Phototropism and Growth through Regulation of Proliferation-to-Differentiation Transition. Plant Cell. 2016;28:1372–87. doi: 10.1105/tpc.15.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noguchi T, et al. Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 2000;124:201–209. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujioka S, Takatsuto S, Yoshida S. An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 2002;130:930–939. doi: 10.1104/pp.008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita S, et al. Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. 2006;45:765–774. doi: 10.1111/j.1365-313X.2005.02639.x. [DOI] [PubMed] [Google Scholar]
- 48.Sekimata K, et al. Brz220 interacts with DWF4, a cytochrome P450 monooxygenase in brassinosteroid biosynthesis, and exerts biological activity. Biosci. Biotechnol. Biochem. 2008;72:7–12. doi: 10.1271/bbb.70141. [DOI] [PubMed] [Google Scholar]
- 49.Nakamoto D, Ikeura A, Asami T, Yamamoto KT. Inhibition of brassinosteroid biosynthesis by either a dwarf4 mutation or a brassinosteroid biosynthesis inhibitor rescues defects in tropic responses of hypocotyls in the arabidopsis mutant nonphototropic hypocotyl 4. Plant Physiol. 2006;141:456–64. doi: 10.1104/pp.105.076273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bancoş S, et al. Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002;130:504–513. doi: 10.1104/pp.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choe S, et al. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim HB, et al. The regulation of DWARF4 expression is likely a critical mechanism in maintaining the homeostasis of bioactive brassinosteroids in Arabidopsis. Plant Physiol. 2006;140:548–557. doi: 10.1104/pp.105.067918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Z, et al. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4. Arabidopsis thaliana. Plant Cell. 2010;22:1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao B, Li J. Regulation of brassinosteroid biosynthesis and inactivation. J. Integr. Plant Biol. 2012;54:746–759. doi: 10.1111/j.1744-7909.2012.01168.x. [DOI] [PubMed] [Google Scholar]
- 55.Chung Y, Choe S. The regulation of brassinosteroid biosynthesis in Arabidopsis. CRC. Crit. Rev. Plant Sci. 2013;32:396–410. doi: 10.1080/07352689.2013.797856. [DOI] [Google Scholar]
- 56.Chaiwanon J, Wang ZY. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 2015;25:1031–1042. doi: 10.1016/j.cub.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaillais Y, Vert G. Brassinosteroid signaling and BRI1 dynamics went underground. Curr. Opin. Plant Biol. 2016;33:92–100. doi: 10.1016/j.pbi.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González-García M-P, et al. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- 59.Hacham Y, et al. Brassinosteroid perception in the epidermis controls root meristem size. Development. 2011;138:839–848. doi: 10.1242/dev.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilarrasa-Blasi J, et al. Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev. Cell. 2014;30:36–47. doi: 10.1016/j.devcel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 61.Chaiwanon J, Wang Z-Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Current Biol. 2015;25:1031–1042. doi: 10.1016/j.cub.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vragović K, et al. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. USA. 2015;112:923–928. doi: 10.1073/pnas.1417947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumakura N, et al. Arabidopsis AtRRP44A is the functional homolog of Rrp44/Dis3, an exosome component, is essential for viability and is required for RNA processing and degradation. PLoS One. 2013;8:e79219. doi: 10.1371/journal.pone.0079219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


