Abstract
Ribosome inactivating proteins (RIPs) are RNA N-glycosidases that depurinate a specific adenine residue in the conserved sarcin/ricin loop of 28S rRNA. These enzymes are widely distributed among plants and their presence has also been confirmed in several bacterial species. Recently, we reported for the first time in silico evidence of RIP encoding genes in metazoans, in two closely related species of insects: Aedes aegypti and Culex quinquefasciatus. Here, we have experimentally confirmed the presence of these genes in mosquitoes and attempted to unveil their evolutionary history. A detailed study was conducted, including evaluation of taxonomic distribution, phylogenetic inferences and microsynteny analyses, indicating that mosquito RIP genes derived from a single Horizontal Gene Transfer (HGT) event, probably from a cyanobacterial donor species. Moreover, evolutionary analyses show that, after the HGT event, these genes evolved under purifying selection, strongly suggesting they play functional roles in these organisms.
Introduction
Ribosome inactivating proteins (RIPs, EC 3.2.2.22) irreversibly modify ribosomes through the depurination of an adenine residue in the conserved alpha-sarcin/ricin loop of 28S rRNA1–4. This modification prevents the binding of elongation factor 2 to the ribosome, arresting protein synthesis5, 6. The occurrence of RIP genes has been experimentally confirmed in a wide range of plant taxa, as well as in several species of Gram positive and Gram negative bacteria7–9. Additionally, the exponential increase of information in databases has suggested the presence of genes coding for RIP–domain containing proteins in lineages of Fungi, Cyanobacteria and Metazoa10–12. Although several of these toxins have been extensively studied at the biochemical level, their biological roles remain open to speculation. In some cases, it seems reasonable to predict their functions. For instance, the high toxicity of the prototypical RIP ricin supports an antifeedant role, whereas bacterial RIPs shiga and shiga-like toxins are strong virulence factors for their harboring bacteria. Antiviral and other defense activities have been postulated for other plant RIPs, but no concluding evidence has been obtained. Recently, the RIP of the symbiotic Spiroplasma (class Mollicutes) in Drosophila neotestacea was shown to play a defensive role in preventing a virulent nematode from infecting this insect13.
In a previous work, we have described that the phylogeny of RIP genes shows incongruence with that of the species. Most of these inconsistencies can be explained by gene duplication, loss and/or lineage sorting11. Another mechanism leading to phylogenetic incongruence is horizontal gene transfer (HGT); namely the non-genealogical transmission of genes among organisms. HGT is accepted as an important force driving prokaryotic genome evolution14, 15. In contrast, its impact on genomes from multicellular eukaryotes, in particular animals, is largely controversial16–18. To be maintained permanently in animal species, heritable changes (i.e. the transferred gene) must be incorporated into germline cells and transmitted to the offspring. Nevertheless, in the particular case of herbivore arthropods and nematodes, HGT has been postulated to play a role in the adaptation to phytophagy, including the efficient assimilation and detoxification of plant metabolites19–21.
Detection of bona fide HGT derived genes is not trivial, and careful data revision is required for its corroboration. Many cases of putative foreign genes have been shown, after further revision, to result from artifacts or misinterpretations, such as contamination of genomic data, incomplete sampling of sequences and/or taxa, incorrect phylogenetic inferences or hidden paralogy. Two emblematic cases illustrating these issues are the initial conclusion that the human genome contained a high percent of bacterial derived genes22, and the recent claim that tardigrade genomes contain significant amounts of foreign DNA23. In both cases, subsequent sounder analyses demonstrated that contamination or incomplete sampling better explained the available data24, 25. Consequently, tidy case-by-case analyses of HGT candidates are required for their efficient detection. To do so, independent evidence and alternative evolutionary scenarios should be taken into account.
Based on the previous finding of in silico evidence for the presence of genes coding putative proteins harboring RIP domains in the genomes of two closely related species of mosquitoes11, we aim to confirm the presence and determine the location of RIP encoding genes in species of the mosquito subfamily Culicinae. Moreover, we provide solid evidence supporting the hypothesis that these genes derive from a single prokaryotic transferred gene.
Results
Culex spp genomes harbor RIP encoding genes
Recently, we found in silico evidence for the presence of genes coding for RIP-containing proteins in two closely related species of Metazoa: Aedes aegypti and Culex quinquefasciatus 11. These intriguing findings led us to design experimental strategies to confirm their presence by ruling out possible database artifacts (i.e. contamination). For this purpose, genomic DNA was obtained from a pool of four mosquitoes of C. quinquefasciatus strain JHB (same strain as the originally sequenced and available in GenBank). Then, two independent PCR experiments were designed to demonstrate the presence of the intronless RIP gene, and to confirm its physical linkage to the predicted neighbor gene, which is an intron-containing metazoan-derived gene (XM_001850822). Figure 1 shows that both PCR products presented the expected size. Also, further cleavage of each amplicon with EcoRI yielded the predicted patterns, confirming their identity. We have also successfully amplified partial coding regions of two putative RIP genes from Ae. aegypti (data not shown).
Figure 1.
