Abstract
Human tissues are sophisticated ensembles of many distinct cell types embedded in the complex, but well-defined, structures of the extracellular matrix (ECM). Dynamic biochemical, physicochemical, and mechano-structural changes in the ECM define and regulate tissue-specific cell behaviors. To recapitulate this complex environment in vitro, dynamic polymer-based biomaterials have emerged as powerful tools to probe and direct active changes in cell function. The rapid evolution of polymerization chemistries, structural modulation, and processing technologies, as well as the incorporation of stimuli-responsiveness, now permit synthetic microenvironments to capture much of the dynamic complexity of native tissue. These platforms are comprised not only of natural polymers chemically and molecularly similar to ECM, but those fully synthetic in origin. Here, we review recent in vitro efforts to mimic the dynamic microenvironment comprising native tissue ECM from the viewpoint of material design. We also discuss how these dynamic polymer-based biomaterials are being used in fundamental cell mechanobiology studies, as well as towards efforts in tissue engineering and regenerative medicine.
Keywords: dynamic cell culture, stimuli-responsive material, hydrogels, shape-memory material, mechanobiology, 4D biology, tissue engineering
1. Introduction
The cellular microenvironment regulates many important biological functions, including adhesion, growth, migration, and differentiation [1, 2]. In addition to the biochemical properties of the extracellular matrix (ECM), mechano-structural cues such as elasticity and topography are of great importance in microenvironmental-based governance of cell function [3, 4]. Growing evidence suggests that mechano-structural cues differentially modulate cell fate in a hierarchical response [5–8]. Efforts to elucidate the effects have primarily centered on static systems where the biochemical and biophysical properties of matrices remain constant over time. Studies focused on the effect of static topography and elasticity of the ECM in artificial materials have enabled the advanced interrogation of cellular mechanotransduction responses to these cues [9–14]. Biochemical cues also play a crucial role in directing cellular function and fate, and the relationship between their ‘static’ effect and cellular responses has been addressed by material-based and molecular biology approaches [15, 16]. However, we know that cellular microenvironments in vivo gradually change their physicochemical properties, as evidenced in cardiomyopathies and in cancer progression [17–19]. This dynamic nature, in turn, is closely related to tissue/organ development, regeneration, wound healing, and disease progression over time [20]. Therefore, in vitro platforms that recapitulate dynamic in vivo signaling may provide for an enhanced understanding of fundamental biological processes, and could lead to eventual advances in tissue engineering and regenerative medicine.
Recently, the scientific community has attempted to mimic dynamic ECM signaling through the development of cell culture platforms with tunable properties. Within this context ‘stimuli-responsive’ or ‘smart’ materials and systems represent useful tools for mechanobiology studies [21, 22]. These material systems can change their properties on demand in response to user-defined triggers (e.g., pH, temperature, light). As we will discuss in further detail, recent examples of dynamic cell culture platforms involve tunable surface properties such as elasticity and topography, spatiotemporal presentation and removal of biochemical signals, and applied force loading against cultured cells. Inspired by dynamic, tissue-dependent microenvironments in vivo, the use of dynamic cell culture platforms to create sophisticated matrices in vitro has been attractive to engineers and biologists in the fields of classic cell biology, tissue engineering, and regenerative medicine.
Although excellent reviews of stimuli-responsive polymers and their biomedical and tissue engineering applications have been published [23–35], few comprehensive reviews summarize how stimuli-responsive polymers and systems enable newfound mechanobiological studies as well as the development of artificial matrices that better mimic the dynamic biophysical aspects of native tissue [21, 22]. In this review, we focus on recent efforts to construct synthetic cell culture microenvironments, discussing the dependence of cell-specific function on specific environmental cues. First, we briefly review dynamic aspects of the human body, motivating the rational designs of in vitro cell culture platforms. We then review different stimuli-responsive polymeric substrates that have been recently developed for dynamic cell-matrix mechanobiology. Lastly, we describe the design of artificial matrices offering four-dimensional (4D) control of material properties and highlight future trends in the field.
2. The Dynamic In Vivo Cellular Microenvironment
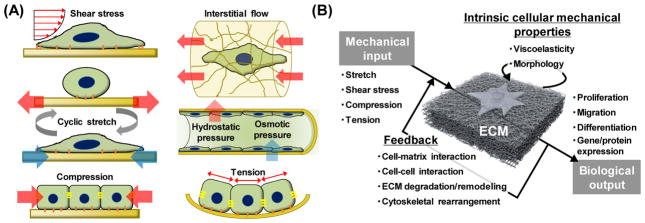
The human body represents a complex collection of dynamic environments where biochemical, physicochemical, and mechano-structural interactions serve to regulate cell behavior and fate [17]. In addition to these environmental cues, various types of regulatory mechanical stimuli exist within the human body (Figure 1A). Cells are constantly subjected to shear flow, stretching, cyclic strain, and generated tensions, where stimuli magnitude is highly dependent on the tissue itself. These tissue-dependent mechanical stimuli ultimately dictate cellular function and fate [36]. Mechanobiology is an emerging field of science interfacing engineering and biology. Understanding mechanotransduction, or how cells of various tissues sense, recognize, and respond to mechanical stimuli, is a major challenge that has become increasingly important in mechanobiology. Here, mechanical stimuli are not limited to externally-imposed forces, such as fluidic shear stress, but also include the intrinsic tensions generated by active cell contraction that occur in the absence of external forces. Thus, the mechanotransduction process can be described as a simple model where mechanical input influences cells’ intrinsic mechanical properties which is then transduced into specific cellular outputs (Figure 1B). Furthermore, the biological output can change the cellular microenvironment, altering the initial mechanical input. In other words, the mechanotransduction process is equipped with a feedback system, which generates a highly complex and dynamic mechanical environment that mechanobiological studies have until recently largely ignored.
Figure 1.
Mechanical forces in our body and their transduction process into biological output. (A) Mechanical stimuli found at the cell, tissue, and organ level inside the body. (B) Mechanotransduction is the process by which cells convert mechanical inputs into biological responses. Mechanotransduction often involves a feedback process, and their mechanical environment is dynamic and complex [36].
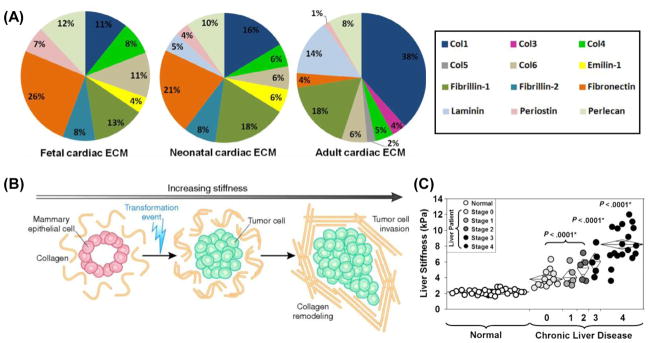
On the other hand, all cell types are in contact with their ECM, a complex and dynamic network of macromolecules with different physicochemical natures. By modulating the production, degradation, and remodeling of its components, the ECM can support organ development, function and repairing [17, 37, 38]. Williams et al. recently reported that the ECM is gradually altered during heart development and demonstrated its importance in cardiac regeneration [39]. They determined ECM composition at different developmental ages – fetal, neonatal and adult – by liquid chromatography tandem mass spectrometry (LC-MS/MS), and found that the most abundant ECM protein in fetal and neonatal hearts is fibronectin, whereas the adult ECM is mostly composed of collagen I (Figure 2A). It is well-known that cardiomyocyte proliferation declines with age [40, 41]; therefore, these findings strongly suggest that observed variations in cardiomyocyte proliferation are a function of dynamic changes in cardiac ECM composition, and may provide a rationale for engineering cardiomyocyte culture platforms. Dynamic changes in ECM structure and mechanical properties have also been implicated with the tumorigenesis process [42]. When mammary epithelial cells (MECs) are grown in Matrigel®, a gelatinous protein mixture secreted by the Engelbreth-Holm-Swarm (EHS) mouse sarcoma cell, they form organized and polarized mammary acini structures. By inducing a transformation event, cells can be triggered to invade the acini lumen and serve as a unique model of tumorigenesis [43]. In this model, transformation can be induced by increasing matrix stiffness alone and without altering biochemical composition (Figure 2B). Here, dynamic alteration in matrix mechanical properties in the form of increased stiffness led to increased cellular growth, loss of polarity, and increased focal adhesion contacts, attributes that are closely related to a malignant phenotype [44].
Figure 2.
Dynamic changes of cellular microenvironment cues in vivo. (A) Changes in rat cardiac ECM composition as characterized by LC-MS/MS at three different developmental stages, fetal, neonatal and adult [39]. (B) Schematic illustrating the remodeling of mammary epithelial cells (MECs) in 3D assays in vitro as a function of stiffness. MECs form organized and polarized acini structures when grown in reconstituted basement membrane in vitro. A transformation event and increasing matrix stiffness results in cell invasion into the lumen [42]. (C) Liver stiffness as a function of fibrosis in patients being evaluated for chronic liver disease. Data was obtained using magnetic resonance elastography [47].
In addition to tumorigenesis in epithelial cells, other diseases are also accompanied with dynamic changes in their cellular microenvironments. For example, liver fibrosis is the excessive accumulation of ECM proteins, including collagen, that occurs in most types of chronic liver diseases [45]. Palmeri et al. observed a significant relationship between liver stiffness and hepatic fibrosis stages [46]. A noninvasive evaluation of hepatic fibrosis using acoustic radiation force-based elasticity measurements in patients with nonalcoholic fatty liver disease was conducted, revealing increased variability in liver stiffness with increased fibrosis. Similar observation was characterized by magnetic resonance-based elastrography (Figure 2C) [47]. Stiffer, more advanced liver fibrosis results in cirrhosis, liver failure, and portal hypertension, and often requires liver transplantation.
These are just a few examples of dynamic cellular microenvironments inside our body, but many other examples undoubtedly exist [17–19]. The instructive cues of the ECM and its surroundings are the result of a number of different biochemical, physicochemical, and mechano-structural characteristics. In addition, all of these aspects are dynamically orchestrated in vivo to maintain an extracellular environment that is optimal for cellular homeostasis and dictates cellular function and fate. Further in vivo studies will not only deepen our understanding of dynamic mechanobiology, but also provide new rationale and design criteria for the fabrication of more sophisticated artificial matrices.
3. Dynamic Polymeric Biomaterial Platforms for In Vitro Culture
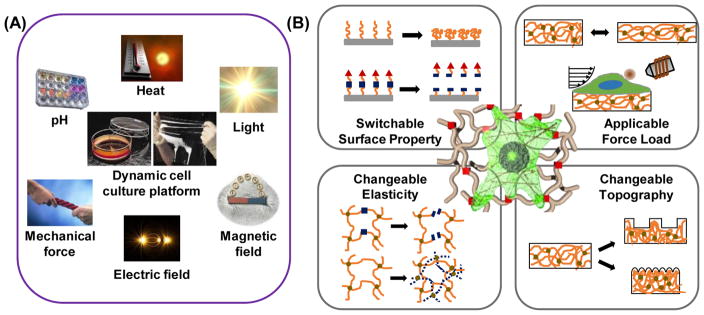
Although dynamic variations in the cellular microenvironment are common in vivo, establishing an understanding of the biological mechanisms that links these changes to alterations in native cell functions has been difficult. To better understand dynamic mechanobiology, the recent development of artificial matrices engineered in vitro, together with the application of in vivo studies, has proven critical to better understand basic cell physiology in the context of dynamic microenvironmental change. Certain synthetic materials that are capable of responding to external or internal stimuli represent one of the most exciting emergent areas of scientific interest [29]. We call this class of materials ‘stimuli-responsive’ or ‘smart’ materials, where some form of energetic input into a system induces changes in the system’s physicochemical properties, and some of those that are especially polymeric materials have contributed to the fields of tissue engineering and mechanobiology (Table 1). Various external stimuli such as heat, light, electrical fields, magnetic fields, mechanical force, active molecules, and environmental pH have been utilized thus far (Figure 3A). Importantly, these external stimuli are used as extremely mild, noninvasive triggers for cells and tissues in optimized conditions. In addition, stimuli-responsive materials can be engineered in various forms including functionalized surfaces, thin films, and 3D gels, which are useful as cellular supports [48]. Therefore, stimuli-responsive materials are powerful tools when designing the substrates of cell culture platforms. These materials can impart various dynamic biochemical, physicochemical, and mechano-structural cues to adhered cells, enabling researchers to assay specific biological outputs for well-defined inputs (Figure 3B).
Table 1.
Overview of commonly used polymers for the designing of dynamic cell culture platforms
| Polymers | Chemical Structures | Characteristics | Application | Reference |
|---|---|---|---|---|
| Poly(N-isopropylacrylamide) (PNIPAAm) |
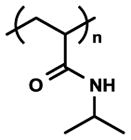
|
Temperature ability responsive | Adhesion-Detachment manipulation | [52, 64, 65, 69, 71] |
| Reversibly changeable hydrophilicity/hydrophobicity or surface energy | Cell sheet engineering | [53, 54, 73] | ||
| Functionalizable | 3D tissue fabrication | [59, 61, 62, 79] | ||
| Poly(ethylene glycol) (PEG) |
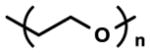
|
Bioinert and Biocompatible | Non-adhesive surface | [87, 88, 115, 116] |
| 3D and 4D tissue culture | [134, 156, 228, 229, 232, 233, 235, 236] | |||
| Polypyrrole |
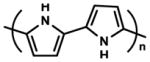
|
Electro-responsive ability Electroconductivity Changeable charge (polarity) |
Adhesion-Detachment manipulation | [105] |
| Poly(3,4-ethylenedioxythiophene) |
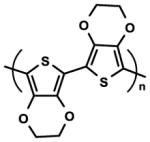
|
Electro-responsive ability Electroconductivity | Adhesion-Detachment manipulation | [106–108] |
| Poly(dimethyl siloxane) (PDMS) |
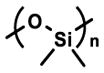
|
Mechanically-responsive ability | Cell assay device | [153, 189, 200, 237] |
| Transparent, Processability Tunable Stiffness (~MPa range) | Organ-on-chip | [197–199] | ||
| Polyacrylamide (PA) |
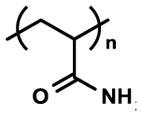
|
Bioinert nature | 2D cell culture | [10, 133, 140] |
| Tunable stiffness (~kPa range) | Cell-material interaction assay | [237, 238] | ||
| Hyaluronic acid (HA) |
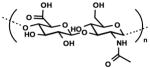
|
Natural ECM | Tissue engineering | [144, 147] |
| Tissue-like stiffness Biocompatible, Biodegradable Modificable | 3D and 4D tissue culture | [218] | ||
| Poly(ε-caprolactone) (PCL) |
|
Biocompatibility and Biodegradability | Tissue engineering | [151, 152] |
| Semicrystalline nature Shape-memory property Changeable shape and stiffness |
Dynamic cell manipulation (Orientation, Movement) | [158–163] |
Figure 3.
Stimuli-responsive polymeric materials for dynamic cell culture platforms. (A) Various external stimuli can be utilized for dynamic biomaterials. By modulating chemical, physicochemical and mechano-structural properties, dynamic cell culture platforms can be generated using these materials. (B) Schematic illustration of dynamic cell culture platforms with: (a) tunable matrix stiffness, (b) variable caging-mediated ligand presentation, (c) cleavage- or desorption-mediated ligand presentation, and (d) applicable forces or switchable topography [22].
In this review, we classify current dynamic cell culture platforms into two different categories which vary by the given property that is dynamically controlled: 1) physicochemical and biochemical cues and 2) mechano-structural cues. Based on these categories, we will address the design criteria of their corresponding materials and systems, as well as how time-dependent dynamic changes in cell-material interactions influence cellular behavior, function, and fate.
4. Dynamic Physicochemical and Biochemical Cues
4.1. Temperature-responsive polymers
Cells in our body interact not only with biochemical signals presented by their surroundings, but also physicochemically with the ECM or adjacent cells through various receptor-ligand interactions [49–51]. Adhesion provides physical support for the cells, regulates cell positioning, and enables microenvironmental sensing. It plays an important role in various physiological processes such as migration, proliferation, differentiation, and death. Therefore, technologies based on dynamic surface properties that exercise spatiotemporal control over cell adhesion, detachment, and migration are desirable for further development of mechanobiology.
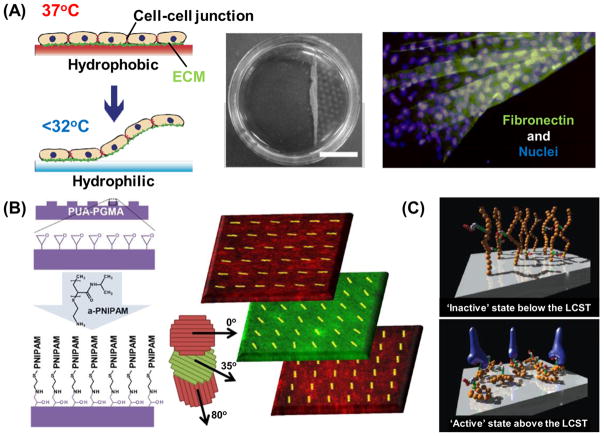
In one foundational approach in the 1990s, Yamada et al. proposed a novel detachment technique for actively adhered cells using poly(N-isopropylacrylamide) (PNIPAAm)-grafted tissue culture polystyrene (TCPS) dishes [52]. They demonstrated that many attachment-dependent cell types that adhered to TCPS modified with a nanometer-thick PNIPAAm-grafted layer at 37°C can spontaneously detach from the surface simply by lowering the temperature below PNIPAAm’s lower critical solution temperature (LCST) of 32°C (Figure 4A). This cellular detachment from the PNIPAAm layer relies on dynamic changes in surface hydrophilicity/hydrophobicity, or energy for polymeric conformational changes, in response to temperature variation, which in turn disrupts cell-substrate interactions [53]. The dynamic modulation of cell adhesion–detachment on surfaces grafted with thermoresponsive PNIPAAm chains was a novel concept because enzymes and chelators were not required to actively detach the adhered cells. The most striking feature of this system is that it allows for the preservation of intact membranes and cell-cell adhesions in that only the interactions between the adhesive proteins on the basal side of cultured cells and grafted PNIPAAm on the surfaces are disrupted. Kushida et al. harvested confluent cultured cells as a contiguous cell sheet from the thermoresponsive surfaces while maintaining cell-cell junctions and basal ECMs [54]. Since the adhesive proteins derived from the cells were also harvested at the same time (Figure 4A), the cell sheets could readily adhere to other surfaces including culture dishes, other cell sheets, and host tissues. This technology is now widely recognized as cell sheet engineering, an area of nanotechnology dedicated to the reconstruction of scaffold-free tissue with potential clinical applications in regenerative medicine.
Figure 4.
Temperature-responsive on-off control of surface physicochemical properties. (A) Cell sheet engineering technology utilizing temperature-responsive PNIPAAm-based substrates. This system allows for temperature controlled cell adhesion-detachment that maintains cell-cell connections and deposited ECM [53]. (B) A temperature-responsive nanofabricated substratum (TNFS) approach combining cell sheet engineering and nanotopographical cues for engineering 3D tissues with oriented layer-by-layer structure. Stacked nanopatterned myoblast sheets maintain individual layer alignment and demonstrate precise control in 3D [59]. (C) Schematic views of cell adhesive peptide-bearing PNIPAAm-based surfaces showing temperature-induced on-off ligand display and control of integrin-peptide binding [67, 68].
Combining cell-patterning techniques with PNIPAAm-grafted surfaces provides a tool for the construction and recovery of functional, tissue-mimicking cell sheet materials for clinical applications. A patterning strategy for grafting polymer and substrate surfaces is required to fabricate in vivo tissue-like cell structures. Idota et al. reported a simple approach for creating dynamic cell culture substrates with adhesive nanopatterns: the direct electron beam lithographic graft-polymerization method [55]. They successfully fabricated PNIPAAm patterns on cell-repellent polyacrylamide (PAAm)-modified surfaces. Their micro- and nano-textured substrates with stripe (3μm and 200nm, groove:ridge = 1:1) patterns resulted in the alignment and elongation along the major axes of the patterns of both bovine aortic endothelial cells (BAECs) and mouse fibroblasts (NIH 3T3) that had been cultured on the surfaces. The aligned confluent cells were recovered as a cell sheet by reducing the temperature from 37°C to 25°C. The cell sheets strongly contracted along the pattern axes, resulting in anisotropic shrinkage and folding. This technique would be useful for constructing muscle cell sheets with efficient shrinkage/relaxation in a given direction, or for the generation of spheroidal 3D cell structures. In a similar approach, Yamashita et al. demonstrated a non-invasive harvest system for living cell clusters cultured in microchannels by combining PNIPAA surface and microelectromechanical systems (MEMS) technologies [56]. They designed and prepared octadecyltrimethoxysilane (ODTMS)- and PNIPAAm-modified separable microchips, then cultured human arterial endothelial cells (HAECs) on the resulting microchannels. After 96 h of cell cultivation, the HAECs were detached from the microchips by temperature reduction and recovered from the microchannels. Intact and viable HAEC sheets formed in this manner could then be cultured on conventional cell culture dishes. This technique may not only be useful for the culture and harvesting of structurally-organized micro-scale tissues, but also for studying the combined effect of geometries and switchable properties of surfaces.
One large, vital challenge encountered in the field of tissue engineering is the replication of complex three-dimensional (3D) structures. The ability to do so is crucial given the huge variety of these structures and the critical role they play in regulating tissue function [57, 58]. Jiao et al. recently developed a simple, versatile platform with a thermoresponsive nanofabricated substratum (TNFS) that incorporates nanotopographic cues for aligning seeded cells with a gel casting method in order to stack and transfer aligned cell sheets for the fabrication of scaffold-free and structurally organized 3D tissues [59]. The TNFS was fabricated through capillary force lithography-based nanopatterning, followed by PNIPAAm surface modification using epoxy-amine chemistry (Figure 4B). This approach enabled the temperature-induced release of the cellular sheets while preserving organizational fidelity. In fact, it was also demonstrated that the platform allowed for the stacking of aligned sheets at specific angles between the layers, potentially enabling the reproducible and robust fabrication of scaffold-free 3D tissues with controllable architecture (i.e., nanoscale control of cell and tissue structure). Importantly, these multilayered tissues retained specific layer organization and integrity in that individual sheets retained their specific alignment and their cells did not mix or migrate between layers. This differs significantly from previous reports of the 3D organization of anisotropic cell sheets using PNIPAAm-based approaches, which often observed a vertical migration within multilayered cell sheets [60–62].
From a molecular design perspective, the introduction of functional groups or biomolecules onto grafted PNIPAAm chains is effective for controlling cell surface adhesion since PNIPAAm itself does not contain any functional groups. To address this issue, Aoyagi et al. successfully synthesized 2-carboxyisopropylacrylamide (CIPAAm) as a novel NIPAAm-based functional monomer which was then linked to a functional carboxyl group, which is useful for bioconjugation [63]. Importantly, the PNIPAAm’s thermoresponsive property was maintained after copolymerization with CIPAAm. They then applied this polymer system as a switchable cell culture substrate. Cell adhesion/detachment was achieved on both P(NIPAAm-co-CIPAAm)-grafted surfaces as well as PNIPAAm surfaces through temperature change [64]. The grafted surfaces allowed for an accelerated rate of BAEC detachment compared to that of conventional PNIPAAm homopolymer surfaces. In this case, the addition of ionized carboxyl groups led to accelerated hydration of the polymer-grafted layer, inducing a rapid swelling and subsequent lifting of the cells from the surface. In addition, carboxyl groups in P(NIPAAm-co-CIPAAm)-grafted surfaces were available for bioconjugation with various bioactive peptides and proteins. The researchers also immobilized a cell-adhesive Arg-Gly-Asp-Ser (RGDS) peptide on grafted P(NIPAAm-co-CIPAAm) via a condensation reaction between an amino terminal in RGDS and a carboxyl group in the copolymer chain [65, 66]. These surfaces even facilitated the adhesion and spreading of both BAECs and human umbilical vein endothelial cells (HUVECs) in non-serum conditions depending on RGDS surface content at 37°C, above the LCST of the copolymer. Alternately, BAECs and HUVECs detached from the surfaces by reducing the temperature below the polymer LCST, where detachment behavior also depended on the immobilized RGDS content.
To further manipulate the affinity between copolymer-grafted surfaces and the cells, Ebara et al. modulated the access of cell integrins to RGDS by tethering a poly(ethylene glycol) (PEG) chain as a spacer [67]. The synergistic effects on cell adhesion of a Pro-His-Ser-Arg-Asn (PHSRN) sequence derived from RGDS using a hexamer glycine (G6) linker were investigated by changing the distance between the peptide and the grafted polymer (Figure 4C) [68]. It was found that the peptide-bearing copolymer distance significantly affected the release time of cells from the surfaces when the temperature was lowered. Specifically, initial cell adhesion was enhanced on PHSRN-G6-RGDS-immobilized surfaces because the synergistic PHSRN was located ca. 3.5 nm away from the RGD loop, corresponding to the chain length of the G6 linker. Importantly, this series of works support the conclusion that a PNIPAAm-based cell culture platform does not just serve as an ‘on-off’ regulator for cellular detachment, but may be useful as a more precise ‘on-off’ display of the surface ligands available for cellular attachment.
The introduction of functional biological ligands is not limited to only cell-adhesive peptides and proteins [65–70]. Functionalization with sugar molecules is also applicable, as these ligands are fascinating candidates for manipulating the interaction between material surfaces and cells. Along these lines, Idota et al. developed smart copolymer brushes that are comprised of NIPAAm and 2-lactobionamidoethyl methacrylate (LAMA), which has a galactose residue that binds specifically to the asialoglycoprotein receptors of hepatocytes, and regulates hepatocyte-selective adhesion through temperature alterations [71]. NIH 3T3 fibroblasts did not adhere to the surface of polymer brushes containing LAMA under serum-free conditions, while the surface promoted human hepatocellular carcinoma cell line (HepG2) adhesion at 37°C through specific interactions with functional sugar moieties. Moreover, the adhered HepG2 successfully detached from the surface of the PNIPAAm-block-PLAMA brush when the temperature was reduced to 25°C. On the other hand, almost all cells remained adhered on the surface of P(NIPAAm-co-LAMA) brushes where the sugar moieties were randomly dispersed in a polymer chain as opposed to uniformly configured in a block polymer. Thus, the molecular architecture of grafted smart polymers is also an important factor in modulating surface properties for the manipulation of dynamic cell adhesion and detachment.
Modification of temperature-responsive polymers is not limited to substrate surfaces for manipulating cell-material interactions. Kim et al. expanded the surface-based, temperature-responsive cell culture platform to a nanofiber web system with dynamically and reversibly tunable scaffold properties based on the temperature-responsivity of PNIPAAm [72]. To obtain stable nanofibers that actuate in an aqueous environment, UV-reactive benzophenone was introduced to a P(NIPAAm-co-CIPAAm) chain, then photo-crosslinked following nanofiber formation by electrospinning. The resulting nanofiber web was quite stable in an aqueous entity and showed the ability to trigger ‘on’ or ‘off’ web properties in response to temperature change. By using this switchability, they were ultimately able to demonstrate the capture, encapsulation, and release of normal human dermal fibroblasts (NHDFs). Since the released NHDFs still showed excellent viability and proliferation behavior following their capture in a nanofiber web, this system is highly promising for biomedical applications such as the separation, purification, preservation, and delivery of cells. To better modulate cell-substrate interactions for further applications in cell sheet engineering, many other functionalization approaches of PNIPAAm-based platforms have been reported, such as co-grafting with polyethylene glycol (PEG) [73]; the introduction of a polyacrylamide (PAAm) layer [74]; the introduction of hydrophobic or charged functional groups [75, 76]; the development of other NIPAAm derivatives with different functionalities [77, 78]; and so on. Lastly, cell sheet manipulation technology has been developed as discussed above. It is currently possible to create functional, vascularized 3D tissue structures similar to those in vivo [59, 79–81]. Cell sheet technology derived from the PNIPAAm-based platform not only enables the creation of complex tissue- or organ-like structures but also has regenerative medicine applications for several different tissues [53, 82, 83]. However, further development of novel functionalization methodologies or system designs would certainly contribute to the achievement of highly functional tissue engineering for clinical applications.
4.2. Photo-responsive polymers
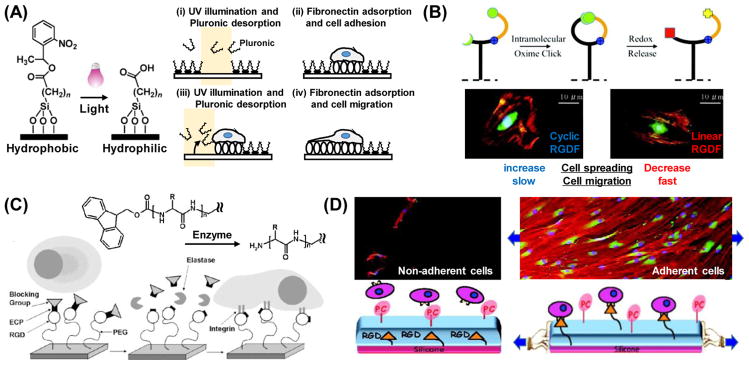
The external stimuli capable of regulating the ‘on-off’ feature of surface properties are not limited to temperature alone. Photoirradiation within certain wavelengths is a prominent example for regulating cellular phenomena because of its unique spatial and temporal controllability. The photochemical reactions that are often used to induce switching in surface physicochemical properties involve two different mechanisms: irreversible photocleavage and reversible photoisomerization. Regarding photocleavage reactions, nitrobenzyl ester derivatives are the most commonly used photocleavable protecting groups. They are especially utilized in organic synthesis, which can efficiently undergo photolysis with an irradiation of near-UV light (~350 nm) [84, 85]. This photocleavable functionality is often applied in phototunable cell culture platforms and allows for the spatiotemporal regulation of cell-surface interactions. Nakanishi et al. developed an example of this methodology in which they prepared a surface modified with an alkylsiloxane having a photocleavable 2-nitrobenzyl functional group that changes hydrophobicity in response to UV light (λ = 365 nm) (Figure 5A) [86]. After a bovine serum albumin (BSA) blocking layer was adsorbed on the surface, specific regions were carboxylate-functionalized by illumination with UV light. The carboxylate-functionalized surface that resulted after UV-irradiation was then coated with fibronectin, whereupon the selective adhesion of human embryonic kidney 293 (HEK 293) cells onto functionalized regions was successfully demonstrated.
Figure 5.
Stimuli-responsive on-off control of surface ligand display. (A) Photochemical switchable surface for defining cell adhesive domains via UV illumination at 365 nm. A schematic illustrates of the controlled placement of single cells followed the induction of cell migration with selective UV exposure and subsequent fibronectin deposition [87]. (B) Redox switchable surface for controlling peptide architecture from cyclic to linear RGD conformation. Cyclic RGD can be linearized by application of mild reduction potential (V=−250 mV) at physiological conditions. Switchable control of RGDF peptide structure is utilized to investigate fibroblast adhesion and migration behaviors [102]. (C) Enzyme responsive switchable surface for controlling cell adhesive ligand display. Bulky Fmoc groups sterically prevent cells from recognizing RGD ligands coupled to the PEG monolayer. After enzymatic degradation and release of Fmoc groups, cells are able to recognize and bind to the exposed RGD moieties [116]. (D) Cyto-mechanoresponsive switchable surface for controlling cell adhesive ligand display that allows for cell interaction with RGD ligands that are exposed from inside a polyelectrolyte multilayer under applied stretching. Fibroblasts dramatically adhered and proliferated on this surface in its stretched state [128].
This type of substrate was also utilized for investigating cell migration in a spatiotemporally-defined manner [87]. In this system, the cell adhesion area was defined by using photochemical reaction-driven surface hydrophilization, followed by fibronectin adsorption. At non-irradiated regions, pluronic layers were physically adsorbed to the hydrophobic surface. Thus, 2D spatial control over cell-adhesive (fibronectin layer) and non-adhesive (pluronic layer) domains was achieved with selective illumination. To create the single-cell migration assay, narrow (5 μm) and wide (25 μm) cell-adhesive paths were optically patterned adjacent to small (25 μm x 25 μm) adhesive squares onto which a single NIH 3T3 fibroblast was seeded. Cell behavior was then monitored over the following 6 hours, and it was clearly revealed that the nature of the formed filopodia was determined by the size and shape of the adhesive path. In addition, switchable substrates can be used for the analysis of not only single-cell migration, but also collective migration. Rolli et al. reported on the collective migration of Madin-Darby canine kidney (MDCK) cells from precisely-controlled adhesion geometries by combining photoswitchable adhesive substrates with photomasks [88]. After seeding cells onto initially-defined areas, the entire substrate was then flood-exposed to UV light, thereby allowing for uninhibited cell movement. Interestingly, it was shown that not only did leader cell appearance and overall migration symmetry depend on the initial size of the cell clusters, but geometry also played a significant role in this process. Recently, Shimizu et al. developed a photoswitchable nanopatterned substrate with tunable cell-ECM ligand interactions [89]. This nanopatterned substrate, onto which gold nanoparticles were arrayed in a regular manner with defined nanometer spacing, was prepared by a block copolymer micelle nanolithography technique. Photocleavable PEG as a shielding layer and cyclic RGD peptides as ECM ligands were immobilized on the surface of the gold nanoparticles. It was observed that there was a loss of collective migration characteristics in HeLa cells cultured on the functionalized nanoparticles, indicating that the degree of interaction between the cells and ECM ligands may play a role in cellular collectivity. The series of studies presented here suggest that the utilization of photocleavable groups offer excellent control over ‘on-off’ cell adhesion and have great potential in the spatiotemporal regulation of cellular behaviors, such as single cell- or collective migration.
Photoisomerization, on the other hand, is a molecular behavior in which structural changes between isomers are caused by photoexcitation. Photoisomerization reactions are used in various applications including actuators [90, 91], variable surface wettability [92] and motional regulation of macroscopic objects such as water [93]. Applying photoisomerization reactions to surface construction creates a powerful cell culture platform. Spiropyrans are well-known photochromic compounds that undergo reversible structural isomerization between a colorless, nonionic structure (closed-ring form) and a colored, zwitterionic structure (open-ring form) via UV irradiation and vice versa via visible light irradiation or heat. This photoisomerization process has been utilized to produce switchable surfaces with photo-tunable cell adhesion. For example, Edashiro et al. synthesized temperature-responsive PNIPAAm by using photo-responsive spiropyran chromophores as side chains [94]. Regions of the culture substrate subjected to UV irradiation saw a significant increase in Chinese hamster ovary (CHO)-K1 cell attachment, which was suggested to be a result of an attractive interaction between the cell and the zwitterionic spiropyran in the polymer chain. As similarly observed in PNIPAAm homopolymer systems, robust cell detachment occurred in the non-irradiated regions upon exposure to low temperatures and washing. Furthermore, complex micropatterns of cell clusters could be generated using this technique in conjunction with a photomask. In light of this demonstration of the reversibility and local control of cell attachment and detachment, a polymer substrate combined with thermo-responsive properties and photoisomerization may be a powerful tool in the field of cell manipulation technology.
In an alternative approach, it is well-known that azobenzene isomerizes from the trans to cis conformation under UV irradiation (340–380 nm) and vice versa (cis to trans conformation) under visible light irradiation (450–490 nm). Auernheimer et al. reported on the possibility of switching cell adhesion properties by changing the distance and orientation of RGD peptide moieties that are presented on the culture substrate [95]. In their study, a set of cyclic RGD peptides containing a photoswitchable 4-[(4-aminiphenyl)azo]benzocarbonyl unit was synthesized, and poly(methyl methacrylate) (PMMA) surfaces were subsequently modified with these peptides. It was found that when these peptides were in their trans conformation, they enhanced mouse osteoblast (MC3T3 E1) adhesion on PMMA disks, whereas adhesion efficiency decreased with UV irradiation at 366 nm and the subsequent conformational change of the peptide. In a similar approach, Liu et al. utilized a self-assembled monolayer (SAM) comprised of alkanethiols containing an azobenzene unit terminated with RGD mixed with other alkanethiols that terminated in a hexa(ethyleneglycol) non-adhesive functional group [96]. The azobenzene component could be photochemically transformed between the trans and cis configurations to either present or shield the RGD peptide, respectively, thereby modulating biospecific cell adhesion to the SAM. Following this principle, the mixed SAM surface allowed for reversible control of NIH 3T3 fibroblast adhesion through changes in azobenzene configurations in response to photoirradiation. Broadly, these techniques make it possible to dynamically modulate patterns of cell adhesion and, in doing so, further investigate various cell behaviors, such as adhesion, proliferation, and migration.
4.3. Electrochemical-responsive polymers
As an alternative to photochemical reactions, electrochemical trigger systems that include the assembly of SAMs and polymers on electrodes have also been used as substrates for a dynamic cell culture platform. In an electrochemically-tunable SAM system, pioneering work was conducted wherein electroactive substrates directly enabled ligand switch activities in response to electrical potential for the dynamic regulation of adhesion and migration [97–100]. They designed a SAM that incorporates an O-silyl hydroquinone moiety in order to present a cell-adhesive RGD peptide [101]. This dynamic substrate involving circular patterns 220 μm in diameter of non-electroactive SAMs allowed for the selective release of Swiss 3T3 fibroblasts through the application of electrical potentials. Because the O-silyl hydroquinone ether is electroactive, electrochemical oxidation via an applied electrical potential to the substrate causes the selective release of the RGD peptide. The RGD peptide was subsequently reattached to the oxidized quinone through a Diels-Alder reaction that enables switching back to a cell-adhesive substrate. Later, Lamb and Yousaf developed a novel strategy for switching an immobilized molecular structure on the substrate surface based on the two sequential, orthogonal click reactions of Huisgen cycloaddition and benzoquinone-oxime chemistry (Figure 5B) [102]. In this system, the oxime linkage played the role of a redox-active site, switching the activity of the RGD peptide presented to the cell from cyclic to linear by changing the peptide conformation in situ. Thus, the switchable surface could modulate various cell behaviors dynamically, including cell adhesion morphology and migration rate. They also prepared electroactive gradient surfaces for spatial and temporal control of mammalian cell behavior [103]. This system combined electroactive and photochemical strategies with chemo-selectively immobilized RGDS ligands in patterns and gradients. They used an inert SAM surface presenting photo-protected and electroactive molecules that could be activated in the presence of cells for directed tissue migration. They successfully demonstrated the controlled morphing of cells from one pattern to another on this dynamic gradient substrate. This electrochemically switchable surface could be used as a model substrate to monitor a range of cellular behaviors, including adhesion, migration and polarization, as well as serve as a tumor invasion model system.