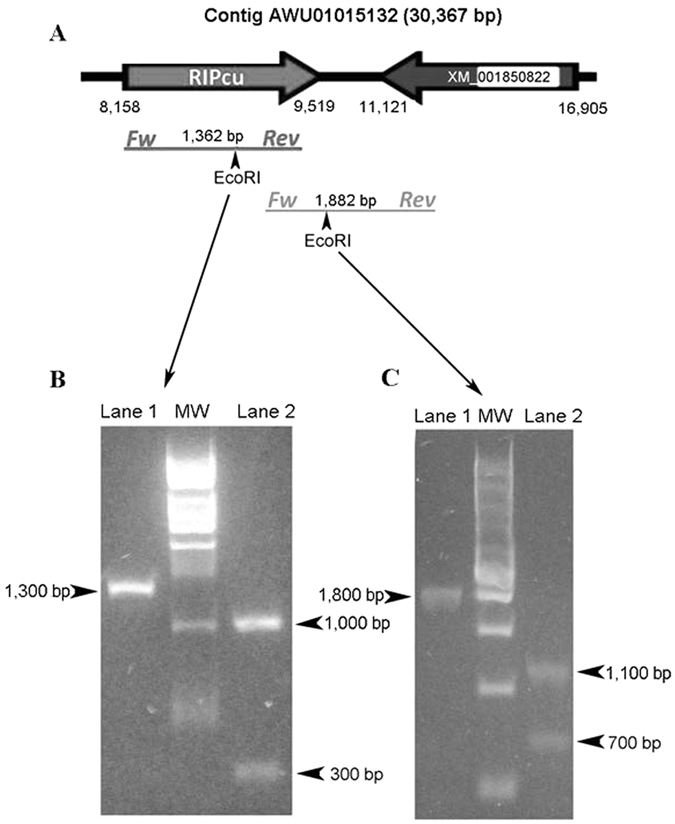
Experimental confirmation of the presence and location of RIP gene in C. quinquefasciatus JHB genome. (A) Schematic representation of a fragment of the contig AAWU01015132 depicting the RIP gene (RIPcu) and its closest neighbor gene (XM_001850822). Expected amplicons and relevant EcoRI restriction sites are also presented. The intron of XM_001850822 is represented with a white box. (B) RIP ORF was amplified by PCR and the product was analyzed by gel electrophoresis before (lane 1) and after EcoRI treatment (lane 2). (C) A fragment of 1,882 bp linking the RIP gene with its neighbor-gene was amplified and electrophoresed before (lane 1) and after EcoRI treatment (lane 2).
Once the presence and location of the RIP sequence in C. quinquefasciatus genome were experimentally confirmed, we successfully amplified the full length RIP coding sequences (~1300 bp) of the closely related species C. pipiens, C. molestus, and C. torrentium (Supplementary Figure 1). PCR products were cloned and sequenced, and the obtained sequences were aligned. As expected, nucleotide sequences showed high similarity (93–97% identity) in relation to the reported sequence in the C. quinquefasciatus genome database. The reference sequence of C. quinquefasciatus JHB (RipCu) obtained from the genome database revealed an in-frame, three-nucleotide (ACC) insertion (encoding an additional Thr residue). Interestingly, the sequence obtained from the C. quinquefasciatus JHB MR4-CDC colony harbors a ten-nucleotide frame-shifting deletion (nt 542–551) generating a premature stop codon disrupting the RIP domain (Supplementary Figure 2). By direct sequencing of PCR products from six individual specimens of C. quinquefasciatus JHB from the MR4-CDC colony, we confirmed all these individuals were homozygous for the deletion, strongly suggesting this null mutation was fixed in this colony (Supplementary Figure 3).
Culicinae RIP genes are monophyletic and syntenic
We have previously shown that RIP encoding genes from particular lineages (e.g. monocots or dicots, bacteria and fungi) are not monophyletic11. Moreover, many RIP clades include sequences belonging to largely distant taxa11, 26. Based on this evidence, we postulated that the evolutionary history of RIP genes is consistent with the existence of several ancient paralogues, followed by multiple lineage-specific gene duplications and losses11. However, metazoan RIP genes are particularly interesting because they are restricted to closely related insects of the subfamily Culicinae (therefore hereafter referred as Culicinae RIPs). As can be seen in Fig. 2, RIPs from C. quinquefasciatus and Ae. aegypti form a well-supported clade [posterior probability (PP): 1, bootstrap (BS): 100%]. Monophyly of Culicinae RIPs along with their apparent narrow taxonomic distribution, suggest that these genes are derived from a rather recent, single ancestral sequence. In order to test this, microsynteny analyses were carried out using scaffolds from Ae. aegypti, C. quinquefasciatus and Anopheles gambiae (the closest relative lacking RIP genes). As expected, partially conserved syntenic blocks were identified in the three species (Fig. 3 and Supplementary Table 1).
Figure 2.