In another electrochemical control approach, Kakegawa et al. developed an electrochemically detachable, biocompatible, and oligopeptide-modified cell culture surface [104]. They used gold-thiolate bonds to form a SAM on gold electrodes, as this type of bond can be reductively cleaved through the application of a negative electrical potential. Cell-repulsive CGGGKEKEKEK and cell-adhesive CGGGKEKEKEKGRGDSP oligopeptides were designed with a terminal cysteine residue to mediate binding to a gold surface. The SAMs spontaneously formed via gold-thiolate bonds and intermolecular, electrostatic interactions. Swiss 3T3 fibroblasts that had adhered to the surface were completely and rapidly (within 2 min) detached due to desorption of the oligopeptide layer following the application of a negative electrical potential. The results of this electrochemical control system illustrate their potential usefulness for tissue-engineering and regenerative medicine applications.
With regard to electrochemically-tunable polymer systems, Wong et al. utilized an electrically conductive polymer known as polypyrrole as a dynamic cell culture substrate which could reversibly change its surface properties (e.g., charge density and wettability) in response to an applied electrical potential [105]. In vitro studies showed that aortic endothelial cells adhered and spread across fibronectin-coated polypyrrole in an oxidized state. However, cell extension and DNA synthesis were inhibited when an electrical potential was applied. This pioneering research showed that electroconductive polymers are not only capable of operating in biological conditions, but can also provide a noninvasive means to dynamically control the shape and function of adhered cells. Other types of conductive polymers, such as thiophene, have also been used to electrochemically regulate cell adhesion and migration. Thin electrode substrates of poly(3,4-ethylenedioxythiophene) (PEDOT) doped with Tosylate were fabricated through vapor-phase polymerization for the culture of neural stem cells (c17.2) [106]. In this system, the binding affinity of human serum albumin (HSA) varied by the oxidized or reduced state of the substrates, ultimately regulating the adhesion property and density of c17.2 cells. This approach may be useful for the electrical control of stem cell function, since both cell adhesion events and cell density are potent parameters for regulating differentiation. Interestingly, Wan et al. demonstrated the electrical control of cellular patterning using PEDOT doped with Tosylate. They successfully created cellular density gradients of both mouse fibroblasts (3T3-L1) and human breast cancer cells (MDA-MB-21) on a surface that carried a redox gradient [107]. Moreover, they found that it was possible to use this substrate to enhance migration speed and guide the migration direction of cultured BAECs [108]. The active regulation of cell adhesion due to the gradient of adsorbed fibronectin depended upon the oxidized or reduced state of the surface, which could be easily manipulated by applying an electrical potential. This ability to control cell adhesion and migration behavior using external electrical stimuli would be valuable in dynamic cell manipulation. However, two later studies showed that the cell adhesion gradients and migration were actively determined by the redox surface state before cell seeding. Therefore, it is difficult to conclude that the cell culture platform is truly dynamic. Although the number of examples of dynamic, electrochemically-based cell culture platforms are noticeably fewer than that of the SAM-based approaches described above, this remains a crucial area of research for the development of additional in vivo applications, including biocompatible, biodegradable, and implantable soft electronic devices [109–111].
4.4. Biomolecular- or mechano-responsive polymers
Although the temperature-, photo-, and electrochemical-responsive polymer-based approaches discussed so far are useful for inducing programmable changes in cultured cells, special equipment and devices are often required to apply the necessary external stimuli to the system. Thus, the utility of these systems for in vivo applications can sometimes be limited. On the other hand, there are many biomolecules, including enzymes, that are involved in a variety of metabolic and biological processes in the body. Since biomolecule-based reactions possess high selectivity and efficiency under physiological conditions, they offer great potential in providing alternative external stimuli to induce programmable change. One conventional approach involves enzymatically- or proteolytically-cleavable substrates. Yeo et al. developed SAMs that transduce enzymatic activities into electrical signals [112, 113]. This type of system is noteworthy because the enzymatic reaction of cutinase, a serine esterase, allowed for efficient removal of an acyl group and conversion of the substrate into a redox-active surface. Ultimately, it provided a way to interface cell behavior with electrochemical approaches like those described above. However, the enzymatic degradation that occurred in this system could not be utilized directly for the dynamic regulation of cell behavior. Todd et al. proposed a more straightforward approach to use enzymatic degradation for dynamic regulation [114]. They utilized poly(ethylene glycol) acrylamide (PEGA) gels as a matrix due to its bioinert nature and ability to easily introduce amino groups [115]. Then, in order to build the desired peptide chains, they applied solid phase peptide synthesis in a stepwise manner to couple Fmoc-protected amino acids. Modification of the integrin-binding peptide RGD rendered the system switchable between a non-adhesive state and an adhesive state through the enzymatic cleaving of the Fmoc group by various enzymes, including proteinase, chymotrypsin, and thermolysin. The switchable capacity of this system allowed for comprehensive cell attachment regulation. Furthermore, they found that grafted polymer monolayers or a brush structure provided a well-defined platform compared to gel systems in terms of their surface properties, such as the density of functional groups. They applied this method using a 2D PEG monolayer system, as opposed to a 3D gel substrate (Figure 5C) [116]. The enzyme-responsive PEG monolayer was formed by coupling the hydroxyl group on PEG chains with epoxy silanes, followed by a stepwise solid phase synthesis. Exposure of the functionalized PEG monolayer to elastase resulted in the activation of cell-binding RGD ligands for primary human osteoblast attachment to the substrate. However, triggering molecules that induce surface property changes are not limited to enzymes alone, and in fact, include other small compounds. For example, boronic acids and their ester derivatives are a class of important molecular-responsive systems that can reversibly interact with diols at a pH value higher than their pKa [117]. This unique interaction is utilized in many biological fields, such as drug delivery [118–120] and the selective capture and release of biomolecules [121]. More recently, Pan et al. demonstrated reversible control of cell adhesion via multivalent, phenylboronic acid/cis-diol polymeric complex formation [122]. This system was composed of two different polymers: phenylboronic acid (PBA) grafted poly(hydroxyethyl methacrylate) (PHEMA-graft-PBA) immobilized on a glass slide and RGD-modified poly(3-gluconamidopropyl methacrylamide) (RGD-PGAPMA). Bioactive RGD peptides were dynamically introduced to the substrate via reversible, multi-covalent interactions between the PBA/cis-diol of the two different polymers. This unique approach was the first to demonstrate reversible control of cell adhesion through dynamic covalent chemistry [123, 124], and it was the first example of a dynamic ‘on-off’ display on a bioactive surface with small, natural biomolecules.
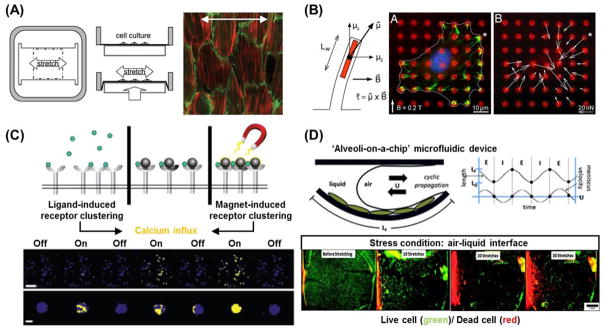
Macroscopic mechanical stimuli have recently attracted much attention because they enable molecular or material manipulation at the nanoscopic and mesoscopic to the macroscopic level [125–127]. In these systems, macroscopic mechanical forces can be utilized to trigger the switching or modulation of substrate surface chemistries and biochemical signal presentation. In a unique approach, Davila et al. developed a mechano-responsive surface that allowed for cell adhesion in response to mechanical stretching [128]. Their grafted surface was comprised of poly(acrylic acid) (PAAc)-bearing RGD peptides deposited under a cell-repellent PAAc-phosphorylcholine (PC)/poly(allylamine hydrochloride) (PAH) multilayer on a silicone substrate. The cell-adhesive RGD groups could be exposed by stretching the film, and the number of exposed ligands from the multilayer could be regulated by stretching the ratio of the substrate (Figure 5D). Furthermore, the density of the adhesion ligands could be gradually changed during cell culture by stretching alone, and there was a clear threshold of stretching ratio (between 1.3 and 1.5), defined as the ratio of length of the substrate after and before stretching, for achieving high cell adherence by mechanically-mediated RGD display. This system shows that macroscopic mechanical stimuli can manipulate nanoscale events of the ligand-receptor interactions between the cells and surface, which are macroscopically observed as the ‘on-off’ regulation of cell adhesion.
While recent examples of biomolecular-responsive approaches for inducing dynamic changes in physicochemical and biochemical properties are relatively few in number, there are wide variety of biomolecules in living organisms that play extremely important roles in cellular function regulation and homeostatic maintenance. Additionally, the expression, distribution, and concentration of these biomolecules varies depending on not only tissue type, but also on disease state and developmental progression. Therefore, the design of dynamic polymers that can spatiotemporally present tissue-specific biomolecular information would benefit the development of next generation dynamic cell culture platforms. Potentially, this new class of culture platforms would be best suited for researchers in the fields of tissue engineering, regenerative medicine, and developmental biology. On the other hand, current mechano-responsive approaches require relatively large forces or macroscopic mechanical actions to induce material property changes, and this limits the applicability of such techniques. However, the overall concept of mechanical-responsive systems and the utilization of mechanical force are crucial, since many physical forces including shear, tension, and compression are generated in the human body. Therefore, the molecular design of dynamic cell culture platforms will continue to face the challenge of achieving more effective, complex control over cell adhesion properties as their development moves forward.
5. Dynamic Mechano-Structural Cues
5.1. Dynamic biomaterials with variable elasticity
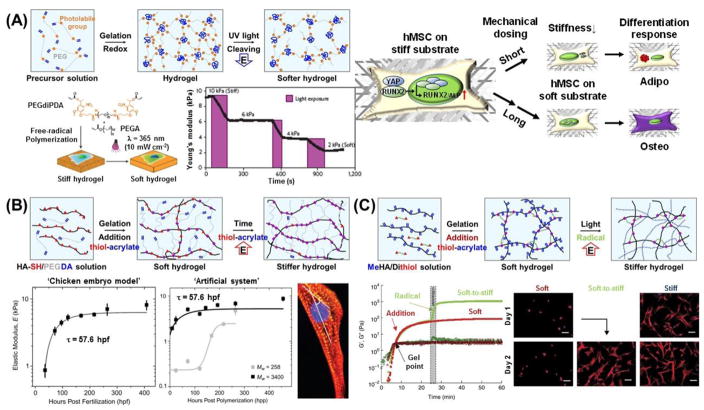
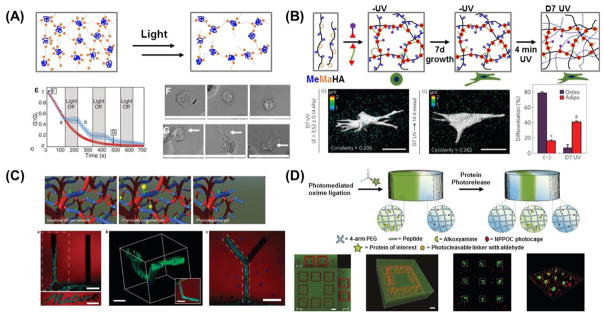
Recent reports have revealed that many types of cells have the capacity to sense and react to mechanical stiffness [10]. For example, stiff gels promote the spreading and scattering of adherent cells, while soft gels promote soft tissue differentiation and tissue-like cell-cell associations [129]. In addition, adherent cells migrate preferentially towards stiffer regions [130–132]. While many studies have shown that the stiffness of synthetic substrates can influence cell fate, research efforts have mostly centered on static effects. However, newer efforts are focused on designing dynamic cell culture substrates or matrices with tunable properties. To allow for the dynamic control of substrate stiffness, Frey and Wang developed a modulatable hydrogel with UV-mediated control of rigidity [133]. The gel was comprised of polyacrylamide (PA) crosslinked with 4-bromomethyl-3-nitrobenzoic acid (BNBA), which contains a nitrobenzyl group that is UV-cleavable. These gels could be softened from 7.2 kPa to 5.5 kPa by UV irradiation, and the spreading of NIH 3T3 fibroblasts was inhibited on softer gels. Interestingly, it was also found that localized softening of the substratum underneath the posterior end of polarized cells caused no apparent effect, which contrasts with the pronounced retraction seen when gel underneath the anterior end was softened. These results potentially shed some light on the asymmetrical sensing capability of many cell types, where rigidity sensing is largely achieved at the frontal region of polarized cells, while mechanosensing is localized to the anterior region. Yang et al. recently investigated whether or not stem cell fate is influenced by culture history – that is, past mechanical memory – using phototunable PEG hydrogels with initially stiff (~10kPa), then soft (~2kPa), rigidity [134]. Activation of the Yes-associated protein (YAP), transcriptional coactivator with the PDZ-binding domain (TAZ) and pre-osteogenic transcription factor RUNX2 in human mesenchymal stem cells (hMSCs) was found to depend on the duration of culture time on substrates of a particular stiffness (Figure 6A). After softening for 10 days, YAP and RUNX2 remained in the nucleus of those cells that had been conditioned on initially stiff, but photodegradable hydrogels for 10 additional days [135]. More interestingly, the mechanical pre-exposure of hMSCs to soft substrates did not prevent the nuclear translocation of YAP or RUNX2 when plated on stiff hydrogels, suggesting the presence of a mechanism for remembering past exposure to stiff, but not soft, environments. These results demonstrate a greater persistence of YAP and RUNX2 activation in the presence of mechanical memory.
Figure 6.
Dynamic hydrogel platforms with tunable elasticity. (A) Photodegradable PEG-based hydrogel in which light irradiation at 365 nm of wavelength cleaves crosslinks and leads to a corresponding decrease in elastic modulus [134]. Mechanical memory in hMSC is revealed by using this substrate. Depending on the previous culture time (mechanical dosing) on stiff substrates, YAP/RUNX2 in the hMSCs on the soft substrates either translocate to the cytoplasm (short duration) or persist in the nucleus (long duration), which determines the differentiation outcome in response to mixed osteogenic/adipogenic media, as indicated by Oil-Red-O-stained lipid droplets (red) and ALP-stained cells (purple) [135]. (B) Hyaluronic acid-based stiffening hydrogel whose elasticity can be increased over time using a slow thiol-ene reaction. This temporal variation in elasticity is similar to that seen in chick myocardial development (left plot). In this example, the kinetics and ultimate moduli can be controlled by the molecular weight of the crosslinker (right plot) [144]. (C) Hyaluronic acid-based hydrogel stiffening through two-step crosslinking of addition reaction and radical polymerization. Rheology profiles (G′, solid and G″, open) showing hydrogel formation via addition reaction only (soft, red) and stiffened hydrogel, addition reaction followed by radical polymerization (soft to stiff, green). Fluorescent images of hMSCs cultured on MeHA substrates for 1 and 2 days with indicated substrate stiffnesses (soft, 3 kPa; stiff, 30 kPa; soft-to-stiff, 3–30 kPa). In situ stiffening of soft gels was performed at day 1 [147].
Emerging concepts are focused on hydrogels that respond to specific biological stimuli, or in other words, molecular-responsive systems. Various types of molecular-responsive hydrogels have been developed as soft, dynamic materials [136–138]. To create a dynamic cell culture platform in this regard, Jiang et al. investigated the effects of dynamic stiffness in the microenvironment on neuronal responses. They used DNA-crosslinked PA hydrogels where DNA strands covalently attached to polymer chains, forming zips [139, 140]. Softening of these substrates was achieved when the complement strand was introduced to the hydrogel to displace the crosslinker DNA out of the gel network. As the substrates became softer, primary dendrite numbers decreased and axonal length increased markedly. These results indicate that neurons are capable of detecting alterations in the mechanical stiffness of their local microenvironment and respond to these alterations, as manifested by changes in neurite outgrowth. These substrates offer a unique opportunity for neural tissue engineering applications where promoting axonal regeneration and growth is desired.
In addition to dynamically-softening materials, current efforts have also targeted dynamically-stiffening materials, since recent studies suggest that changes in tissue stiffness are related to specific disease characteristics. For example, liver fibrosis as a result of chronic inflammatory reactions leads to the stiffening of liver tissue and may result in cirrhosis, portal hypertension, and hepatocellular carcinoma [46, 47, 141]. Recently, the effects of fibrosis progression on cellular behavior has been demonstrated by using dynamically-stiffening substrates [142]. The stiffening from 1.75 kPa (corresponding to the stiffness of healthy liver) to 33.0 kPa (corresponding to that of fibrotic liver) of methacrylated hyaluronic acid (HA)-based hydrogels in response to visible-light irradiation resulted in myofibroblast activation, which is a common feature of pathological fibrosis. They also revealed that the timing of substrate stiffening strongly influenced cell fate and delayed stiffening further promoted myofibroblast activation. In a different study using photodegradable PEG-based hydrogels, results showed that myofibroblasts could be deactivated by substrate softening [143]. These results strongly suggest that dynamically tunable cell culture platforms are of crucial importance for not only understanding the molecular mechanisms behind stiffness-mediated cellular processes, but also for potentially developing material-based treatment approaches for tissue fibrosis.
These substrate stiffening systems have also been applied to explore many other cellular functions in response to dynamic shifts in tissue moduli. For embryonic chicken hearts, maturation from mesoderm to adult myocardium results in a 9-fold stiffening that originates, in part, from a change in collagen expression and localization (Figure 6B). To mimic this temporal stiffness change in vitro, Young and Engler synthesized thiolated-hyaluronic acid (HA) hydrogels crosslinked with poly(ethylene glycol) diacrylate [144]. It was found that the HA hydrogels stiffened from 1.9 to 8.2 kPa over several days, with a time constant of 69.6 hours post-polymerization due to a Michael-type addition reaction. When pre-cardiac cells were cultured on collagen-coated HA hydrogels for a period of 2 weeks, they exhibited a 3-fold increase in mature cardiac-specific markers and formed up to 60% more maturing muscle fibers compared to that of their growth on compliant, but static, polyacrylamide hydrogels. Jiang et al. also utilized the aforementioned DNA-crosslinked PA hydrogel system to investigate the effects of dynamic stiffening on fibroblast growth [145]. Two types of fibroblasts, L929 and GFP, were subjected to alterations in substrate rigidity. Unlike the substrate softening system based on the exchange reaction of the DNA chain [146], substrate stiffness was increased by incorporating DNA strands into the DNA gel network, resulting in changes in crosslinking density. The range of mechanical stiffness achieved in the resulting gels was 5.9–22.9 kPa. Both cell types shared specific responses in common, but their projection areas and polarities responded differently. Guvendiren and Burdick developed a stiffening hydrogel system that provides fast dynamic changes, long-term stability, and structural uniformity [147]. By using a model system that permitted osteoblast and adipocyte differentiation in a bipotential media, they investigated the short-term (minutes-to-hours) and long-term (days-to-weeks) hMSC response to substrate stiffening from soft (~3 kPa) to stiff (~30 kPa) (Figure 6C). First, methacrylated HA was crosslinked with dithiothreitol via Michael-type reactions. The resulting hydrogel could be stiffened within minutes by radical polymerization of the remaining methacrylate groups. Therefore, this two-step crosslinking process enabled the dynamic stiffening of the substrate in a soft-to-stiff stiffness transition. Further, hMSCs were selectively differentiated based on temporal changes in substrate stiffness while in culture. For example, earlier stiffening favored osteogenic differentiation, whereas later stiffening favored adipogenic differentiation.
A dynamic cell culture platform with reversibly tunable stiffness opens up new ways to understand dynamic mechanobiology as well. Yoshikawa et al. reported that thin, micellar hydrogels based on ABA-type triblock copolymers composed of pH-sensitive poly(2-(diisopropylamino)ethyl methacrylate) as A blocks were capable of a 30-fold reversible modulation of Young’s modulus during a modest pH change of 7.0–8.0 [148]. As hydrogel elasticity increased, cultured C2C12 mouse myoblasts became more flattened and increased formation of stress fibers. They also observed sharp, reversible transitions between a rounded and stretched morphology corresponding to the soft (1.4 kPa at pH 7.0) and stiff (40 kPa at pH 8.0) nature of the hydrogel, respectively. This suggests that the reversible tunability of the hydrogel allows for the dynamic modulation of cell-substrate contacts. Interestingly, this result was significantly different from that of a simulation based on a full dynamic model that predicted the rate-dependent hysteresis that would occur when cells are exposed to dynamic changes in substrate stiffness [149]. Recently, Rosales et al. developed a PEG-based hydrogel system in which matrix elasticity could be reversibly modulated with both UV and visible light due to the use of azobenzene incorporation into peptide crosslinkers [150]. Azobenzene is a photoisomer that does not require an additional initiator, and will transition from the trans to cis isomer with UV irradiation. Inducing this transition resulted in a change in gel structure and corresponding decrease in storage modulus, and reversing the transition with visible light then led to a restoration of the gel’s modulus. While some thermal relaxation of azobenzene back to the more energetically stable trans state was found to occur even in the absence of light, this process was quite gradual, with relaxation reported to complete in 18 hours at 37°C after a 5 min exposure to UV.
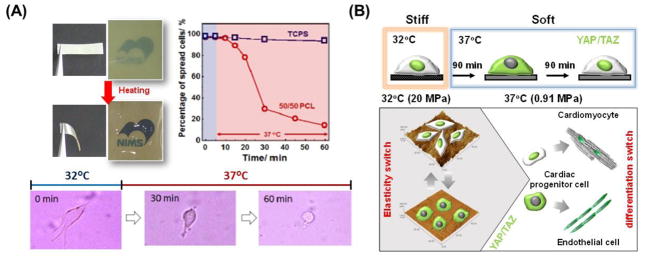
Along with hydrogels, semicrystalline polymers have been used as a temperature-responsive softening substrate. Uto et al. recently developed a semicrystalline, polymer-based cell culture platform with dynamically tunable elasticity using temperature-responsive poly(ε-caprolactone) (PCL) films [151]. While the crosslinked films were relatively stiff (50 MPa) below their melting temperature (Tm), they quickly become soft (1 MPa) above Tm (Figure 7A). Correspondingly, myoblasts cultured on the film became rounded as temperature suddenly increased above Tm. On the other hand, significant changes in cell morphology were not observed in fibroblasts. These results indicate that while cells are capable of sensing dynamic changes in the surrounding environment, their sensitivity differs based on cell type. These temperature-responsive PCL films were also used to investigate how dynamic changes in substrate stiffness regulates the intracellular localization of YAP/TAZ in cardiac progenitor cells [152]. Following a substrate stiffness decrease, a significant decrease in the nuclear expression of YAP/TAZ could be detected after 90 minutes (Figure 7B). However, the percentage of nuclear YAP-positive cells returned to their original values after 180 min. This indicates that from a biological perspective, YAP/TAZ transcriptional coactivators act as intracellular, mechanical rheostats that mediate cellular response to dynamic changes in substrate stiffness. It also illustrates that cells are able to dynamically sense the mechanical properties of their surroundings, and can adapt to new environments in order to retain their original function and fate.
Figure 7.
Semi-crystalline polymer platforms with tunable elasticity. (A) Temperature-induced softening of semi-crystalline crosslinked PCL substrates. More than 70% of primary myoblasts cultured on the film that were initially flattened in morphology became rounded when the elasticity transition was triggered. Representative phase contrast images of myoblasts on crosslinked PCL illustrate the changes observed after heat treatment from 32 to 37°C [151]. (B) YAP/TAZ intracellular localization is dynamically regulated in cardiac progenitor cells as a function of PCL substrate stiffness. While the crosslinked films are relatively stiff (20 MPa) when below the polymer melting temperature (Tm), they suddenly become soft (1 MPa) above the Tm. Following the stiffness switch, a significant decrease in nuclear expression of YAP/TAZ (green) could be detected after 90 minutes. However, the percentage of YAP nuclear positive cells returned to original values after 180 min [152].
In this section, we discussed how dynamic cell culture platforms with tunable elasticities have gradually provided new insights that were difficult to obtain from traditional static systems and have enabled greater manipulation of cellular functions. Although our attention thus far has been directed only at two-dimensional culture systems, these types of platforms can also be extended to three-dimensional systems, which will be discussed later in this review.
5.2. Dynamic biomaterials with switchable topography
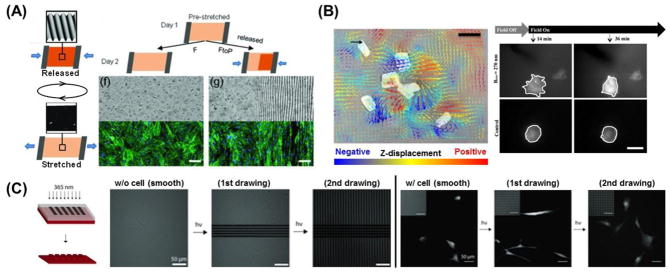
In addition to matrix stiffness, topographical cues also play an integral role in influencing cell fate. Current efforts, however, are centered on static patterns. The scientific community has recently shown an increased interest in developing surfaces with tunable topographies. Lam et al. first developed a reconfigurable, microtopographical system customized for cell culture that consists of reversible wavy microfeatures on poly(dimethylsiloxane) (PDMS) [153]. The microfeatures were created by first oxidizing the PDMS substrate with plasma to create a brittle thin film before applying a compressive force on it. To return the substrate to a flat topography, the compression was simply removed. The average amplitude of the features created using this technique was approximately 670 nm. By reversibly generating and removing the microfeatures, C2C12 cells were able to align, unalign, and realign themselves repeatedly while being cultured on the same substrate. While the topography could be controlled reversibly with this platform, pattern complexity and versatility was relatively simple and limited. Guvendiren and Burdick expanded this idea by designing strain-responsive buckling patterns with more complex structures [154]. By combining ultraviolet/ozone (UVO) exposure to induce the formation of a thin oxide layer on PDMS and a systematic method of masking and substrate stretching, they were able to both spatially control pattern generation and pattern dimension. Additionally, the generated patterns could be reversibly removed, thereby demonstrating for the first time the ability to spatially control reversible cellular alignment. By disrupting the organization of cultured hMSCs, preferential cellular alignment was reversibly repeated for at least 8 cycles depending on the response of surface morphology to the mechanical stretching/releasing of the substrate (Figure 8A). They also presented a biaxial stretching system where cellular orientation angle and order were dynamically controlled. This technology is interesting as it further demonstrates a culture system with a degree of spatially- and dynamically- controlled structural complexity similar to that found in development in vivo.
Figure 8.
Dynamic cell culture platforms with tunable topography. (A) Strain responsive [154] exhibiting reversible surface topography (left). This system is utilized to spatially control of dynamic surface topography by utilizing masks during UVO treatment. Schematic of spatially defined dynamic topography switching on substrates half exposed to UVO (right). Representative bright field images of the substrate surface and fluorescent images of hMSCs cultured on corresponding substrates for 2 days, where substrates were maintained under strain or released to reveal patterned topography after 1 day [154]. (B) Magnetic field-responsive nickel microwire-embedded polyacrylamide gels. (left) Displacement was induced by microwires in a 0.3 T magnetic field. Arrows indicate planar displacements whereas the color scale indicates z-displacements tracked by traction force microscopy. Warm (red) and cool (blue) colors correspond to positive and negative changes in the z-axis relative to the undeformed surface. Scale bar = 20 μm. Black arrow = 5 μm displacement. (right) Cell behavior on the substrate (top) with or (bottom) without magnetic field. White lines have been added to highlight the cell perimeter. Scale bar is 30 μm [155]. (C) Photolabile PEG-based hydrogels (red) with tunable microtopography. Fluorescently labeled, photodegradable PEG-based hydrogels demonstrate the ability to introduce unique topographies spatially and temporally (left). Representative images of hMSC morphology on sequentially-presented dynamic microtopographies. hMSCs were initially seeded onto smooth surfaces, which were in turn patterned using photolithography (365 nm, 10 mW/cm2, 250 s) into an anisotropic (∞:1) channel pattern and then an isotropic (1:1) square pattern. Cells exhibited a more rounded morphology on the smooth pattern, were elongated along the feature major axis on the channels, and returned to a more rounded morphology upon exposure to the isotropic squares topography [156].
Hydrogels can also be applied to the design of dynamic cell culture platforms with tunable surface topography. Kiang et al. demonstrated the effects of dynamic and reversible surface topographies on vascular smooth muscle cell (VSMCs) morphology [155]. They synthesized a soft (E ~ 1kPa) PA hydrogel with magnetic nickel microwires homogeneously distributed across the material’s surface that were around 5 μm in diameter and 20 μm in length. By applying various magnetic fields to the hydrogel, reversible changes in surface roughness ranging from 0.05 to 0.70 mm were induced due to the movement of the microwires inside the gel (Figure 8B). Cultured VSMCs responded to the rapid and dynamic topographical changes; however, oscillating topography over a prolonged period did not produce significant changes in cell morphology. This result implies that there is a difference between an acute versus chronic cellular response to dynamic topographic changes. Alternatively, Kirschner and Anseth demonstrated real-time cell morphology control through the controlled erosion of a photolabile, PEG-based hydrogel system [156]. Photolithographic techniques were employed to generate dynamic topographies featuring variable dimensions (~5 to 40 μm) and aspect ratios of 1:1 to infinity. hMSCs were seeded onto initially smooth surfaces which were then patterned sequentially so that the microtopography underneath the cells were dynamically shifting over time (Figure 8C). During this process, it was found that cell morphology and alignment could be drastically altered to reflect changes in topographical cues. For example, when the presented pattern was comprised of anisotropic channels, there was an overall increase cell aspect ratio and alignment along the pattern. When the pattern was subsequently changed to a series of isotropic squares, the cells returned to a more rounded morphology similar to that seen when they were cultured on smooth substrates. Using this platform and technique, it would be possible to investigate the influence of dynamic changes on cellular anisotropy in hMSCs, and how these changes may affect eventual lineage determination in these cells.
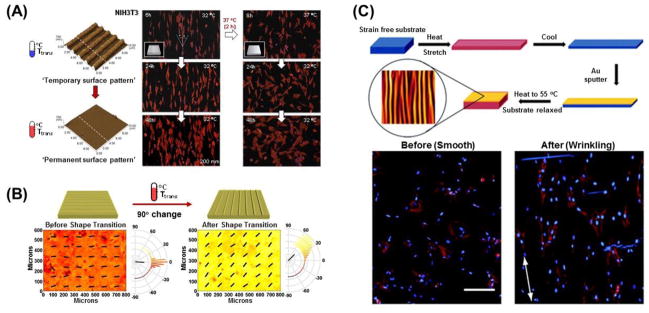
Recently, dynamic regulation of cell morphology has been achieved by utilizing substrates comprised of shape-memory polymers (SMPs), a class of “smart” materials which have the unique ability to recover their original shape upon exposure to specific external stimuli, and Davis et al. were one of the first to demonstrate this [157]. Specifically, they used the commercially available, non-cytotoxic polymer system known as Norland Optical Adhesive 63 (NOA-63), which is polyurethane-based and thiol-ene-crosslinked, as a dynamic cell culture substrate. They successfully embossed parallel microgrooves with an amplitude of 25.6 ± 0.8 μm into the substrate, which were then “erased” by heating the substrate to the glass transition temperature (Tg) of approximately 30–37°C. This transition from a microgrooved to smooth topography significantly changed the alignment and cytoskeletal organization of C3H/10T1/2 mouse embryonic fibroblasts, clearly demonstrating that SMP-based substrates potentially allow for a high degree of control over topographically-induced cellular behavior. To enable subcellular resolution control with SMP-mediated variable surface geometries, Le et al. developed thermally responsive PCL-SMP surfaces to dynamically probe cell-topography interactions [158]. Thermomechanical programming of this substrate was accomplished by casting a methacrylate end-functionalized PCL pre-polymer into a mold for the primary, or permanent, shape, and subsequent photocuring with diethoxyacetophenone. The substrate was then mechanically deformed at 130°C with a mold for the secondary, or temporary, shape before being cooled to −78°C. Afterwards, the substrate could then transition between the secondary and primary surface topographies by exposure to a 40°C environment. In this study, hMSC morphology switched from being highly aligned to stellate-shaped in direct response to a topographical transformation from a 3 μm×5 μm anisotropic groove array to a flat surface.
In a study that sought to dynamically control the alignment of NIH 3T3 fibroblasts, Ebara et al. utilized PCL-SMP substrates with which topographical changes could be induced in response to much narrower temperature variations, specifically between 32°C and 37°C [159]. They successfully demonstrated topography-dependent changes in cell orientation over time with a PCL-SMP substrate capable of a nanotopographical transition from a temporary pattern to an original and permanent pattern with heating. Substrates featured a grooved surface, with grooves 1 μm in width, 300 nm in height and spaced 2 μm apart, that could transition to a flat surface, and it was found that cells lost a significant degree of alignment post-transition (Figure 9A). They then utilized the same polymer to create substrates in which the direction of the grooves would abruptly transition orthogonally to the initial orientation [160]. When the topographical shift was induced, the aligned fibroblasts situated along the temporary pattern direction gradually re-aligned in a perpendicular direction. Eventually, 70% of the cells were aligned in the direction of the permanent pattern approximately 48 hours after shape-memory transition. They have since focused on the interlude between topographic transitions and the corresponding cellular response on shape-memory surfaces [161]. A holographic microscope revealed that the application of heat to PCL-SMP induced a complete transition of the substrate’s temporary surface pattern to the permanent pattern in as little as 30 seconds. However, it took more than 2 hours for cells on a substrate with 500 nm grooves to rotate 90 degrees in orientation, and more than 8 hours for cells on a substrate with 2000 nm grooves to do the same. The difference in alignment behavior can be explained by differences in adhesion strength and the reorganization of cytoskeletal proteins on nano- vs. micropatterns. Given the selective intracellular localization of YAP/TAZ in response to stiffness changes as previously described in cardiac progenitor cells, it was posited that these proteins could also be sensitive to dynamic changes in surface nanotopography [152]. Mengsteab et al. recently reported that these PCL-SMP-based nanotopographic cues are capable of serving as instructive signals that dynamically regulate the structural and contractile properties of neonatal rat ventricular myocytes (NRVMs) into a well-defined anisotropic monolayer [162]. Indeed, the orthogonal transition of anisotropic nanogrooves could be used to directly manipulate the contractile direction of cardiac cell sheets (Figure 9B). Importantly, they found that the contractile direction of NRVM cell sheets was reorganized by the dynamic transition of their underlying nanotopography. This result suggests that not only can cardiac cellular function be dynamically manipulated, but surface topography can influence tissue-level properties. Moreover, the dynamic modulation of rat bone marrow mesenchymal stem cell (rBMSC) differentiation using a dynamic microgroove surface made of a similar PCL SMP polymer has also been reported [163].
Figure 9.
Shape memory polymer platforms with tunable topography. (A) Crosslinked PCL-based surface shape memory platform with erasable topography. Changes in topography of crosslinked PCL substrates as observed by atomic force microscopy (AFM) before and after shape-memory transition induced by a 37°C heat treatment. Shape-memory transition from a temporary nanopattern to the permanent flat surface induced dynamic cell reorientation from being aligned to being randomly organized [159]. (B) Crosslinked PCL-based shape memory platform designed for cardiac cell culture with variable surface nanopattern orientation. Contraction analysis of cultured cardiac cell monolayer reorganization caused by an orthogonal shift in the orientation of nanogrooves. The kurtosis of contraction direction after shape transition decreases, and a 49 degree shift of the contraction angle peak is observed (kurtosis = 2.45, 0.31 before and after, respectively. *p < 0.001, n = 3, paired t-test) [162]. (C) Shape memory assisted dynamic microwrinkle display system. (top) Process for fabricating the wrinkled material. Stretching, gold coating, and relaxing the substrate to form wrinkles due to shape memory effect. (bottom) Cell morphology before (a) and after (b) wrinkling. Phalloidin (cytoskeleton) and DAPI (nuclear) stains were performed on cells seeded (a) at 30 °C for 5 h and (b) at 30 °C for 5 h followed by 37 °C for 24 h. Scale bar is 200 μm. Cells show no orientation prior to wrinkle formation and subsequently align along the wrinkle direction after wrinkle formation is triggered [166].
The surface wrinkling of materials provides a simple and versatile approach to fabricate cell culture substrates with highly-ordered topographies to investigate cell mechanobiology, as discussed above, and shape-memory phenomena have the potential to assist in the formation of these surface wrinkle structures [164, 165]. Yang et al. fabricated a coated SMP bilayer substrate platform that would be capable of forming micron and sub-micron-sized wrinkles under physiological conditions [166]. The substrate was created by first mixing tert-butyl acrylate (tBA) and butyl acrylate (BA) at varying weight percentages to tune the Tg. Triethylene glycol dimethacrylate (TEGDMA) and a photoinitiator (2,2-dimethoxy-2-phenyl acetophenone; DMPA) were then added to the monomer mixture and UV-irradiated to induce crosslinking. A gold coating (33 nm in thickness) was then sputter-coated onto a SMP film that had pre-strained anisotropically (2–23%) and the entire bilayer substrate would then be cooled to allow for thermal recovery and compressive buckling (Figure 9C). The compressive buckling of the gold coating induced the formation of wrinkles ranging from 2–6 μm in wavelength and 200–500 nm in amplitude. The response of human adipose-derived stem cells (hASCs) cultured on static pre-wrinkled static substrates that had been fabricated by varying the uniaxial pre-straining of the SMP substrates, were examined, revealing cell alignment along wrinkle direction. A similar hASC response was observed when cultured on actively wrinkling substrates. One advantage of a Tg-based SMP system is that it can be used to study how dynamic topographical changes elicit transient effects in cells in real-time, and these platforms may serve as a feasible starting point for studying the kinetics of cellular responses to topographical changes.