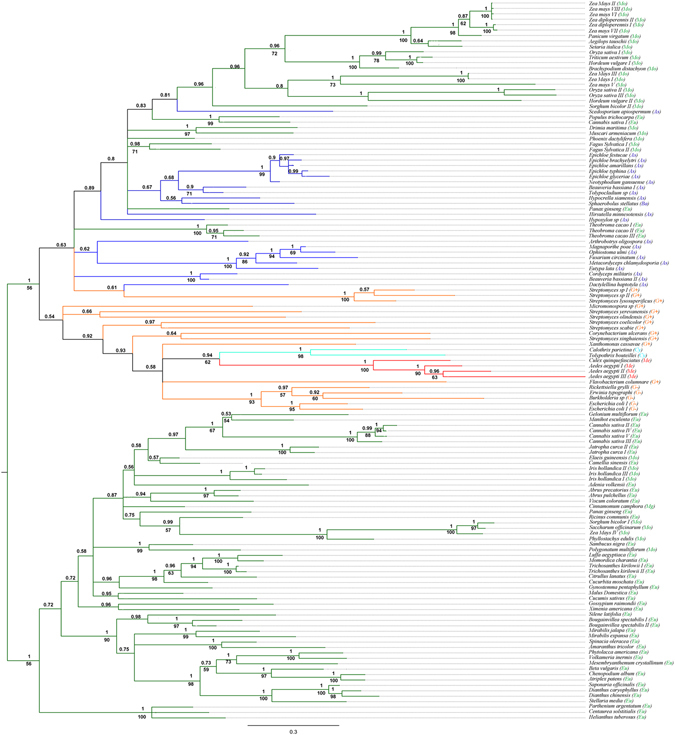
Phylogenetic tree of RIP proteins family. Bayesian tree topology based on a matrix analysis of 133 proteins sequences with 209 informative sites is presented. Numbers above branches indicate PP support values. Bootstrap values (BS) >50% are shown below branches for nodes where topology of ML analysis was coincident with Bayesian inference. Lineages are indicated by different colors as follows: green (Plantae, including Mo: monocots; Eu: eudicots and Mg: magnoliids), blue (Fungi, including As: Ascomycota and Ba: Basidiomycota), red (Me: Metazoa), orange (Bacteria, including G+: Gram positive, G-: Gram negative, and excluding Cyanobacteria) and turquoise (Cy: Cyanobacteria). The clade of Culicinae RIPs is emphasized with red branches. Information related to the sequences used to infer these trees is available in Supplementary Table 2.
Figure 3.
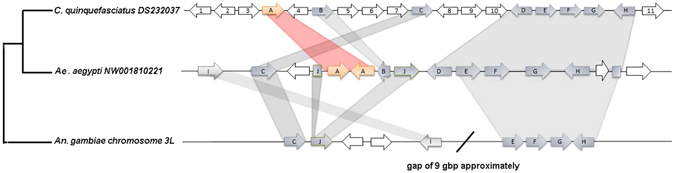
Schematic representation of the shared genomic context between C. quinquefasciatus (DS232037), Aedes aegypti (NW001810221) and Anopheles gambiae (chromosome 3L). Grey shadows link conserved syntenic ORFs. RIP genes are represented with orange arrows. The ORF 4 is equivalent to XM_001850822 and is absent in other scaffolds. Additional information about each gene is available in Supplementary Table 1.
Culicinae RIP genes are derived from a single HGT event
Sequence similarity searches further confirmed that metazoan RIP genes are restricted to the subfamily Culicinae. In addition to the above described genes, we found in silico seven RIP genes in Ae. albopictus (GenBank: KXJ78156, KXJ78155, KXJ78158, KXJ73132, KXJ78157, KXJ73133 and KXJ73764). A more detailed analysis on the evolutionary history of mosquito RIPs (including synteny analysis and hypothesis on gene duplications and losses) can be found in the Supplementary Data File 1. In addition, two transcriptomic sequences from another Culicinae mosquito; Armigeres subalbatus, partially covering the ORFs (GenBank: EU212208, EU211398) were found. In light of these findings, two alternative hypotheses were postulated:
-
(i)
These genes have been vertically inherited from the metazoan cenancestor, and were purged from other metazoan genomes by a number of independent gene loss events.
-
(ii)
These genes are derived from a unique HGT event which took place in the common ancestor of the Culex and Aedes species.
Regarding the first alternative, the minimal number of independent gene losses required was determined in order to evaluate the plausibility of the vertical transmission hypothesis (see Supplementary Data File 2 for details). Following a conservative approach, at least 15 gene losses should be postulated, from Bilateria to Culicinae, to explain the narrow taxonomic distribution of RIP encoding genes in Metazoa. The HGT hypothesis involves the loss of the ancestral RIP genes before the origin of Metazoa, followed by a single recent HGT event to the ancestor of Culex and Aedes, yielding a more parsimonious evolutionary scenario.