A cell culture platform with tunable topography may be a potent tool for exploring cellular response to complex mechano-structural microenvironments and, in turn, advancing our understanding of dynamic mechanobiology. Additionally, these systems have been shown to regulate not only cellular alignment but cell function as well, as illustrated in the cases of cardiomyocyte contraction and stem cell differentiation, suggesting that such platforms could also be useful for tissue engineering and regenerative medicine applications.
5.3. Dynamic biomaterials with applicable force loads
External mechanical forces such as shear stress, the extension/contraction of blood vessels and lungs, and the beating of the heart influence functions on both the cellular and tissue level. Therefore, cell culture platforms with a force load application are crucial from a biomimetic standpoint [167]. So far, a wide variety of apparatuses have been developed to achieve the mechanical stimulation of cell and tissue cultures. These can generally be classified under the following types in the context of force loading modalities: longitudinal stretch [168], bending [169], out-of-plane substrate distention [170], in-plane substrate distention [171, 172], fluid shear stress [173, 174], or combinations therein [175, 176]. Undoubtedly, rational material design is necessary to achieve dynamic mechanical stimulation of cells, and requires soft and flexible substrates like rubber, which are superior to stiff, brittle substrates like those in direct or remote actuation. Among numerous materials, polydimethylsiloxane (PDMS) has been widely used in this approach. It offers significant advantages as a cell culture substrate, including a high optical transparency and easy manipulation of stiffness and surface coating. Elastomers that include PDMS are able to deform their dimensions under applied forces. This mechanical deformation then becomes a trigger for the application of force loading against cells on the substrate. The fabrication of this device and its combination with loading apparatuses are essential in achieving a dynamic applicable force platform.
In vivo, endothelial cells (ECs) that cover the luminal surface of blood vessels are exposed to two major mechanical forces: fluidic shear stress, produced by blood circulation, and the periodic stretching and relaxing that occurs as a result of the diameter oscillations produced by blood pulsation [36]. As early reports suggest, a cyclic uniaxial stretch induces perpendicular orientation of the stress fibers, relative to the principal direction of the stretch, in the endothelial cells [177]. To further elucidate the molecular contribution of mechanical stress, Kaunas et al. designed a system that is capable of producing a cyclic uniaxial stretch (Figure 10A) [178]. An elastic silicone rubber membrane secured on a square frame was pushed up by an I-shaped teflon indenter that resulted in the principal stretch forces being applied along the long axis of the indenter. They demonstrated that in bovine aortic ECs, the activity of Rho pathways play a critical role in determining the direction and extent of the uniaxial, stretch-induced stress fiber orientation. In conjunction with other results, these findings show that a cyclic mechanical stimulus not only influences microscopic cytoskeleton structure and macroscopic cell behavior (e.g., cellular alignment), but also contributes to molecular events and signaling [179–181].
Figure 10.
Dynamic platforms for applying forces to cultured cells. (A) Cyclic uniaxial stretching device designed stretch chamber and indenter geometries. The extensions at the corners of the indenter increase the uniformity of the strain field over the indenter, resulting in a virtually uniaxial stretch. Actin stress fibers (red) orient perpendicular to the direction of cyclic stretch (white double arrow) in cultured arterial endothelial cells with normal Rho activity. Cell borders are indicated by β-catenin staining (green) [178]. (B) Magnetic post actuation device using electromagnets. Representative immunofluorescent micrograph of focal adhesions (green), microposts (red), and nucleus (blue) after force actuation. The direction and magnitude of the field are shown. The cell is outlined, and the location of the magnetic post is marked by the asterisk (*). FA proteins recruit to site of external force application [189]. (C) Nanomagnetic manipulation of receptor signal transduction. The biochemical mechanism that stimulates downstream signaling (left) involves the binding of multi-valent ligands (small green hexagons) that induce oligomerization of individual IgE/FceRI receptor complexes. Monovalent ligand-coated magnetic nanobeads (dark grey circles) bind individual IgE/Fc1RI receptor complexes without inducing receptor clustering (center). However, applying a magnetic field that magnetizes the beads and pulls them into tight clusters (right) rapidly switches on receptor oligomerization and calcium signaling. Fluorescence photographs: Low (top; scale bar, 100 μm) and high (bottom; scale bar, 10 μm) magnification views of pseudocoloured microfluorimetric images showing repeated on- and off-switching of calcium signaling in groups of cells (top) and individual cells (bottom) that precisely coincide with activation and deactivation of the magnetic field, respectively [192]. (D) Schematic of a microfluidic “alveoli-on-a-chip” that incorporates cyclic propagation of a meniscus over a flexible, PDMS membrane to recreate combined stresses found in vivo, thereby providing a more physiologic recreation of the stresses occurring in a variety of surface tension diseases. Fluorescence photographs show an impact of mechanical stress type on A549 cell morphology. Low magnification fluorescent micrographs enable visualization of cell death (red stain) and cell detachment on a membrane-wide scale. Cell detachment is visible after as few as 10 cycles of stretch, and significant cell death begins as early as 20 cycles [200].
Mechanical stress exerted at cell-substrate and cell-cell interfacial boundaries is involved in the regulation of a variety of physiological processes. Microfabricated silicone elastomeric posts have been used to study cell adhesion [182–185] and stem cell differentiation [186–188]. As the mechanical properties of the microposts can be manipulated by altering geometric variables such as their diameter and height, various mechanical environments for adhered cells can and have been created by changing these parameters while keeping material composition constant. These systems can be utilized to study cellular response to external forces, as well as forces cells apply to their environment. Sniadecki et al. designed microposts containing magnetic materials which acted not only as a sensor for cellular traction force, but also as a device for exerting forces onto cells (Figure 10B) [189]. They constructed magnetic and nonmagnetic, PDMS-based post arrays with a diameter of 3 μm, length of 10 μm, and center-to-center spacing of 9 μm. Magnetic cobalt nanowires with a diameter of 350 nm and length of 5–7 μm were embedded in the elastomeric posts to endow the posts with magnetic properties. A step force applied by the magnetic micropost array led to an increase in local focal adhesion size at the site of application without increasing focal adhesion sizes at the sites of nearby nonmagnetic posts. This dynamic platform provided remarkable results showing how adaptive mechanical changes within the cells are potentially important to understanding the transduction of external forces into biochemical regulators of cell function. Furthermore, it underscores the need for deeper insight into the interactions between the cell’s external and internal forces, which cannot be obtained from static micropost systems.
In conjunction with microelectromechanical system (MEMS) technology, microdevices containing microelectrode arrays have been developed for such applications as cell culture devices. While this type of system targets electroactive cells such as neurons and myocytes [190, 191], it cannot be used with non-electroactive cells and therefore lacks versatility in this regard. However, magnetic actuation has the potential to be an alternative to electrical actuation. Mannix et al. focused on magnetic properties by developing a strategy to rapidly and reversibly control a wide range of transmembrane signal transduction pathways in a non-invasive manner using superparamagnetic nanoparticles (Figure 10C) [192]. They first prepared 30-nm superparamagnetic nanoparticles modified with monovalent ligands, which included dinitrophenyl (DNP) antigens targeting FcεRI receptors that were located on the IgE expressed on mast (RBL-2H3) cells [193, 194]. The prepared nanoparticles then interacted with the cells by binding to their transmembrane receptors. Application of a magnetic field induced particle aggregation and subsequent receptor oligomerization, which in turn induced a physiological cellular output (i.e., calcium signaling in the mast cells). In this way, external magnetic input, in conjunction with the nanoparticles, acted as a dynamic nanomagnetic cellular switch. By combining surface functionalization with various electrochemical stimuli that have been developed so far, nanoscale magnetic actuation mechanisms and spatiotemporal controllability are broadly relevant to a vast array of cell types and receptor systems. Moreover, nanoscale size scales are crucial because this trait makes it possible to manipulate individual receptors in contrast to the activity of microscale magnetic particles on baseline receptors [195, 196]. The ability to apply controlled forces to the cell membrane can help researchers elucidate signal transduction mechanisms and pathways that are activated by dynamic physical stimuli.
The ‘organ-on-a-chip’ concept encompasses a microfluidic cell culture device created with microchip manufacturing methods that contains continuously perfused chambers inhabited by living cells and arranged to simulate tissue- and organ-level physiology [197–199]. Undoubtedly, organ-on-a-chip technology has a high impact on the development of dynamic cell culture platforms in light of its ability to recapitulate native tissue architectures, interfaces and functionality in the presence of vascular-like perfusion. Relevant studies are useful to the areas of drug screening and regenerative medicine research, as well as mechanobiology. In particular, elastomeric, PDMS-based devices in which cultured cells are mechanically loaded can achieve much greater complexity in recapitulating tissue or organ geometry and function. Ventilator-induced lung injury (VILI) results from a combination of solid mechanical stresses, which occur as alveolar epithelial cells cyclically stretch and relax, and fluid mechanical stresses, which occur as an air-liquid interface propagates over airway and alveolar epithelial cells. It is related to a variety of surface tension-related diseases including acute respiratory distress syndrome (ARDS) and neonatal respiratory distress syndrome (NRDS) [200]. Based on this knowledge, Douville et al. designed a new alveoli-on-a-chip system that provides a physiologic model of the stresses contributing to the occurrence of VILI. Although alveolar models had previously been presented [201–203], this new system showed a cyclic propagation of a meniscus over a flexible and stretchable PDMS membrane to recreate combined stresses within the device (Figure 10D). Cell populations that experienced a combination of solid and fluid mechanical stresses demonstrated significantly increased cell detachment and death compared to those experiencing a solid stretch with minimal or non-existent fluid stresses. This study offers the first in vitro technique to systematically study complex mechanical stimuli like those found inside the body, highlighting the important role that both solid and fluid mechanical stresses play in disease pathophysiology and progression.
As discussed above, dynamic cell culture platforms that feature applicable force load are typically constructed with simple materials such as elastomeric material or nanoparticles. However, its combination with MEMS and functionalization technology allows for much more complex mechanical environments and cellular manipulation. Additionally, various ‘organ-on-a-chip’ models have been established. The simultaneous stimulation of solid and fluid mechanical stresses in these platforms better mimics tissue- or organ-specific environments, and provides greater knowledge of dynamic mechanobiology.
6. Towards Fourth Dimensional Biomaterials
Thus far, we have primarily focused on various two-dimensional (2D) dynamic cell culture platforms where a given substrate has established contact and interacted with the basal surfaces of cells. However, the cellular microenvironments provided by 2D systems are quite different from those in vivo, where cells are surrounded on all sides by other cells and by ECM, and understanding the differences in biological response to these environments is crucial to the design and engineering of novel culture platforms and materials. Bissell and co-workers examined the significant differences in cellular morphology that occur when normal and malignant human breast epithelial cells are cultured in 2D and 3D. In this seminal study, reconstituted basement membrane material derived from Engelbreth-Holm-Awarm (EHS) mouse tumors (also known as Matrigel) was used as the 3D cell culture matrix [204]. Normal breast epithelial cells embedded in Matrigel differentiated into polarized multicellular spherical colonies similar to acini found in vivo. Interestingly, in 3D matrices, it was generally observed that the loss of epithelial cell polarity was associated with tumor progression [205]. The mechanism behind these differences in cell shape determination and structural organization is hypothesized to be associated with how cell surface integrins interact with their surroundings in 2D and 3D [206]. For example, in 3D conditions, α6β4-integrins play an important role in maintaining acinar polarity, while β1-integrin activity stimulates loss of polarity associated with tumor progression [205]. In addition to mediating cell adhesion events, down-stream signaling as a result of specific integrin interactions in 3D environments can lead to an in vivo-like regulation of cellular function and fate. It has also been proposed that epithelial morphogenesis is driven by the pursuit of maintaining apical, lateral, and basal surfaces that in 2D leads to the formation of simple monolayers, but in 3D leads to the formation of luminal structures [207]. Additionally, epithelial cysts exhibit physiologically-relevant responses to growth factors in 3D as evidenced by the induction of tubular growth when exposed to hepatocyte growth factor (HGF).
Other significant differences have been found when comparing 2D and 3D cell culture systems in terms of cell adhesion and migration [208], gene expression [209], morphogenesis [207], and tumorigenesis [206, 210]. With this knowledge, researchers have set out to design a variety of 3D matrices using either biological or synthetic materials, and it has been observed that cells indeed behave more physiologically when cultured in these 3D environments [35, 58, 211–213]. Therefore, applying chemistries and system designs for dynamic cell culture platforms in 3D cellular matrices could be done to enable the regulation and study of dynamic cellular response phenomena under more physiologically-relevant conditions. Consequently, the development of 3D dynamic cell culture platforms that mimic the functions of human tissues and organs, known as in vitro 4D biology, is presently significantly attractive to the field [20].
Kloxin et al. demonstrated that 3D cell-material interactions could be dynamically and externally directed by photodegradable hydrogels [214]. Acrylated nitrobenzyl ether-derived moieties that are susceptible to two-photon photolysis are combined with PEG to create the photolytically degradable hydrogels whose mechanical and biochemical properties could be precisely and spatiotemporally modulated with light. They successfully demonstrated the ability to pattern the hydrogels in both 2D and 3D using masked flood irradiation and two-photon laser scanning microscopy, respectively. Additionally, the morphology of encapsulated of hMSCs could be dynamically regulated through changes in the gel’s crosslinking density due to the photodegradation of polymer networks (Figure 11A). Using this photodegradable hydrogel system, it was shown that the time-dependent presentation of the integrin-specific ligand RGDS significantly enhanced the differentiation of hMSCs into chondrocytes compared to hMSCs cultured with persistent RGDS signaling. Thus, both mechanical and biochemical cues within this hydrogel platform could be dynamically manipulated with photoirradiation, although these cues were only tuned independently of one another in this study. They also further developed a dynamic platform that can manipulate the geometry and connectivity of the 3D cell culture hydrogel using similar chemistry [215]. Stowers et al. recently reported a near-infrared (NIR) light-triggered mechanism to increase or decrease the stiffness of 3D hydrogels in a spatiotemporal manner [216]. Alginate was selected as the base component of this platform due to its ability to crosslink in the presence of divalent cations such as calcium. Thus, temporal modulation of gel stiffness could be achieved through the controlled introduction of calcium or calcium chelators, and in this case, thermo-responsive liposomes containing gold nanorods and calcium were used for this purpose. Upon irradiation with NIR light, the lipid bilayer of the liposomes were heated above its transition temperature and the encapsulated calcium or calcium chelator (diethylenetriaminepentaacetic acid; DTPA) could be released, leading to a corresponding increase or decrease in alginate cross-linking density. NIR light gradients were capable of patterning regions of varying gel stiffnesses in vitro, and using mice, they demonstrated dynamic regulation of fibroblast morphology in vivo through local transdermal irradiation.
Figure 11.
Fourth-dimensional cell biology platforms with three-dimensionally tunable mechanical and biochemical properties. (A) Photodegradable hydrogel for tuning three-dimensional gel mechanical properties. (left) Gel degradation was modulated by either continuous or periodic irradiation with 365 nm light at 10 mW/cm2. hMSCs encapsulated within dense hydrogels exhibit a rounded morphology (right upper), while those seeded within degraded hydrogels exhibit a spreading morphology (right bottom). Scale bar, 50 μm [214]. (B) Sequentially crosslinkable and proteolytic degradable HA-based hydrogels. Schematic of delayed UV exposure following 7 d culture of hMSCs within gels. Representative TFM image of a single hMSC within MeMaHA hydrogels following D7 UV exposure and an additional 14 d mixed-media incubation. Differentiation of hMSCs within D7 UV hydrogels following 14 d mixed-media incubation changed dramatically compared to those not exposed to UV [218]. (C) Cytocompatible click-based PEG hydrogels with biochemical and biophysical properties tunable through orthogonal photoconjugation and photocleavage reactions. A fibrin clot containing 3T3 fibroblasts was encapsulated within the click hydrogel formulation. Chemical channels of RGD, a cell-adhesive fibronectin motif, as well as physical channels of user-defined shape were created radially out of the roughly spherical clot. The combination of having a physical space to spread into, as well as chemical moieties to bind to, was required for collective cell migration [228]. (D) PEG-based hydrogel with the capability for the photoreversible patterning of biochemical cues. (top) Schematic of the photoreversible patterning strategy, in which protein tethering is achieved within the hydrogels through photomediated oxime ligation and subsequently removed on further light exposure. (bottom left) Dual-protein spatial patterning (green- and red-labeled BSA) imaged by fluorescence confocal microscopy. Scale bars, 50 μm. (bottom right) Reversible differentiation of hMSCs to osteoblasts in a spatially-defined manner. For hydrogels containing patterned vitronectin (VTN), osteocalcin-positive staining was confined to VTN-functionalized portions of the material (indicated by dashed rectangles). On patterned removal of the adhesive protein, staining for osteocalcin was observed only in VTN islands. Scale bar, 200 μm [233].
In addition to light-modulated platforms, enzymatically degradable systems for triggering the induction of dynamic environmental changes in 3D have also been developed. Salinas and Anseth reported on the enhancement of chondrogenic differentiation of hMSCs encapsulated in PEG gels through enzyme-triggered release of RGD [217]. In this work, a peptide sequence containing the major cleavage sites for matrix metalloproteinase-13 (MMP-13) found on aggrecan was covalently bound to PEG such that RGD groups would become available as encapsulated hMSCs began the chondrogenesis process. After 21 days of culture, hMSCs in cleavable RGD gels produced 10 times more glycosaminoglycan (GAG) than those seeded in gels with uncleavable RGD. Although no direct comparison was made in this study between cells cultured in this enzymatically degradable PEG platform and those cultured in gels with persistent RGD signaling, prior studies have found overstimulation of hMSCs with RGD to be detrimental to chondrogenesis. Khetan et al. found that hMSC differentiation could also be directed by degradation-mediated cellular traction in HA hydrogels that used enzymatically cleavable crosslinks [218]. HA was functionalized with both methacrylate and maleimide groups and was then formed into gels using Michael-type reactions between the maleimides and thiols on bi-functional, MMP-degradable peptides. Finally, the gel was incubated with a photoinitiator and exposed to UV light to initiate the free-radical photopolymerization of the methacrylates. In gels without UV, hMSCs spread, deformed the surrounding matrix, and primarily underwent osteogenesis. Cells within a UV-irradiated gel, however, underwent adipogenesis (Figure 11B). More interestingly, while delayed crosslinking by UV light did not affect cell morphology inside gel, it significantly increased the level of adipogenesis relative to osteogenesis. Therefore, they demonstrated that regardless of their morphology, hMSCs’ ability to degrade and interact with this type of gel during differentiation can ultimately dictate cellular fate.