Based on the strong evidence supporting that Culicinae RIPs are derived from an HGT event, a search for possible donors was conducted. The phylogeny shows that Culicinae RIPs form a well-supported monophyletic group (PP: 1, BS: 100%) embedded within bacterial sequences (Fig. 2), suggesting a prokaryotic origin. The lack of introns in Culicinae RIPs also supports this fact. Moreover, homology searches using DELTA-BLAST tool (in Bacteria taxa) and using one of the Ae. aegypti RIPs (XP_001650164) as query, yielded sequences belonging to Calothrix parietina (Cyanobacteria), and Xanthomonas cassavae (Xanthomonadales). In a second iteration using these sequences, additional bacterial sequences are retrieved, including those belonging to Tolypothrix bouteillei (Cyanobacteria) and Spiroplasma poulsonii (Tenericutes). In order to perform a more robust study we focused on Culicinae, including the recently reported sequences from Ae. albopictus, and on bacterial RIPs. Phylogenetic analysis showed that Culicinae RIPs form a monophyletic group (PP: 0.93, BS: 59%) with sequences from Cyanobacteria. This clade also groups with Spiroplasma spp sequences (PP: 0.96) although with low bootstrap support (BS: 30%) (Fig. 4). Thus, these organisms −or others closely related- could have been potential donors.
Figure 4.
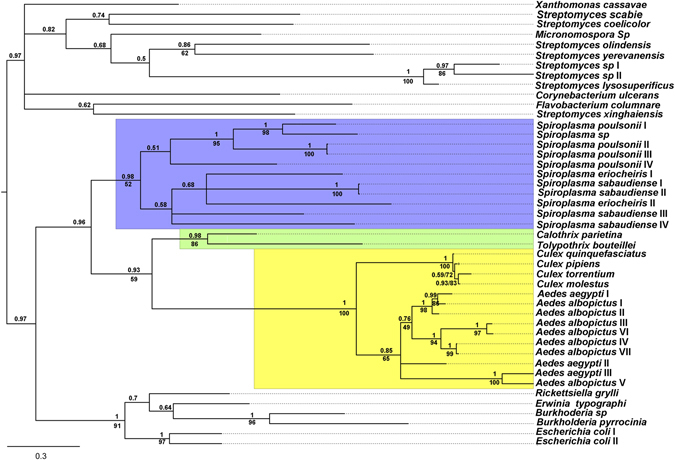
Phylogenetic relationships among Culicinae and bacterial RIPs. Bayesian tree topology based on matrix analysis of 45 proteins sequences with 275 informative sites is presented. Numbers above branches indicate PP support values. Bootstrap values (BS) >50% are shown below branches for nodes where topology of ML analysis was coincident with Bayesian inference. Light blue, green and yellow backgrounds indicate Spiroplasma, Cyanobacteria and mosquito RIPs, respectively. Information related to the sequences used to infer these trees is available in Supplementary Table 2.
Horizontally acquired RIP genes evolve under purifying selection pressure
Except for a few well characterized potent toxins (e.g. ricin, shiga and shiga-like), the physiological role of most RIPs remains unknown27. The defensive role demonstrated for Spiroplasma RIP genes in Drosophila neotestacea 13 induced us to postulate a biological function for the foreign RIP genes in mosquitoes. In addition, searches on transcriptomic databases revealed that RIP genes from C. quinquefasciatus, Ae. aegypti and Ae albopictus are expressed at the RNA level (see Supplementary Data File 1). Besides, as mentioned above, two transcriptomic sequences from Armigeres subalbatus have also been deposited (GenBank: EU212208, EU211398). However, mRNA expression is needed for, but not proof of biological significance. Therefore, we searched for traces of selective pressure (i.e. impact on the fitness) on HGT-derived sequences, as reliable evidence of functionality28. To do this, all RIP encoding sequences from C. quinquefasciatus, C. molestus, C. pipiens, C. torrentium, Ae. Aegypti, and Ae. albopictus were aligned. Interestingly, the observed INDELs were always multiple of three nucleotides, strongly suggesting that frame-shifting mutations were actively purged by negative selection pressure (except for the RIP sequence of C. quinquefasciatus JHB MR4 mentioned above; see discussion below). Figure 5 shows a logo representation of the MSA, where higher conservation of first and second codon position seems to be the rule. Moreover, an integrative analysis of synonymous vs. nonsynonymous substitutions employing three different methods; SLAC, FEL and REL (see Materials and Methods) showed purifying (negative) selection for 64 codons, including most of the amino acids forming the active site (Fig. 5 and Supplementary Table 3). Additionally, the global nonsynonymous (Ka)/synonymous (Ks) rate (ω value) was calculated for whole coding sequences of Culicinae RIPs. This yielded a ω = 0.23877, being values of less than 1 indicative of purifying selection. Overall, our results suggested that Culicinae RIP genes have been under purifying (negative) selection, supporting the idea that these genes have biological significance.
Figure 5.
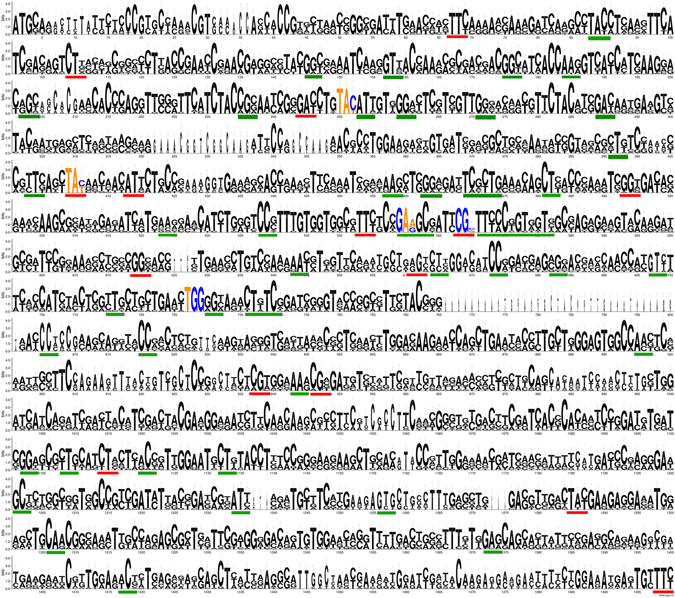
Analyses of the synonymous vs. nonsynonymous substitution rates. Codons forming the active site are indicated by colored nucleotides (A and T: orange, G and C: blue). Codons under significant purifying (negative) selection determined by the three tests (SLAC, FEL and REL), or by two out of the three tests, are underlined in red or green color, respectively.
Discussion
Ribosome inactivating proteins form a very interesting protein family displaying a patchy taxonomic distribution. As it was mentioned above, in a previous report we have found in silico evidence of the presence of RIP genes in two closely related species of mosquitoes11. Due to the uniqueness of this finding, in this work we have experimentally confirmed the presence and location of RIP genes in the C. quinquefasciatus genome, as well as in other Culex species. Moreover, new exhaustive searches on metazoan databases, revealed the presence of additional homologous genes in Ae. albopictus and Armigeres subalbatus, confirming the RIP gene family is taxonomically restricted to the Culicinae subfamily (Supplementary Data Files 1 and 2).
Currently, there is not a “gold standard” methodology for automatic and reliable detection of HGT. Moreover, several reports claiming the presence of foreign genes have been undermined after more thorough analyses due to artifacts or misinterpretations22, 29–31. Therefore, careful integration of information derived from taxonomic distribution, phylogenetic inferences and biological information is needed to detect bona fide horizontally acquired genes. The evidence obtained in this work shows that the most plausible origin of Culicinae RIPs is a single HGT event to the cenancestor of Culex, Aedes and Armigeres genera. This model is supported by the monophyly of metazoan RIPs and their very narrow taxonomic distribution, the gathering into a clade along with prokaryotic sequences and the shared genomic context among Culicinae species.
According to phylogenetic inferences (Figs 2 and 4), the donor of the RIP gene was, most likely a prokaryotic organism. An obvious donor candidate for insects is Wolbachia spp, since several HGT events between these bacteria and arthropods have been clearly documented32–34. It is expected that animal genomes are marginally affected by HGT because of the separation of the germline from somatic cells. This barrier to HGT -known as Weismann barrier- is not present in the case of bacteria infecting germline cells as Wolbachia spp, which is consistent with the relatively high number of Wolbachia to insect HGT events16. However, no RIP encoding sequences can be found in any of the Wolbachia spp databases (including 27 fully sequenced genomes). Interestingly, HMMER searches showed hypothetical proteins harboring the RIP domain on Tenericutes class, specifically in Spiroplasma species. The fact that Spiroplasma species lack a cell wall and are frequent endosymbionts of arthropods makes them logical donor candidates. However, considering Spiroplasma sp. as donor involves two major drawbacks. Spiroplasma coding sequences harbor very low GC content (around 23%), whereas Culicinae RIP genes range from 39.8% to 55.6% (Table 1). Secondly, Spiroplasma spp. and some species of Millicutes use a non-universal UGA tryptophan codon. This variation in the genetic code is presumed to have occurred in the early divergence of these genera (dating 250 mya approximately)35 while the Culicinae subfamily has diverged more recently (between 51 to 204 mya)36, 37. Therefore, the transferred genes containing the non-universal UGA tryptophan codons would be read as a stop. Although GC content of a transferred functional gene could be gradually modified by amelioration, the reversion of several nonsense codons to Trp does not seem plausible. Therefore, these pieces of evidence lead us to reject that Culicinae RIP genes are derived from Spiroplasma.
Table 1.
GC content analysis.
| Organism | RIP name | GenBank | GC % | Average GC* |
|---|---|---|---|---|
| Culex quinquefasciatus | RIPcu | XM_001850821.1 | 53.7 | 51.07% |
| Culex pipiens | RIPpi | KX674697 | 55.6 | 52.48% |
| Aedes aegypti | RIPaeI | XM_001650114.1 | 50.7 | 50.65% |
| RIPaeI LIKE | AAGE02007824 | 44.8 | ||
| RIPaeII | XM_001658803.1 | 39.8 | ||
| Aedes albopictus | RIPalI | JXUM01048494 | 50.9 | 53.2% |
| RIPalII | JXUM01048494 | 50.2 | ||
| RIPalIII | JXUM01048511 | 46.5 | ||
| RIPalIV | JXUM01048511 | 46.9 | ||
| RIPalV | JXUM01085134 | 45.5 | ||
| RIPalVI | JXUM01090901 | 46.8 | ||
| RIPalVII | JXUM01090901 | 47.2 | ||
| Spiroplasma poulsonii | JHEG02000036.1 | 26.3 | 24.90% | |
| JTLV01000005.1 | 25.3 | |||
| Tolypothrix sp | JHEG02000036.1 | 38.1 | 41.73% | |
| Calothrix sp | CP003610.1 | 35.8 | 45.97% | |
| Ancestral RIP Aedes | 51.1 | |||
| Ancestral RIP Culex | 54.5 | |||
| Ancestral RIP metazoan | 55.4 |
The GC content of RIP genes was calculated using DNA/RNA GC Content Calculator (http://www.endmemo.com/bio/gc.php). *The average of GC content in coding regions was obtained from the codon usage database (http://www.kazusa.or.jp/codon/).