Cell migration plays an essential role in a wide spectrum of biological processes, including embryogenesis, tissue generation, wound healing and cancer metastasis [219, 220]. In terms of cell migration behavior, cells from various tissues respond differently based on a diverse array of factors, such as gradients of electrical fields (electrotaxis) [221], light intensity (phototaxis) [222], gravitational potential (geotaxis) [223] and dissolved (chemotaxis) or surface-immobilized (haptotaxis) chemicals [224–226]. In addition to physicochemical or biochemical cues, substrate rigidity (durotaxsis or mechanotaxis) and topography (topotaxis) also influence cell migration [11, 130–132, 227]. These findings have mainly been derived from 2D systems of cellular migration on a substrate surface, whereas 3D regulation of cell migration is a major challenge. However, DeForest and Anseth successfully created a 3D system that allowed for the dynamic spatiotemporal regulation of biochemical and mechano-structural cues that influence cell migration [228, 229]. They designed PEG-based click-functionalized hydrogels that allow for the photocleavage of crosslinks (mechano-structural cues) and photoconjugation of pendant functionalities (biochemical cues) (Figure 11C). The hydrogel polymer backbone was formed by means of a copper-free alkyne-azide reaction. With visible light irradiation, thiol-containing biomolecules could be covalently bonded to vinyl functionalities in the hydrogel using thiol-ene reactions. In the presence of UV light, however, nitrobenzyl ether moieties are cleaved, resulted in the photodegradation of the network. Both photocleavage and photoconjugation reactions could be used to orthogonally, or independently, regulate the mechanical and biochemical properties of the hydrogels. This system allowed for real-time manipulation of cell function within a simplified synthetic microenvironment, and the successful on-demand regulation of 3T3 fibroblast migration and outgrowth behavior within the hydrogel was demonstrated via three-dimensional functionalized channels. Moreover, these studies also illustrate the ability of such click-based chemistries to spatiotemporally define microenvironmental niches that could potentially recapitulate those found in vivo.
Collectively, these studies are representative of the emerging field of 4D cellular biology, and other promising approaches have been utilized to control various cellular functions in a dynamic and spatially-defined manner [230, 231]. However, these systems lack the capability to introduce and subsequently remove biochemical cues, or in other words, to be reversible. Such platforms would allow for the dynamic presentation of signaling biomolecules often found in in vivo cellular environments. DeForest and Anseth first demonstrated that the combination of two orthogonal, biocompatible photoreactions based on the combination of the two shown in Figure 11C enables the reversible spatial presentation of biological cues [228, 232]. Biomolecules were synthesized to contain both a thiol group for photocoupling and o-nitrobenzyl for photocleavage such that the respective light-mediated chemistries could be utilized to attach and remove the biomolecule from the hydrogel network on-demand. They successfully demonstrated spatiotemporal regulation of the reversible presentation of biochemical cues, as well as selective attachment and detachment of NIH 3T3 fibroblasts on the surface of these hydrogels. In addition, DeForest and Tirrell proposed a new strategy in which the presentation of functional proteins could be reversibly patterned within a 3D space by combining three bio-orthogonal chemistries [233]. Specifically, they utilized strain-promoted azide-alkyne cycloaddition, photodeprotection of oxime-ligation groups, and photoscission of o-nitrobenzyl ester moieties for gel crosslinking, protein tethering, and protein removal, respectively. Introduction and subsequent removal of biochemical cues in a spatially-defined manner was first demonstrated with aldehyde-functionalized proteins (Figure 11D). Not only were specific regions of protein introduction achieved, but concentration gradients could also be produced based on a dose-dependent response to light irradiation. Furthermore, since the chemistries used for protein tethering and excision can be done using the same light source, intricate patterns comprised of multiple proteins can be created in 3D space. Interestingly, when vitronectin was selected as a protein to dynamically control the adhesion and subsequent osteogenesis of hMSCs in a PEG-based network, it was found that reversible differentiation in a user-directed and spatially-controlled manner could be accomplished. Therefore, this approach may prove useful in expanding our understanding of how cells respond to transient extracellular biochemical cues, as well serve as an intriguing new scaffold design for tissue engineering and regenerative medicine applications.
7. Conclusion and Future Perspectives
Although our understanding of the role of biochemical, physicochemical, and mechano-structural cues in the cellular microenvironment has constantly improved, much of the findings have been rooted in static biology thus far. Few examples exist of systems in which those cues are dynamically controlled. In this review, we highlighted recent advances in the design of dynamic cell culture platforms where the biochemical, physicochemical, and mechano-structural properties of matrices were modulated over time in a user-defined manner to better recapitulate in vivo cellular environments. In this regard, stimuli-responsive cell culture platforms are becoming one of the most powerful tools available for leading a new generation of dynamic systems capable of responding to external stimuli with temporal and spatial precision. Various stimuli, including temperature, light, magnetic fields, electrical potential, environmental pH, mechanical force and enzymes have been used as triggers to induce dynamic changes in material properties or cell surroundings such as surface physicochemical properties (e.g., surface energy such as hydrophilicity/hydrophobicity), mechanical properties (e.g., elasticity) and topography. With the incorporation of MEMS technology, dynamic cell culture systems that can apply forces against cells have also been developed. Collectively, these platforms address a major biological question of how cell function and fate are influenced by continual changes in cellular niches. The field is rapidly evolving, and studies of cellular interactions within 2D flat biology, 3D complex biology, and 4D real biology have been conducted in order to elucidate cellular behavior from a single-cell to tissue and organ level. Moreover, these dynamic cell culture platforms, which have the ability to incorporate a wide range of biologically-relevant molecules, could potentially provide useful therapeutic benefits due to the fact that, to date, it has been difficult to produce highly-organized constructs with tunable properties in non-synthetic biomaterials.
However, there are many challenges yet to overcome in developing 4D real biology and realizing its clinical applications from a materials standpoint. In particular, the concept of ‘reversibility’ in dynamic changes is a novel idea for recapitulating cellular environments within the human body and is one of the most interesting ways to mimic in vivo activity a synthetic environment. Nevertheless, this type of system is quite simplified compared to those inside our body. Additionally, while we know that physicochemical, biochemical, and mechano-structural cues jointly influence cells to decide their function and fate, there are few examples that showcase the controlled, simultaneous regulation of these cues within a single material. The integration between artificial, dynamic 3D matrices and other functional materials (i.e., a multi-materials-combined system), as well as its combination with MEMS technology, may open up a new way to address 4D real biology. Thus, moving forward, new developments in the independent, dynamic regulation of multiple parameters across tissue formation will be essential. Materials will be designed with spatial heterogeneity and hierarchy to replicate properties in native tissue structures. Since it is an ongoing challenge to control stem cell differentiation, novel strategies for designing new materials or dynamic cell culture platforms will facilitate the development of new classes of biomaterials that have the capacity to contribute to our basic understanding of cell and developmental biology, as well as tissue engineering and regenerative medicine.
Acknowledgments
This work was supported by an American Heart Association (AHA) Scientist Development Grant (13SDG14560076 to DHK), a National Institutes of Health R21 Grant (R21AR064395 to DHK), a Muscular Dystrophy Association Research Grant (MDA255907 to DHK) and International Collaborative R&D Program through KIAT grant funded by the MOTIE (N0000894 to DHK). The authors are grateful to the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (to KU). A subset of the topics discussed in this review have been previously highlighted in the NIMS Monograph, “Smart Biomaterials” by M. Ebara, K. Uto et al. [234].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane SW, Williams DA, Watt FM. Modulating the stem cell niche for tissue regeneration. Nat Biotech. 2014;32:795–803. doi: 10.1038/nbt.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 3.Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13:547–57. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watt FM, Huck WTS. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14:467–73. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 5.Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Mechanical properties of cellularly responsive hydrogels and their experimental Determination. Adv Mater. 2010;22:3484–94. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kshitiz, Kim DH, Beebe DJ, Levchenko A. Micro- and nanoengineering for stem cell biology: the promise with a caution. Trends Biotechnol. 2011;29:399–408. doi: 10.1016/j.tibtech.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levental I, Georges PC, Janmey PA. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 8.Ross AM, Jiang Z, Bastmeyer M, Lahann J. Physical aspects of cell culture substrates: Topography, roughness, and elasticity. Small. 2012;8:336–55. doi: 10.1002/smll.201100934. [DOI] [PubMed] [Google Scholar]
- 9.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Kim D-H, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012;197:351–60. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kshitiz??, Park J, Kim P, Helen W, Engler AJ, Levchenko A, Kim DH. Control of stem cell fate and function by engineering physical microenvironments. Integr Biol. 2012;4:1008–18. doi: 10.1039/c2ib20080e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo ROC, Dalby MJ. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 2011;10:637–44. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 14.McNamara LE, McMurray RJ, Biggs MJP, Kantawong F, Oreffo ROC, Dalby MJ. Nanotopographical control of stem cell differentiation. J Tissue Eng. 2010;1:120623/1–13. doi: 10.4061/2010/120623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingal PCDP, Discher DE. Combining insoluble and soluble factors to steer stem cell fate. Nat Mater. 2014;13:532–7. doi: 10.1038/nmat3997. [DOI] [PubMed] [Google Scholar]
- 16.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotech. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 17.Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim Biophys Acta Gen Subj. 2014;1840:2506–19. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Leinwand LA, Anseth KS. Cardiac valve cells and their microenvironment–insights from in vitro studies. Nat Rev Cardiol. 2014;11:715–27. doi: 10.1038/nrcardio.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibbitt MW, Anseth KS. Dynamic microenvironments: The fourth dimension. Sci Transl Med. 2012;4:160ps24/1–4. doi: 10.1126/scitranslmed.3004804. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi A, Ling QD, Kumar SS, Chang Y, Kao TC, Munusamy MA, Alarfaj AA, Hsu ST, Umezawa A. External stimulus-responsive biomaterials designed for the culture and differentiation of ES, iPS, and adult stem cells. Prog Polym Sci. 2014;39:1585–613. [Google Scholar]
- 22.Kim J, Hayward RC. Mimicking dynamic in vivo environments with stimuli-responsive materials for cell culture. Trends Biotechnol. 2012;30:426–39. doi: 10.1016/j.tibtech.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Aizawa Y, Owen SC, Shoichet MS. Polymers used to influence cell fate in 3D geometry: New trends. Prog Polym Sci. 2012;37:645–58. [Google Scholar]
- 24.Brun-Graeppi AKAS, Richard C, Bessodes M, Scherman D, Merten OW. Thermoresponsive surfaces for cell culture and enzyme-free cell detachment. Prog Polym Sci. 2010;35:1311–24. [Google Scholar]
- 25.Fonseca KB, Granja PL, Barrias CC. Engineering proteolytically-degradable artificial extracellular matrices. Prog Polym Sci. 2014;39:2010–29. [Google Scholar]
- 26.Higuchi A, Ling QD, Kumar SS, Munusamy M, Alarfajj AA, Umezawa A, Wu GJ. Design of polymeric materials for culturing human pluripotent stem cells: Progress toward feeder-free and xeno-free culturing. Prog Polym Sci. 2014;39:1348–74. [Google Scholar]
- 27.Kim BS, Park IK, Hoshiba T, Jiang HL, Choi YJ, Akaike T, Cho CS. Design of artificial extracellular matrices for tissue engineering. Prog Polym Sci. 2011;36:238–68. [Google Scholar]
- 28.Lih E, Oh SH, Joung YK, Lee JH, Han DK. Polymers for cell/tissue anti-adhesion. Prog Polym Sci. 2015;44:28–61. [Google Scholar]
- 29.Liu F, Urban MW. Recent advances and challenges in designing stimuli-responsive polymers. Prog Polym Sci. 2010;35:3–23. [Google Scholar]
- 30.Nandivada H, Ross AM, Lahann J. Stimuli-responsive monolayers for biotechnology. Prog Polym Sci. 2010;35:141–54. [Google Scholar]
- 31.Rodda AE, Meagher L, Nisbet DR, Forsythe JS. Specific control of cell–material interactions: Targeting cell receptors using ligand-functionalized polymer substrates. Prog Polym Sci. 2014;39:1312–47. [Google Scholar]
- 32.Yamato M, Akiyama Y, Kobayashi J, Yang J, Kikuchi A, Okano T. Temperature-responsive cell culture surfaces for regenerative medicine with cell sheet engineering. Prog Polym Sci. 2007;32:1123–33. [Google Scholar]
- 33.Kharkar PM, Kiick KL, Kloxin AM. Designing degradable hydrogels for orthogonal control of cell microenvironments. Chem Soc Rev. 2013;42:7335–72. doi: 10.1039/c3cs60040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehmann MS, Kloxin AM. Tunable and dynamic soft materials for three-dimensional cell culture. Soft Matter. 2013;9:6737–46. doi: 10.1039/c3sm50217a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeForest CA, Anseth KS. Advances in bioactive hydrogels to probe and direct cell fate. Annu Rev Chem Biomol Eng. 2012;3:421–44. doi: 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- 36.Polacheck WJ, Li R, Uzel SGM, Kamm RD. Microfluidic platforms for mechanobiology. Lab Chip. 2013;13:2252–67. doi: 10.1039/c3lc41393d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–33. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3:a005058/1–24. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams C, Quinn KP, Georgakoudi I, Black LD., III Young developmental age cardiac extracellular matrix promotes the expansion of neonatal cardiomyocytes in vitro. Acta Biomater. 2014;10:194–204. doi: 10.1016/j.actbio.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 41.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;4:165–78. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver VM, Fischer AH, Peterson OW, Bissell MJ. The importance of the microenvironment in breast cancer progression: recapitulation of mammary tumorigenesis using a unique human mammary epithelial cell model and a three-dimensional culture assay. Biochem Cell Biol. 1996;74:833–51. doi: 10.1139/o96-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–18. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmeri ML, Nightingale KR. Acoustic radiation force-based elasticity imaging methods. Interface Focus. 2011;1:553–64. doi: 10.1098/rsfs.2011.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin M, Talwalkar JA, Glaser KJ, Manduca A, Grimm RC, Rossman PJ, Fidler JL, Ehman RL. Assessment of hepatic fibrosis with magnetic resonance elastography. Clin Gas Hep. 2007;5:1207–1213. doi: 10.1016/j.cgh.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuart MAC, Huck WTS, Genzer J, Muller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, Winnik F, Zauscher S, Luzinov I, Minko S. Emerging applications of stimuli-responsive polymer materials. Nat Mater. 2010;9:101–13. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 49.Morgan MR, Humphries MJ, Bass MD. Synergistic control of cell adhesion by integrins and syndecans. Nat Rev Mol Cell Biol. 2007;8:957–69. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat Rev Mol Cell Biol. 2011;12:189–97. doi: 10.1038/nrm3068. [DOI] [PubMed] [Google Scholar]
- 51.Berrier AL, Yamada KM. Cell–matrix adhesion. J Cell Physiol. 2007;213:565–73. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 52.Yamada N, Okano T, Sakai H, Karikusa F, Sawasaki Y, Sakurai Y. Thermo-responsive polymeric surfaces; control of attachment and detachment of cultured cells. Makromol Chem Rapid Commun. 1990;11:571–6. [Google Scholar]
- 53.Matsuda N, Shimizu T, Yamato M, Okano T. Tissue engineering based on cell sheet technology. Adv Mater. 2007;19:3089–99. [Google Scholar]
- 54.Kushida A, Yamato M, Konno C, Kikuchi A, Sakurai Y, Okano T. Decrease in culture temperature releases monolayer endothelial cell sheets together with deposited fibronectin matrix from temperature-responsive culture surfaces. J Biomed Mater Res. 1999;45:355–62. doi: 10.1002/(sici)1097-4636(19990615)45:4<355::aid-jbm10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 55.Idota N, Tsukahara T, Sato K, Okano T, Kitamori T. The use of electron beam lithographic graft-polymerization on thermoresponsive polymers for regulating the directionality of cell attachment and detachment. Biomaterials. 2009;30:2095–101. doi: 10.1016/j.biomaterials.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita T, Tanaka Y, Idota N, Sato K, Mawatari K, Kitamori T. Cultivation and recovery of vascular endothelial cells in microchannels of a separable micro-chemical chip. Biomaterials. 2011;32:2459–65. doi: 10.1016/j.biomaterials.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 57.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–24. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 58.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 59.Jiao A, Trosper NE, Yang HS, Kim J, Tsui JH, Frankel SD, Murry CE, Kim DH. Thermoresponsive nanofabricated substratum for the engineering of three-dimensional tissues with layer-by-layer architectural Control. ACS Nano. 2014;8:4430–9. doi: 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kino-oka M, Ngo TX, Nagamori E, Takezawa Y, Miyake Y, Sawa Y, Saito A, Shimizu T, Okano T, Taya M. Evaluation of vertical cell fluidity in a multilayered sheet of skeletal myoblasts. J Biosci Bioeng. 2012;113:128–31. doi: 10.1016/j.jbiosc.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi H, Shimizu T, Nakayama M, Yamato M, Okano T. The use of anisotropic cell sheets to control orientation during the self-organization of 3D muscle tissue. Biomaterials. 2013;34:7372–80. doi: 10.1016/j.biomaterials.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi H, Shimizu T, Nakayama M, Yamato M, Okano T. Anisotropic cellular network formation in engineered muscle tissue through the self-organization of neurons and endothelial cells. Adv Healthc Mater. 2015;4:356–60. doi: 10.1002/adhm.201400297. [DOI] [PubMed] [Google Scholar]
- 63.Aoyagi T, Ebara M, Sakai K, Sakurai Y, Okano T. Novel bifunctional polymer with reactivity and temperature sensitivity. J Biomater Sci Polym Ed. 2000;11:101–10. doi: 10.1163/156856200743526. [DOI] [PubMed] [Google Scholar]
- 64.Ebara M, Yamato M, Hirose M, Aoyagi T, Kikuchi A, Sakai K, Okano T. Copolymerization of 2-carboxyisopropylacrylamide with N-isopropylacrylamide accelerates cell detachment from grafted surfaces by reducing temperature. Biomacromolecules. 2003;4:344–9. doi: 10.1021/bm025692t. [DOI] [PubMed] [Google Scholar]
- 65.Ebara M, Yamato M, Aoyagi T, Kikuchi A, Sakai K, Okano T. Temperature-responsive cell culture surfaces enable “on–off” affinity control between cell integrins and RGDS ligands. Biomacromolecules. 2004;5:505–10. doi: 10.1021/bm0343601. [DOI] [PubMed] [Google Scholar]
- 66.Ebara M, Yamato M, Aoyagi T, Kikuchi A, Sakai K, Okano T. Immobilization of cell-adhesive peptides to temperature-responsive surfaces facilitates both serum-free cell adhesion and noninvasive cell harvest. Tissue Eng. 2004;10:1125–35. doi: 10.1089/ten.2004.10.1125. [DOI] [PubMed] [Google Scholar]
- 67.Ebara M, Yamato M, Aoyagi T, Kikuchi A, Sakai K, Okano T. The effect of extensible PEG tethers on shielding between grafted thermo-responsive polymer chains and integrin– RGD binding. Biomaterials. 2008;29:3650–5. doi: 10.1016/j.biomaterials.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 68.Ebara M, Yamato M, Aoyagi T, Kikuchi A, Sakai K, Okano T. A Novel approach to observing synergy effects of PHSRN on integrin–RGD binding using intelligent surfaces. Adv Mater. 2008;20:3034–8. [Google Scholar]
- 69.Kobayashi J, Hayashi M, Ohno T, Nishi M, Arisaka Y, Matsubara Y, Kakidachi H, Akiyama Y, Yamato M, Horii A, Okano T. Surface design of antibody-immobilized thermoresponsive cell culture dishes for recovering intact cells by low-temperature treatment. J Biomed Mater Res Part A. 2014;102:3883–93. doi: 10.1002/jbm.a.35064. [DOI] [PubMed] [Google Scholar]
- 70.Arisaka Y, Kobayashi J, Yamato M, Akiyama Y, Okano T. Switching of cell growth/detachment on heparin-functionalized thermoresponsive surface for rapid cell sheet fabrication and manipulation. Biomaterials. 2013;34:4214–22. doi: 10.1016/j.biomaterials.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 71.Idota N, Ebara M, Kotsuchibashi Y, Narain R, Aoyagi T. Novel temperature-responsive polymer brushes with carbohydrate residues facilitate selective adhesion and collection of hepatocytes. Sci Technol Adv Mater. 2012;13:064206/1–9. doi: 10.1088/1468-6996/13/6/064206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YJ, Ebara M, Aoyagi T. A Smart nanofiber web that captures and releases Cells. Angew Chem Int Ed. 2012;51:10537–41. doi: 10.1002/anie.201204139. [DOI] [PubMed] [Google Scholar]
- 73.Hyeong Kwon O, Kikuchi A, Yamato M, Okano T. Accelerated cell sheet recovery by co-grafting of PEG with PIPAAm onto porous cell culture membranes. Biomaterials. 2003;24:1223–32. doi: 10.1016/s0142-9612(02)00469-6. [DOI] [PubMed] [Google Scholar]
- 74.Akiyama Y, Kikuchi A, Yamato M, Okano T. Accelerated cell-sheet recovery from a surface successively grafted with polyacrylamide and poly(N-isopropylacrylamide) Acta Biomater. 2014;10:3398–408. doi: 10.1016/j.actbio.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 75.Nagase K, Hatakeyama Y, Shimizu T, Matsuura K, Yamato M, Takeda N, Okano T. Thermoresponsive cationic copolymer brushes for mesenchymal stem cell separation. Biomacromolecules. 2015;16:532–40. doi: 10.1021/bm501591s. [DOI] [PubMed] [Google Scholar]
- 76.Nagase K, Hatakeyama Y, Shimizu T, Matsuura K, Yamato M, Takeda N, Okano T. Hydrophobized thermoresponsive copolymer brushes for cell separation by multistep temperature change. Biomacromolecules. 2013;14:3423–33. doi: 10.1021/bm4006722. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida T, Aoyagi T, Kokufuta E, Okano T. Newly designed hydrogel with both sensitive thermoresponse and biodegradability. J Polym Sci Part A Polym Chem. 2003;41:779–87. [Google Scholar]
- 78.Maeda T, Kanda T, Yonekura Y, Yamamoto K, Aoyagi T. Hydroxylated poly(N-isopropylacrylamide) as functional thermoresponsive materials. Biomacromolecules. 2006;7:545–9. doi: 10.1021/bm050829b. [DOI] [PubMed] [Google Scholar]
- 79.Haraguchi Y, Shimizu T, Sasagawa T, Sekine H, Sakaguchi K, Kikuchi T, Sekine W, Sekiya S, Yamato M, Umezu M, Okano T. Fabrication of functional three-dimensional tissues by stacking cell sheets in vitro. Nat Protoc. 2012;7:850–8. doi: 10.1038/nprot.2012.027. [DOI] [PubMed] [Google Scholar]
- 80.Sakaguchi K, Shimizu T, Horaguchi S, Sekine H, Yamato M, Umezu M, Okano T. In vitro engineering of vascularized tissue surrogates. Sci Rep. 2013;3:1316/1–7. doi: 10.1038/srep01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, Kobayashi E, Umezu M, Okano T. In vitro fabrication of functional three-dimensional tissues with perfusable blood vessels. Nat Commun. 2013;4:1399/1–10. doi: 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang Z, Okano T. Recent development of temperature-responsive surfaces and their application for cell sheet engineering. Regener Biomater. 2014;1:91–102. doi: 10.1093/rb/rbu011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elloumi-Hannachi I, Yamato M, Okano T. Cell sheet engineering: a unique nanotechnology for scaffold-free tissue reconstruction with clinical applications in regenerative medicine. J Intern Med. 2010;267:54–70. doi: 10.1111/j.1365-2796.2009.02185.x. [DOI] [PubMed] [Google Scholar]
- 84.Kahl JD, Greenberg MM. Introducing structural diversity in oligonucleotides via photolabile, convertible C5-substituted nucleotides. J Am Chem Soc. 1999;121:597–604. [Google Scholar]
- 85.Bai X, Li Z, Jockusch S, Turro NJ, Ju J. Photocleavage of a 2-nitrobenzyl linker bridging a fluorophore to the 5′ end of DNA. Proc Natl Acad Sci USA. 2003;100:409–13. doi: 10.1073/pnas.242729099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakanishi J, Kikuchi Y, Takarada T, Nakayama H, Yamaguchi K, Maeda M. Spatiotemporal control of cell adhesion on a self-assembled monolayer having a photocleavable protecting group. Anal Chim Acta. 2006;578:100–4. doi: 10.1016/j.aca.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 87.Nakanishi J, Kikuchi Y, Inoue S, Yamaguchi K, Takarada T, Maeda M. Spatiotemporal control of migration of single cells on a photoactivatable cell microarray. J Am Chem Soc. 2007;129:6694–5. doi: 10.1021/ja070294p. [DOI] [PubMed] [Google Scholar]
- 88.Rolli CG, Nakayama H, Yamaguchi K, Spatz JP, Kemkemer R, Nakanishi J. Switchable adhesive substrates: Revealing geometry dependence in collective cell behavior. Biomaterials. 2012;33:2409–18. doi: 10.1016/j.biomaterials.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 89.Shimizu Y, Boehm H, Yamaguchi K, Spatz JP, Nakanishi J. A Photoactivatable nanopatterned substrate for analyzing collective cell migration with precisely tuned cell-extracellular matrix ligand interactions. PLoS One. 2014;9:e91875/1–10. doi: 10.1371/journal.pone.0091875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu Y, Nakano M, Ikeda T. Photomechanics: Directed bending of a polymer film by light. Nature. 2003;425:145–5. doi: 10.1038/425145a. [DOI] [PubMed] [Google Scholar]
- 91.Hosono N, Kajitani T, Fukushima T, Ito K, Sasaki S, Takata M, Aida T. Large-area three-dimensional molecular ordering of a polymer brush by onestep processing. Science. 2010;330:808–11. doi: 10.1126/science.1195302. [DOI] [PubMed] [Google Scholar]
- 92.Wagner N, Theato P. Light-induced wettability changes on polymer surfaces. Polymer. 2014;55:3436–53. [Google Scholar]
- 93.Ichimura K, Oh SK, Nakagawa M. Light-driven motion of liquids on a photoresponsive surface. Science. 2000;288:1624–6. doi: 10.1126/science.288.5471.1624. [DOI] [PubMed] [Google Scholar]
- 94.Edahiro Ji, Sumaru K, Tada Y, Ohi K, Takagi T, Kameda M, Shinbo T, Kanamori T, Yoshimi Y. In situ control of cell adhesion using photoresponsive culture surface. Biomacromolecules. 2005;6:970–4. doi: 10.1021/bm0493382. [DOI] [PubMed] [Google Scholar]
- 95.Auernheimer J, Dahmen C, Hersel U, Bausch A, Kessler H. Photoswitched cell adhesion on surfaces with RGD peptides. J Am Chem Soc. 2005;127:16107–10. doi: 10.1021/ja053648q. [DOI] [PubMed] [Google Scholar]
- 96.Liu D, Xie Y, Shao H, Jiang X. Using azobenzene-embedded self-assembled monolayers to photochemically control cell adhesion reversibly. Angew Chem Int Ed. 2009;48:4406–8. doi: 10.1002/anie.200901130. [DOI] [PubMed] [Google Scholar]
- 97.Hodneland CD, Mrksich M. Biomolecular surfaces that release ligands under electrochemical control. J Am Chem Soc. 2000;122:4235–6. [Google Scholar]
- 98.Yeo WS, Hodneland CD, Mrksich M. Electroactive monolayer substrates that selectively release adherent cells. ChemBioChem. 2001;2:590–3. doi: 10.1002/1439-7633(20010803)2:7/8<590::AID-CBIC590>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 99.Yousaf MN, Houseman BT, Mrksich M. Using electroactive substrates to pattern the attachment of two different cell populations. Proc Natl Acad Sci USA. 2001;98:5992–6. doi: 10.1073/pnas.101112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yousaf MN, Houseman BT, Mrksich M. Turning on cell migration with electroactive substrates. Angew Chem Int Ed. 2001;40:1093–6. [PubMed] [Google Scholar]
- 101.Yeo WS, Yousaf MN, Mrksich M. Dynamicinterfaces between cells and surfaces: Electroactive substrates that sequentially release and attach Cells. J Am Chem Soc. 2003;125:14994–5. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]
- 102.Lamb BM, Yousaf MN. Redox-switchable surface for controlling peptide structure. J Am Chem Soc. 2011;133:8870–3. doi: 10.1021/ja203198y. [DOI] [PubMed] [Google Scholar]
- 103.Luo W, Yousaf MN. Tissue morphing control on dynamic gradient surfaces. J Am Chem Soc. 2011;133:10780–3. doi: 10.1021/ja204893w. [DOI] [PubMed] [Google Scholar]
- 104.Kakegawa T, Mochizuki N, Sadr N, Suzuki H, Fukuda J. Cell-adhesive and cell-repulsive zwitterionic oligopeptides for micropatterning and rapid electrochemical detachment of cells. Tissue Eng Part A. 2012;19:290–8. doi: 10.1089/ten.tea.2011.0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wong JY, Langer R, Ingber DE. Electrically conducting polymers can noninvasively control the shape and growth of mammalian cells. Proc Natl Acad Sci USA. 1994;91:3201–4. doi: 10.1073/pnas.91.8.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saltó C, Saindon E, Bolin M, Kanciurzewska A, Fahlman M, Jager EWH, Tengvall P, Arenas E, Berggren M. Control of neural stem cell adhesion and density by an electronic polymer surface switch. Langmuir. 2008;24:14133–8. doi: 10.1021/la8028337. [DOI] [PubMed] [Google Scholar]
- 107.Wan AMD, Brooks DJ, Gumus A, Fischbach C, Malliaras GG. Electrical control of cell density gradients on a conducting polymer surface. Chem Commun. 2009:5278–80. doi: 10.1039/b911130a. [DOI] [PubMed] [Google Scholar]
- 108.Gumus A, Califano JP, Wan AMD, Huynh J, Reinhart-King CA, Malliaras GG. Control of cell migration using a conducting polymer device. Soft Matter. 2010;6:5138–42. [Google Scholar]
- 109.Reeder J, Kaltenbrunner M, Ware T, Arreaga-Salas D, Avendano-Bolivar A, Yokota T, Inoue Y, Sekino M, Voit W, Sekitani T, Someya T. Mechanically adaptive organic transistors for implantable electronics. Adv Mater. 2014;26:4967–73. doi: 10.1002/adma.201400420. [DOI] [PubMed] [Google Scholar]
- 110.Xu L, Gutbrod SR, Bonifas AP, Su Y, Sulkin MS, Lu N, Chung HJ, Jang KI, Liu Z, Ying M, Lu C, Webb RC, Kim JS, Laughner JI, Cheng H, Liu Y, Ameen A, Jeong JW, Kim GT, Huang Y, Efimov IR, Rogers JA. 3D multifunctional integumentary membranes for spatiotemporal cardiac measurements and stimulation across the entire epicardium. Nat Commun. 2014;5:3329/1–10. doi: 10.1038/ncomms4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hwang SW, Lee CH, Cheng H, Jeong JW, Kang SK, Kim JH, Shin J, Yang J, Liu Z, Ameer GA, Huang Y, Rogers JA. Biodegradable elastomers and silicon nanomembranes/nanoribbons for stretchable, transient electronics, and biosensors. Nano Lett. 2015;15:2801–8. doi: 10.1021/nl503997m. [DOI] [PubMed] [Google Scholar]
- 112.Yeo WS, Mrksich M. Self-assembled monolayers that transduce enzymatic activities to electrical signals. Angew Chem Int Ed. 2003;42:3121–4. doi: 10.1002/anie.200250862. [DOI] [PubMed] [Google Scholar]
- 113.Nayak S, Yeo WS, Mrksich M. Determination of kinetic parameters for interfacial enzymatic reactions on self-assembled monolayers. Langmuir. 2007;23:5578–83. doi: 10.1021/la062860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Todd SJ, Farrar D, Gough JE, Ulijn RV. Enzyme-triggered cell attachment to hydrogel surfaces. Soft Matter. 2007;3:547–50. doi: 10.1039/b618256a. [DOI] [PubMed] [Google Scholar]
- 115.Zourob M, Gough JE, Ulijn RV. A Micropatterned hydrogel platform for chemical synthesis and biological analysis. Adv Mater. 2006;18:655–9. [Google Scholar]
- 116.Todd SJ, Scurr DJ, Gough JE, Alexander MR, Ulijn RV. Enzyme-activated RGD ligands on functionalized poly(ethylene glycol) monolayers: surface analysis and cellular response. Langmuir. 2009;25:7533–9. doi: 10.1021/la900376h. [DOI] [PubMed] [Google Scholar]
- 117.Bull SD, Davidson MG, van den Elsen JMH, Fossey JS, Jenkins ATA, Jiang YB, Kubo Y, Marken F, Sakurai K, Zhao J, James TD. Exploiting the reversible covalent bonding of boronic acids: Recognition, sensing, and assembly. Acc Chem Res. 2013;46:312–26. doi: 10.1021/ar300130w. [DOI] [PubMed] [Google Scholar]
- 118.Matsumoto A, Yamamoto K, Yoshida R, Kataoka K, Aoyagi T, Miyahara Y. A totally synthetic glucose responsive gel operating in physiological aqueous conditions. Chem Commun. 2010;46:2203–5. doi: 10.1039/b920319b. [DOI] [PubMed] [Google Scholar]
- 119.Deshayes S, Cabral H, Ishii T, Miura Y, Kobayashi S, Yamashita T, Matsumoto A, Miyahara Y, Nishiyama N, Kataoka K. Phenylboronic acid-installed polymeric micelles for targeting sialylated epitopes in solid tumors. J Am Chem Soc. 2013;135:15501–7. doi: 10.1021/ja406406h. [DOI] [PubMed] [Google Scholar]
- 120.Naito M, Ishii T, Matsumoto A, Miyata K, Miyahara Y, Kataoka K. A Phenylboronate-functionalized polyion complex micelle for ATP-triggered release of siRNA. Angew Chem Int Ed. 2012;51:10751–5. doi: 10.1002/anie.201203360. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Kotsuchibashi Y, Uto K, Ebara M, Aoyagi T, Liu Y, Narain R. pH and glucose responsive nanofibers for the reversible capture and release of lectins. Biomater Sci. 2015;3:152–62. doi: 10.1039/c4bm00269e. [DOI] [PubMed] [Google Scholar]
- 122.Pan G, Guo B, Ma Y, Cui W, He F, Li B, Yang H, Shea KJ. Dynamic introduction of cell adhesive factor via reversible multicovalent phenylboronic acid/cis-diol polymeric complexes. J Am Chem Soc. 2014;136:6203–6. doi: 10.1021/ja501664f. [DOI] [PubMed] [Google Scholar]
- 123.Maeda T, Otsuka H, Takahara A. Dynamic covalent polymers: Reorganizable polymers with dynamic covalent bonds. Prog Polym Sci. 2009;34:581–604. [Google Scholar]
- 124.Otsuka H. Reorganization of polymer structures based on dynamic covalent chemistry: polymer reactions by dynamic covalent exchanges of alkoxyamine units. Polym J. 2013;45:879–91. [Google Scholar]
- 125.Davis DA, Hamilton A, Yang J, Cremar LD, Van Gough D, Potisek SL, Ong MT, Braun PV, Martinez TJ, White SR, Moore JS, Sottos NR. Force-induced activation of covalent bonds in mechanoresponsive polymeric materials. Nature. 2009;459:68–72. doi: 10.1038/nature07970. [DOI] [PubMed] [Google Scholar]
- 126.Brantley JN, Wiggins KM, Bielawski CW. Unclicking the click: Mechanically facilitated 1,3-dipolar cycloreversions. Science. 2011;333:1606–9. doi: 10.1126/science.1207934. [DOI] [PubMed] [Google Scholar]
- 127.Ariga K, Mori T, Hill JP. Mechanical control of nanomaterials and nanosystems. Adv Mater. 2012;24:158–76. doi: 10.1002/adma.201102617. [DOI] [PubMed] [Google Scholar]
- 128.Davila J, Chassepot A, Longo J, Boulmedais F, Reisch A, Frisch B, Meyer F, Vogel JC, Mésini PJ, Senger B, Metz-Boutigue M-H, Hemmerlé J, Lavalle P, Schaaf P, Jierry L. Cyto-mechanoresponsive polyelectrolyte multilayer films. J Am Chem Soc. 2012;134:83–6. doi: 10.1021/ja208970b. [DOI] [PubMed] [Google Scholar]
- 129.Reinhart-King CA, Dembo M, Hammer DA. Cell-cell mechanical communication through compliant substrates. Biophys J. 2008;95:6044–51. doi: 10.1529/biophysj.107.127662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lo CM, Wang HB, Dembo M, Wang Yl. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kidoaki S, Matsuda T. Microelastic gradient gelatinous gels to induce cellular mechanotaxis. J Biotechnol. 2008;133:225–30. doi: 10.1016/j.jbiotec.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 132.Kidoaki S, Sakashita H. Rectified cell migration on saw-like micro-elastically patterned hydrogels with asymmetric gradient ratchet teeth. PLoS One. 2013;8:e78067/1–10. doi: 10.1371/journal.pone.0078067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Frey MT, Wang Yl. A photo-modulatable material for probing cellular responses to substrate rigidity. Soft Matter. 2009;5:1918–24. doi: 10.1039/b818104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13:645–52. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eyckmans J, Chen CS. Stem cell differentiation: Sticky mechanical memory. Nat Mater. 2014;13:542–3. doi: 10.1038/nmat3989. [DOI] [PubMed] [Google Scholar]
- 136.Miyata T, Asami N, Uragami T. A reversibly antigen-responsive hydrogel. Nature. 1999;399:766–9. doi: 10.1038/21619. [DOI] [PubMed] [Google Scholar]
- 137.Miyata T, Uragami T, Nakamae K. Biomolecule-sensitive hydrogels. Adv Drug Deliv Rev. 2002;54:79–98. doi: 10.1016/s0169-409x(01)00241-1. [DOI] [PubMed] [Google Scholar]
- 138.Miyata T, Jige M, Nakaminami T, Uragami T. Tumor marker-responsive behavior of gels prepared by biomolecular imprinting. Proc Natl Acad Sci USA. 2006;103:1190–3. doi: 10.1073/pnas.0506786103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin DC, Yurke B, Langrana NA. Inducing reversible stiffness changes in DNA-crosslinked gels. J Mater Res. 2005;20:1456–64. [Google Scholar]