The phylogenetically closest sequences to Culicinae RIPs belong to two cyanobacteria (Tolypothrix bouteillei and Calothrix parietina). Cyanobacteria constitute a significant fraction of the microbiota at breeding sites of mosquitoes. Remarkably, cyanobacterial species account for 40% of the bacterial midgut content of larval and pupal stages in An. gambiae 38. Moreover, Calothrix sp. has been detected in the midgut bacterial flora of An. stephensi in larval stage39. In addition, the GC content of cyanobacterial genomes is closer to the Culicinae RIPs (Table 1). Altogether, the presented phylogenetic inferences, the shared ecological niches between these bacteria and insects, and the colonization of mosquitoes in their early developmental stages38 strongly suggest that Cyanobacteria is the most plausible donor lineage of RIP genes to the subfamily Culicinae via HGT. In line with Huang16 we postulate that Weismann barrier is, if not absent, markedly weakened in the egg, pupal, and larval stages of mosquitoes. Consequently, these early developmental stages could be particularly prone to the acquisition of heritable foreign genes by environmental bacteria.
An important question about the HGT derived genes is their fate. In other words, the issue at stake is whether “foreign” genes will have an impact on fitness, and if so, to what extent these genes will be affected by natural selection and/or genetic drift. Probably, mosquito RIP genes display a defensive role, which has helped them to be fixed by natural selection. However, the fact that individuals from the C. quinquefasciatus MR4 colony are homozygous for a null mutation in the RIP gene shows it is not essential for viability under laboratory conditions. On the other hand, evolutionary analyses of mosquito sequences revealed evidence of purifying (negative) selection both at the whole-sequence level (ω < 1) and at several codons, which is also reflected by the occurrence of the majority of changes at the third codon position (Fig. 5). According to the nearly-neutral evolutionary theory, slightly deleterious mutations can be fixed in populations with low effective size, as in the case of laboratory colonies. On the contrary, these mutations are efficiently purged from larger populations, such as natural populations40. This seems to be the case of Culicinae RIPs, since null mutant C. quinquefasciatus are viable in captivity, while clear evidence of selection pressure on coding sequences in wild specimens has been found.
Materials and Methods
PCR experiments
PCR experiments were conducted to confirm the presence of RIP encoding sequences in selected organisms. Individuals of C. quinquefasciatus strain JHB were obtained from the MR4 colony (Malaria Research and Reference Reagent Resource Center) of the Center for Diseases Control and Prevention (CDC). Genomic DNA from these specimens was extracted employing the protocol previously described by Collins41. Genomic DNA from wild specimens of C. molestus, C. pipiens and C. torrentium was kindly provided by Dr. Stefanie Becker (Bernhard Nocht Institute for Tropical Medicine, Hamburg)42. Primer sets were designed to amplify the full-length RIP coding sequence of C. quinquefasciatus and also to identify the predicted neighbor gene (Supplementary Table 4). High fidelity Phusion DNA polymerase (New England Biolabs) was used under the following PCR conditions: initial denaturation for 30 s at 98 °C, followed by 35 cycles of denaturation (10 s at 98 °C), annealing (30 s at a particular Ta for each primer pair, see Supplementary Table 4) and extension (30 s at 72 °C), and a final extension at 72 °C for 10 min. PCR products were cloned into the pGEMT-easy® vector (Promega) following standard methods, and sequenced. Alternatively, PCR products from individual specimens belonging to the MR4-CDC colony were directly sequenced. The obtained sequences are available under the following GenBank accession numbers: KX674699, KX674697, KX644696, and KX674698.
Data collection and Multiple Sequence Alignments (MSAs)
We used a previously reported alignment (Data Set S1 in ref. 11) as a matrix to perform searches on reference proteomes using the hmmsearch tool (http://www.ebi.ac.uk/Tools/hmmer/search/hmmsearch)43 under default parameters. Although this search strategy is powerful, it is designed to explore only protein databases. Therefore, each one of the retrieved sequences was used as query to perform tBLASTn searches under default parameters against different nucleotide (WGS, ESTs, nr/nt, refseq-rna) databases. In order to confirm the absence of RIP genes in relevant lineages (e.g. Archaea, metazoan other than Culicinae) searches were performed with taxonomic restriction. All retrieved sequences were curated by confirming the presence of a canonical RIP domain (PF00161) using the Pfam server (http://pfam.xfam.org/) and those amino acids predicted to form the active site. The conserved region from new sequences was selected and used for constructing MSAs using the MAFFT 7 online server (http://mafft.cbrc.jp/alignment/server). A few regions where the alignment showed rare insertions exclusively found in a group of sequences with high sequence identity among them were manually removed by blocks. Even though the alignment was difficult as a consequence of the high divergence of the sequences, we confirmed that residues predicted to form the active sites were correctly aligned.
Phylogenetic inferences
The MSAs obtained were used to perform phylogenetic analysis using Bayesian and Maximum Likelihood inference methods. The substitution matrix and gamma distribution model with invariable sites were calculated using ProtTest3.444 with WAG selected always as the best model. PhyML45 was run using the algorithm Tree-Bisection-Reconnection (TBR) with 5 random starting trees. To estimate the robustness of the phylogenetic inference, we ran 100 bootstrap (BS) replicates. Bayesian inferences were performed using Mr. Bayes 3.2 software46. A mixed amino acid substitution model was set up, and WAG was retrieved as the best fit model, 4 gamma categories and a proportion of invariable sites were considered. The analyses were concluded after 2,000,000 generations when the split frequency was <0.02. FigTree 1.4.2 software was used to visualize and edit the trees. Sequence alignments (in Fasta format) and phylogenetic trees (in Newick format) used for constructing Figs 2 and 4 are in Supplementary Data File 3.
Genomic context analyses
The complete sequence of the scaffolds containing RIPs of C. quinquefasciatus (DS232037), Ae. aegypti (NW001810221) and An. gambiae (chromosome 3 L) were subjected to BLASTx searches on different protein databases to identify individual genes upstream and downstream of the RIP open reading frame (ORF). The retrieved sequences were subjected to reciprocal BLASTp and tBLASTn searches in order to determine putative orthologous sequences. Orthologs were confirmed using the comparative genomic tool available at VectorBase website (https://www.vectorbase.org/).
Evolutionary analyses
Nucleotide encoding RIP sequences found in mosquitoes were aligned by codons, using PAL2NAL algorithm47. Then, the codons under purifying (negative) selection were estimated employing the Single Likelihood Ancestor Counting (SLAC), Fixed Effects Likelihood (FEL), and Random Effects Likelihood (REL) tests, available at the Datamonkey Package48, 49. The omega (ω) value represented the global ratio between nonsynonymous and synonymous mutations and was assessed using the ML method of CodeML in PAML50, 51. This analysis was performed under the one ratio model (M0). Finally, a sequence logo was constructed employing the codon alignment using the Weblogo server52 and codons under purifying selection were highlighted.
Electronic supplementary material
Acknowledgements
M.J.A. and M.V.S.P. are members of the CONICET Research Career. CONICET fellowships to W.J.L. and M.L.M. are also acknowledged. The DNA samples of C. molestus, C. pipiens and C. torrentium were kindly gifted by Dr. Stefanie Becker. The authors acknowledge the Malaria Research and Reference Reagent Resource Center (MR4, CDC) for providing the following mosquito strain through BEI Resources, NIAID, NIH: Culex quinquefasciatus, strain JHB, NR-43025. The authors are also grateful to Dr. Jimena Juri Ayub for her technical support. We would also like to thank GAECI for their services. The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC).
Author Contributions
Conceived and designed the experiments: W.J.L., P.L.M., M.L.M. and M.J.A. Performed the experiments: W.J.L., P.L.M. and M.L.M., Analyzed the data: W.J.L., P.L.M., M.L.M., M.V.S.P and M.J.A. Contributed reagents/materials/analysis tools: W.J.L., P.L.M. and M.J.A. Wrote the paper: W.J.L., P.L.M., M.L.M., M.V.S.P. and M.J.A.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01859-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Walter J. Lapadula, Email: wlapadula@gmail.com
Maximiliano Juri Ayub, Email: mjuriayub@hotmail.com.
References
- 1.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. The Journal of biological chemistry. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 2.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. The Journal of biological chemistry. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 3.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. The Journal of biological chemistry. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 4.Rajamohan F, Ozer Z, Mao C, Uckun FM. Active center cleft residues of pokeweed antiviral protein mediate its high-affinity binding to the ribosomal protein L3. Biochemistry. 2001;40:9104–9114. doi: 10.1021/bi002851p. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson L, Nygard O. The mechanism of the protein-synthesis elongation cycle in eukaryotes. Effect of ricin on the ribosomal interaction with elongation factors. European journal of biochemistry/FEBS. 1986;161:111–117. doi: 10.1111/j.1432-1033.1986.tb10130.x. [DOI] [PubMed] [Google Scholar]
- 6.Sperti S, Montanaro L, Mattioli A, Stirpe F. Inhibition by ricin of protein synthesis in vitro: 60 S ribosomal subunit as the target of the toxin. The Biochemical journal. 1973;136:813–815. doi: 10.1042/bj1360813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girbés T, Ferreras JM, Arias FJ, Stirpe F. Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini reviews in medicinal chemistry. 2004;4:461–476. doi: 10.2174/1389557043403891. [DOI] [PubMed] [Google Scholar]
- 8.Reyes AG, et al. The Streptomyces coelicolor genome encodes a type I ribosome-inactivating protein. Microbiology. 2010;156:3021–3030. doi: 10.1099/mic.0.039073-0. [DOI] [PubMed] [Google Scholar]
- 9.Sandvig K. Shiga toxins. Toxicon. 2001;39:1629–1635. doi: 10.1016/S0041-0101(01)00150-7. [DOI] [PubMed] [Google Scholar]
- 10.Di Maro A, Citores L, Russo R, Iglesias R, Ferreras JM. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant molecular biology. 2014;85:575–588. doi: 10.1007/s11103-014-0204-y. [DOI] [PubMed] [Google Scholar]
- 11.Lapadula WJ, Sanchez Puerta MV, Juri Ayub M. Revising the taxonomic distribution, origin and evolution of ribosome inactivating protein genes. PLoS One. 2013;8:e72825. doi: 10.1371/journal.pone.0072825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peumans, W. J. & Van Damme, E. J. In Toxic plant proteins 1-26 (Springer, 2010).