- 140.Lin DC, Yurke B, Langrana NA. Mechanical properties of a reversible, DNA-crosslinked polyacrylamide hydrogel. J Biomech Eng. 2004;126:104–10. doi: 10.1115/1.1645529. [DOI] [PubMed] [Google Scholar]
- 141.Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol. 2007;293:G1147–G54. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 142.Caliari SR, Perepelyuk M, Cosgrove BD, Tsai SJ, Lee GY, Mauck RL, Wells RG, Burdick JA. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci Rep. 2016;6:21387/1–10. doi: 10.1038/srep21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kloxin AM, Benton JA, Anseth KS. In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials. 2010;31:1–8. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Young JL, Engler AJ. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32:1002–9. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiang FX, Yurke B, Schloss RS, Firestein BL, Langrana NA. The relationship between fibroblast growth and the dynamic stiffnesses of a DNA crosslinked hydrogel. Biomaterials. 2010;31:1199–212. doi: 10.1016/j.biomaterials.2009.10.050. [DOI] [PubMed] [Google Scholar]
- 146.Jiang FX, Yurke B, Schloss RS, Firestein BL, Langrana NA. Effect of dynamic stiffness of the substrates on neurite outgrowth by using a DNA-crosslinked hydrogel. Tissue Eng Part A. 2010;16:1873–89. doi: 10.1089/ten.TEA.2009.0574. [DOI] [PubMed] [Google Scholar]
- 147.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792–9. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 148.Yoshikawa HY, Rossetti FF, Kaufmann S, Kaindl T, Madsen J, Engel U, Lewis AL, Armes SP, Tanaka M. Quantitative evaluation of mechanosensing of cells on dynamically tunable hydrogels. J Am Chem Soc. 2011;133:1367–74. doi: 10.1021/ja1060615. [DOI] [PubMed] [Google Scholar]
- 149.Besser A, Schwarz US. Hysteresis in the cell response to time-dependent substrate stiffness. Biophys J. 2010;99:L10–L2. doi: 10.1016/j.bpj.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rosales AM, Mabry KM, Nehls EM, Anseth KS. Photoresponsive elastic properties of azobenzene-containing poly(ethylene-glycol)-based hydrogels. Biomacromolecules. 2015;16:798–806. doi: 10.1021/bm501710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Uto K, Ebara M, Aoyagi T. Temperature-responsive poly(ε-caprolactone) cell culture platform with dynamically tunable nano-roughness and elasticity for control of myoblast morphology. Int J Mol Sci. 2014;15:1511–24. doi: 10.3390/ijms15011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, Nakanishi J, Taniguchi A, Franzese O, Di Nardo P, Goumans MJ, Traversa E, Pint-do-Ó P, Aoyagi T, Forte G. Hippo pathway effectors control cardiac progenitor cell fate by acting as dynamic sensors of substrate mechanics and nanostructure. ACS Nano. 2014;8:2033–47. doi: 10.1021/nn4058984. [DOI] [PubMed] [Google Scholar]
- 153.Lam MT, Clem WC, Takayama S. Reversible on-demand cell alignment using reconfigurable microtopography. Biomaterials. 2008;29:1705–12. doi: 10.1016/j.biomaterials.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guvendiren M, Burdick JA. Stem cell response to spatially and temporally displayed and reversible surface topography. Adv Healthc Mater. 2013;2:155–64. doi: 10.1002/adhm.201200105. [DOI] [PubMed] [Google Scholar]
- 155.Kiang JD, Wen JH, del Álamo JC, Engler AJ. Dynamic and reversible surface topography influences cell morphology. J Biomed Mater Res Part A. 2013;101:2313–21. doi: 10.1002/jbm.a.34543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kirschner CM, Anseth KS. In situ control of cell substrate microtopographies using photolabile hydrogels. Small. 2013;9:578–84. doi: 10.1002/smll.201201841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Davis KA, Burke KA, Mather PT, Henderson JH. Dynamic cell behavior on shape memory polymer substrates. Biomaterials. 2011;32:2285–93. doi: 10.1016/j.biomaterials.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 158.Le DM, Kulangara K, Adler AF, Leong KW, Ashby VS. Dynamic topographical control of mesenchymal stem cells by culture on responsive poly(ε-caprolactone) surfaces. Adv Mater. 2011;23:3278–83. doi: 10.1002/adma.201100821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ebara M, Uto K, Idota N, Hoffman JM, Aoyagi T. Shape-memory surface with dynamically tunable nano-geometry activated by body heat. Adv Mater. 2012;24:273–8. doi: 10.1002/adma.201102181. [DOI] [PubMed] [Google Scholar]
- 160.Ebara M, Uto K, Idota N, Hoffman JM, Aoyagi T. The taming of the cell: shape-memory nanopatterns direct cell orientation. Int J Nanomed. 2014;9(Suppl 1):117–26. doi: 10.2147/IJN.S50677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ebara M, Akimoto M, Uto K, Shiba K, Yoshikawa G, Aoyagi T. Focus on the interlude between topographic transition and cell response on shape-memory surfaces. Polymer. 2014;55:5961–8. [Google Scholar]
- 162.Mengsteab PY, Uto K, Smith AS, Frankel S, Fisher E, Nawas Z, Macadangdang J, Ebara M, Kim DH. Spatiotemporal control of cardiac anisotropy using dynamic nanotopographic cues. Biomaterials. 2016;86:1–10. doi: 10.1016/j.biomaterials.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gong T, Zhao K, Yang G, Li J, Chen H, Chen Y, Zhou S. The control of mesenchymal stem cell differentiation using dynamically tunable surface microgrooves. Adv Healthc Mater. 2014;3:1608–19. doi: 10.1002/adhm.201300692. [DOI] [PubMed] [Google Scholar]
- 164.Fu CC, Grimes A, Long M, Ferri CGL, Rich BD, Ghosh S, Ghosh S, Lee LP, Gopinathan A, Khine M. Tunable nanowrinkles on shape memory polymer sheets. Adv Mater. 2009;21:4472–6. [Google Scholar]
- 165.Xie T, Xiao X, Li J, Wang R. Encoding localized strain history through wrinkle based structural colors. Adv Mater. 2010;22:4390–4. doi: 10.1002/adma.201002825. [DOI] [PubMed] [Google Scholar]
- 166.Yang P, Baker RM, Henderson JH, Mather PT. In vitro wrinkle formation via shape memory dynamically aligns adherent cells. Soft Matter. 2013;9:4705–14. [Google Scholar]
- 167.Brown TD. Techniques for mechanical stimulation of cells in vitro: a review. J Biomech. 2000;33:3–14. doi: 10.1016/s0021-9290(99)00177-3. [DOI] [PubMed] [Google Scholar]
- 168.Leung DYM, Glagov S, Mathews MB. A new in vitro system for studying cell response to mechanical stimulation: Different effects of cyclic stretching and agitation on smooth muscle cell biosynthesis. Exp Cell Res. 1977;109:285–98. doi: 10.1016/0014-4827(77)90008-8. [DOI] [PubMed] [Google Scholar]
- 169.Bottlang M, Simnacher M, Schmitt H, Brand RA, Claes L. A cell strain system for small homogeneous strain applications. Biomed Tech. 1997;42:305–9. doi: 10.1515/bmte.1997.42.11.305. [DOI] [PubMed] [Google Scholar]
- 170.Hasegawa S, Sato S, Saito S, Suzuki Y, Brunette DM. Mechanical stretching increases the number of cultured bone-cells synthesizing DNA and alters their pattern of protein-synthesis. Calcif Tissue Int. 1985;37:431–6. doi: 10.1007/BF02553714. [DOI] [PubMed] [Google Scholar]
- 171.Schaffer JL, Rizen M, L’Italien GJ, Benbrahim A, Megerman J, Gerstenfeld LC, Gray ML. Device for the application of a dynamic biaxially uniform and isotropic strain to a flexible cell culture membrane. J Orthop Res. 1994;12:709–19. doi: 10.1002/jor.1100120514. [DOI] [PubMed] [Google Scholar]
- 172.Hung CT, Williams JL. A method for inducing equi-biaxial and uniform strains in elastomeric membranes used as cell substrates. J Biomech. 1994;27:227–32. doi: 10.1016/0021-9290(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 173.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol Rev. 1995;75:519–60. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Topper JN, Cai J, Qiu Y, Anderson KR, Xu YY, Deeds JD, Feeley R, Gimeno CJ, Woolf EA, Tayber O, Mays GG, Sampson BA, Schoen FJ, Gimbrone MA, Falb D. Vascular MADs: Two novel MAD-related genes selectively inducible by flow in human vascular endothelium. Proc Natl Acad Sci USA. 1997;94:9314–9. doi: 10.1073/pnas.94.17.9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Owan I, Burr DB, Turner CH, Qiu J, Tu Y, Onyia JE, Duncan RL. Mechanotransduction in bone: osteoblasts are more responsive to fluid forces than mechanical strain. Am J Physiol. 1997;273:C810–C5. doi: 10.1152/ajpcell.1997.273.3.C810. [DOI] [PubMed] [Google Scholar]
- 176.Ayajiki K, Kindermann M, Hecker M, Fleming I, Busse R. Intracellular pH and tyrosine phosphorylation but not calcium determine shear stress–induced nitric oxide production in native endothelial cells. Circ Res. 1996;78:750–8. doi: 10.1161/01.res.78.5.750. [DOI] [PubMed] [Google Scholar]
- 177.Dartsch PC, Betz E. Response of cultured endothelial cells to mechanical stimulation. Basic Res Cardiol. 1989;84:268–81. doi: 10.1007/BF01907974. [DOI] [PubMed] [Google Scholar]
- 178.Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc Natl Acad Sci USA. 2005;102:15895–900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Naruse K, Yamada T, Sokabe M. Involvement of SA channels in orienting response of cultured endothelial cells to cyclic stretch. Am J Physiol. 1998;274:H1532–H8. doi: 10.1152/ajpheart.1998.274.5.H1532. [DOI] [PubMed] [Google Scholar]
- 180.Yano Y, Saito Y, Narumiya S, Sumpio BE. Involvement of rho p21 in cyclic strain-inducedtyrosine phosphorylation of focal adhesion kinase (pp125FAK), morphological changes and migration of endothelial cells. Biochem Biophys Res Commun. 1996;224:508–15. doi: 10.1006/bbrc.1996.1057. [DOI] [PubMed] [Google Scholar]
- 181.Birukov KG, Jacobson JR, Flores AA, Ye SQ, Birukova AA, Verin AD, Garcia JGN. Magnitude-dependent regulation of pulmonary endothelial cell barrier function by cyclic stretch. Am J Physiol. 2003;285:L785–L97. doi: 10.1152/ajplung.00336.2002. [DOI] [PubMed] [Google Scholar]
- 182.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 183.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–9. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sochol RD, Higa AT, Janairo RRR, Li S, Lin L. Unidirectional mechanical cellular stimuli via micropost array gradients. Soft Matter. 2011;7:4606–9. [Google Scholar]
- 185.Weng S, Fu J. Synergistic regulation of cell function by matrix rigidity and adhesive pattern. Biomaterials. 2011;32:9584–93. doi: 10.1016/j.biomaterials.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–6. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells. Nat Mater. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ahn EH, Kim Y, Kshitiz, An SS, Afzal J, Lee S, Kwak M, Suh KY, Kim DH, Levchenko A. Spatial control of adult stem cell fate using nanotopographic cues. Biomaterials. 2014;35:2401–10. doi: 10.1016/j.biomaterials.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sniadecki NJ, Anguelouch A, Yang MT, Lamb CM, Liu Z, Kirschner SB, Liu Y, Reich DH, Chen CS. Magnetic microposts as an approach to apply forces to living cells. Proc Natl Acad Sci USA. 2007;104:14553–8. doi: 10.1073/pnas.0611613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fromherz P, Offenhausser A, Vetter T, Weis J. A neuron-silicon junction: a Retzius cell of the leech on an insulated-gate field-effect transistor. Science. 1991;252:1290–3. doi: 10.1126/science.1925540. [DOI] [PubMed] [Google Scholar]
- 191.Young TH, Chen CR. Assessment of GaN chips for culturing cerebellar granule neurons. Biomaterials. 2006;27:3361–7. doi: 10.1016/j.biomaterials.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 192.Mannix RJ, Kumar S, Cassiola F, Montoya-Zavala M, Feinstein E, Prentiss M, Ingber DE. Nanomagnetic actuation of receptor-mediated signal transduction. Nat Nanotechnol. 2008;3:36–40. doi: 10.1038/nnano.2007.418. [DOI] [PubMed] [Google Scholar]
- 193.Lu-Kuo JM, Joyal DM, Austen KF, Katz HR. gp49B1 inhibits IgE-initiated mast cell activation through both immunoreceptor tyrosine-based inhibitory motifs, recruitment ofsrc homology 2 domain-containing phosphatase-1, and suppression of early and late calcium mobilization. J Biol Chem. 1999;274:5791–6. doi: 10.1074/jbc.274.9.5791. [DOI] [PubMed] [Google Scholar]
- 194.Segal DM, Taurog JD, Metzger H. Dimeric immunoglobulin-Eserves as a unit signal for mast-cell degranulation. Proc Natl Acad Sci USA. 1977;74:2993–7. doi: 10.1073/pnas.74.7.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Glogauer M, Ferrier J, McCulloch CA. Magnetic fields applied to collagen-coated ferric oxide beads induce stretch-activated Ca2+ flux in fibroblasts. Am J Physiol. 1995;269:C1093–104. doi: 10.1152/ajpcell.1995.269.5.C1093. [DOI] [PubMed] [Google Scholar]
- 196.Meyer CJ, Alenghat FJ, Rim P, Fong JHJ, Fabry B, Ingber DE. Mechanical control of cyclic AMP signalling and gene transcription through integrins. Nat Cell Biol. 2000;2:666–8. doi: 10.1038/35023621. [DOI] [PubMed] [Google Scholar]
- 197.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–8. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotech. 2014;32:760–72. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 199.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–54. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Douville NJ, Zamankhan P, Tung YC, Li R, Vaughan BL, Tai CF, White J, Christensen PJ, Grotberg JB, Takayama S. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip. 2011;11:609–19. doi: 10.1039/c0lc00251h. [DOI] [PubMed] [Google Scholar]
- 201.Tschumperlin DJ, Margulies SS. Equibiaxial deformation-induced injury of alveolar epithelial cells in vitro. Am J Physiol. 1998;275:L1173–L83. doi: 10.1152/ajplung.1998.275.6.L1173. [DOI] [PubMed] [Google Scholar]
- 202.Tschumperlin DJ, Oswari J, Margulies ASS. Deformation-induced injury of alveolar epithelial cells. Am J Respir Crit Care Med. 2000;162:357–62. doi: 10.1164/ajrccm.162.2.9807003. [DOI] [PubMed] [Google Scholar]
- 203.Huh D, Fujioka H, Tung YC, Futai N, Paine R, Grotberg JB, Takayama S. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc Natl Acad Sci USA. 2007;104:18886–91. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–8. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–45. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–10. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 207.O’Brien LE, Zegers MMP, Mostov KE. Building epithelial architecture: Insights from three-dimensional culture models. Nat Rev Mol Cell Biol. 2002;3:531–7. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- 208.Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3d matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ghosh S, Spagnoli GC, Martin I, Ploegert S, Demougin P, Heberer M, Reschner A. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: A high density oligonucleotide array study. J Cell Physiol. 2005;204:522–31. doi: 10.1002/jcp.20320. [DOI] [PubMed] [Google Scholar]
- 210.Takai A, Fako V, Dang H, Forgues M, Yu Z, Budhu A, Wang XW. Three-dimensional organotypic culture models of human hepatocellular carcinoma. Sci Rep. 2016;6:21174/1–11. doi: 10.1038/srep21174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–63. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Khademhosseini A, Langer R. Microengineered hydrogels for tissue engineering. Biomaterials. 2007;28:5087–92. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 214.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kloxin AM, Lewis KJR, DeForest CA, Seedorf G, Tibbitt MW, Balasubramaniam V, Anseth KS. Responsive culture platform to examine the influence of microenvironmental geometry on cell function in 3D. Integr Biol. 2012;4:1540–9. doi: 10.1039/c2ib20212c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stowers RS, Allen SC, Suggs LJ. Dynamic phototuning of 3D hydrogel stiffness. Proc Natl Acad Sci USA. 2015;112:1953–8. doi: 10.1073/pnas.1421897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Salinas CN, Anseth KS. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29:2370–7. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12:458–65. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9:628–38. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- 220.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–80. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 221.Erickson CA, Nuccitelli R. Embryonic fibroblast motility and orientation can be influenced by physiological electric fields. J Cell Biol. 1984;98:296–307. doi: 10.1083/jcb.98.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Saranak J, Foster KW. Rhodopsin guides fungal phototaxis. Nature. 1997;387:465–6. doi: 10.1038/387465a0. [DOI] [PubMed] [Google Scholar]
- 223.Lowe B. The role of Ca2+ in deflection-induced excitation of motile, mechanoresponsive balancer cilia in the ctenophore statocyst. J Exp Biol. 1997;200:1593–606. doi: 10.1242/jeb.200.11.1593. [DOI] [PubMed] [Google Scholar]
- 224.Carter SB. Principles of Cell Motility: The direction of cell movement and cancer invasion. Nature. 1965;208:1183–7. doi: 10.1038/2081183a0. [DOI] [PubMed] [Google Scholar]
- 225.Carter SB. Haptotaxis and the mechanism of cell motility. Nature. 1967;213:256–60. doi: 10.1038/213256a0. [DOI] [PubMed] [Google Scholar]
- 226.Aznavoorian S, Stracke ML, Krutzsch H, Schiffmann E, Liotta LA. Signal transduction for chemotaxis and haptotaxis by matrix molecules in tumor cells. J Cell Biol. 1990;110:1427–38. doi: 10.1083/jcb.110.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials. 2009;30:5433–44. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.DeForest CA, Anseth KS. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat Chem. 2011;3:925–31. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat Mater. 2009;8:659–64. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat Mater. 2011;10:799–806. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 231.Aizawa Y, Wylie R, Shoichet M. Endothelial cell guidance in 3D patterned scaffolds. Adv Mater. 2010;22:4831–5. doi: 10.1002/adma.201001855. [DOI] [PubMed] [Google Scholar]
- 232.DeForest CA, Anseth KS. Photoreversible patterning of biomolecules within click-based hydrogels. Angew Chem Int Ed. 2012;51:1816–9. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.DeForest CA, Tirrell DA. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat Mater. 2015;14:523–31. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- 234.Ebara M, Kotsuchibashi Y, Narain R, Idota N, Kim YJ, Hoffman JM, Uto K, Aoyagi T. Smart biomaterials in NIMS monographs. Tokyo: Springer Japan; 2014. p. 373. [Google Scholar]
- 235.Adzima BJ, Tao Y, Kloxin CJ, DeForest CA, Anseth KS, Bowman CN. Spatial and temporal control of the alkyne–azide cycloaddition by photoinitiated Cu(II) reduction. Nat Chem. 2011;3:256–9. doi: 10.1038/nchem.980. [DOI] [PubMed] [Google Scholar]
- 236.Mosiewicz KA, Kolb L, van der Vlies AJ, Martino MM, Lienemann PS, Hubbell JA, Ehrbar M, Lutolf MP. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat Mater. 2013;12:1072–8. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 237.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–9. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 238.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–87. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]