- 13.Hamilton PT, Peng F, Boulanger MJ, Perlman SJ. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:350–355. doi: 10.1073/pnas.1518648113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 15.Treangen TJ, Rocha EP. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J. Horizontal gene transfer in eukaryotes: the weak-link model. BioEssays: news and reviews in molecular, cellular and developmental biology. 2013;35:868–875. doi: 10.1002/bies.201200182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku C, Martin WF. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70% rule. BMC biology. 2016;14:89. doi: 10.1186/s12915-016-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ku C, et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 19.Danchin EG, et al. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17651–17656. doi: 10.1073/pnas.1008486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wybouw N, Pauchet Y, Heckel DG, Van Leeuwen T. Horizontal Gene Transfer Contributes to the Evolution of Arthropod Herbivory. Genome biology and evolution. 2016;8:1785–1801. doi: 10.1093/gbe/evw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haegeman A, Jones JT, Danchin EG. Horizontal gene transfer in nematodes: a catalyst for plant parasitism? Molecular plant-microbe interactions: MPMI. 2011;24:879–887. doi: 10.1094/MPMI-03-11-0055. [DOI] [PubMed] [Google Scholar]
- 22.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 23.Boothby TC, et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proceedings of the National Academy of Sciences. 2015;112:15976–15981. doi: 10.1073/pnas.1510461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salzberg SL, White O, Peterson J, Eisen JA. Microbial genes in the human genome: lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 25.Richards TA, Monier A. A tale of two tardigrades. Proceedings of the National Academy of Sciences. 2016;113:4892–4894. doi: 10.1073/pnas.1603862113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapadula WJ, Sanchez-Puerta MV, Ayub MJ. Convergent evolution led ribosome inactivating proteins to interact with ribosomal stalk. Toxicon. 2012;59:427–432. doi: 10.1016/j.toxicon.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Puri M, Kaur I, Perugini MA, Gupta RC. Ribosome-inactivating proteins: current status and biomedical applications. Drug discovery today. 2012;17:774–783. doi: 10.1016/j.drudis.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Hotopp JCD. Horizontal gene transfer between bacteria and animals. Trends in Genetics. 2011;27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koutsovoulos G, et al. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proceedings of the National Academy of Sciences. 2016;113:5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurland CG, Canback B, Berg OG. Horizontal gene transfer: a critical view. Proceedings of the National Academy of Sciences. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurence M, Hatzis C, Brash DE. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PloS one. 2014;9:e97876. doi: 10.1371/journal.pone.0097876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunning Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 33.Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. Bmc Genomics. 2009;10:1. doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woolfit M, Iturbe-Ormaetxe I, McGraw EA, O’Neill SL. An ancient horizontal gene transfer between mosquito and the endosymbiotic bacterium Wolbachia pipientis. Molecular biology and evolution. 2009;26:367–374. doi: 10.1093/molbev/msn253. [DOI] [PubMed] [Google Scholar]
- 35.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiology and Molecular Biology Reviews. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC evolutionary biology. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foley DH, Bryan JH, Yeates D, Saul A. Evolution and systematics of Anopheles: insights from a molecular phylogeny of Australasian mosquitoes. Molecular phylogenetics and evolution. 1998;9:262–275. doi: 10.1006/mpev.1997.0457. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Gilbreath TM, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PloS one. 2011;6:e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasites & vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohta T. The nearly neutral theory of molecular evolution. Annual Review of Ecology and Systematics. 1992;23:263–286. doi: 10.1146/annurev.es.23.110192.001403. [DOI] [Google Scholar]
- 41.Collins FH, et al. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. The American journal of tropical medicine and hygiene. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- 42.Rudolf M, et al. First nationwide surveillance of Culex pipiens complex and Culex torrentium mosquitoes demonstrated the presence of Culex pipiens biotype pipiens/molestus hybrids in Germany. PloS one. 2013;8:e71832. doi: 10.1371/journal.pone.0071832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 46.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic acids research. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delport W, Poon AF, Frost SD, Pond SLK. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pond SLK, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular biology and evolution. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 50.Xu B, Yang Z. PAMLX: a graphical user interface for PAML. Molecular biology and evolution. 2013;30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 52.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


