Abstract
Background
B cell chronic lymphocytic leukemia (B-CLL) is the most common adult leukemia in the Western world. Although therapeutic advances have notably improved the outcome for many patients, B-CLL remains an incurable disease. The purpose of this study was to search for therapeutic drugs based on altered pathways in individual patients.
Material/Methods
Genes from microarray data were mapped to 300 Homo sapiens-related pathways. Individual pathway aberrance analysis was used to identify altered pathways. Drug data, obtained from connectivity map (cMAP), were subjected to drug-set enrichment analysis. To analyze the relations between drug-induced pathways and disease-induced altered pathways in individuals, Pearson correlation analysis was applied.
Results
The disease-induced pathways with P-values <0.05 in individual samples were recorded and presented in a heatmap. Drug-induced pathways were analyzed in the 104 samples. After Pearson correlation analysis between altered pathways and drug, the 20 top-ranked drugs that were most relevant to disease were obtained. There were 9 drugs with positive scores and 11 with negative scores.
Conclusions
With this method, we identified the 20 top-ranked drugs that were most relevant to disease. The drugs with negative scores may play therapeutic roles in B-CLL.
MeSH Keywords: Abnormalities, Drug-Induced; Critical Pathways; Leukemia, Lymphocytic, Chronic, B-Cell
Background
B cell chronic lymphocytic leukemia (B-CLL) is the most common adult leukemia in the Western world [1], and it targets mainly elderly adults [2]. CLL is a monoclonal expansion of small mature B lymphocytes accumulating in the peripheral blood, bone marrow, and secondary lymphoid organs. Although recent therapeutic advances have notably improved the outcome for many patients, B-CLL remains an incurable disease [3].
Genetics and molecular genetics have contributed to illuminating the biological bases of the clinical heterogeneity of CLL [4]. In recent years, advances in massively parallel sequencing technologies have dramatically enhanced our understanding of the CLL genome and epigenome [5]. In 1990 Juliusson et al. reported that more than half of CLL patients had clonal chromosomal changes, despite its technological limitations [6]. In 2000, chromosomal abnormalities were detected in over 80% of patients by using interphase fluorescence in situ hybridization [7]. With the development of next-generation sequencing technologies, new genes implicated in CLL were revealed. In 2009, a study showed that the constitutive NOTCH signaling activation is implicated in CLL cell survival and apoptosis resistance [8]. SF3B1 is almost always found to cause mutations in CLL, with missense substitutions affecting the HEAT domains [9]. In addition, ATM, TP53, and BIRC3 were identified to be closely related to CLL. Novel small-molecule inhibitors that target BCR signaling, including the Bruton’s tyrosine kinase inhibitor ibrutinib and the phosphoinositide-3-kinase delta inhibitor idelalisib, have become the most successful new therapeutics in CLL [10].
Our knowledge of the molecular genetics of chronic lymphocytic leukemia has significantly broadened, which may offer new clinical implications.
Previous studies have proved that many critical genes and pathways are dysregulated during cancer initiation and progression [11]. Gene-based pathway analysis in a tumor is becoming an important method for understanding disease mechanisms. Pathways that are commonly deregulated across all cancer patients may be useful in identifying cancer from unknown samples. The analysis of altered pathways in an individual cancer patient may help to understand disease status and suggest customized anticancer therapies [12]. It was revealed that therapies targeting the B cell receptor signaling pathway are changing the treatment paradigms of many B cell lymphoproliferative disorders [13]. Thus, understanding CLL altered pathways may help find break-outs of B-CLL therapy.
Although significant progress has been generated based on critical genes in the treatment of chronic lymphocytic leukemia [14], B-CLL remains incurable. Immunochemotherapy with monoclonal antibodies and, recently, small molecules significantly improved survival. The drug used in CLL treatment for many years is chlorambucil, which is still recommended for elderly patients with numerous comorbidities [14]. Rituximab or obinutuzumab is usually combined with chlorambucil and improves overall survival [15]. Bendamustine, rituximab (BR regimen), FCR-Lite, Q-Lite regimens, or reduced doses of purine analogs are used for treatment [16–18]. Since CLL patients are burdened with numerous comorbidities, the optimum combination of drugs for therapy highlights the importance of personalized pathway analysis. The purpose of this study was to search for therapeutic drugs according to altered pathways in individual patients.
In this study, we introduced individual pathway aberrance analysis (iPAS) to analyze altered pathways in individual CLL patient. Thousands of drugs underwent drug-set enrichment analysis (DSEA) in individuals. We tried to find the corresponding therapeutic drugs by analyzing the relationships between drug data and disease.
Material and Methods
Disease-induced pathways analysis
Dataset
Microarray data of E-GEOD-39411 [19] used in this study were downloaded from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/experiments/E-GEOD-39411/). Two groups of data were included: a normal group (healthy B cell, n=48) and a disease group (chronic lymphocytic leukemia B cell, n=104).
According to the annotation files, probes were mapped to gene symbols. If more than 1 probe was mapped to a single gene, the levels of probes were averaged as the final gene expression value. In total, 20 389 genes were obtained.
Pathways of Homo sapiens used in this study were downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/) [20]. In total, 300 pathways including 6919 genes were obtained. Genes of expression profiling were mapped to pathways.
Pathway analysis
Pathway analysis has become an important method to capture clinical information. An individualized pathway aberrance score (iPAS) model was introduced to discover pathways in individuals [12]. The method included the following procedures:
Gene-level statistics
The gene expression levels were normalized with quantile normalization in the preprocessCore package [21] and the mean and standard deviation of each gene in normal samples were calculated. Genes in disease samples were normalized with quantile normalization function after combining a disease sample with all normal samples, generating gene-level statistics of each gene in an individual disease sample. The formula used was:
| (1) |
where Zi symbolizes the standardized expression value of i-th gene and n represents the number of genes belonging to a pathway.
Pathway level statistics
For a specific pathway, gene-level statistics of all genes belonging to a pathway were calculated. Pathway level statistics were calculated by averaging gene-level statistics. The formula was:
| (2) |
where Zi represents gene-level statistics of the i-th gene and n represents the number of genes belonging to a pathway.
With this procedure, pathway statistics of all pathways in normal samples and disease samples were obtained.
Statistical analysis
According to the distribution characteristics of pathway statistics in normal samples, pathway-level statistics of disease samples were analyzed for normal distribution. After being corrected for false discovery rate [22], the P-value of each pathway was calculated in individuals. By ranking pathways with P-values in disease group, pathways with P-values <0.05 were obtained, which were considered significantly dysregulated pathways. The frequency of altered pathways is presented in the heatmap.
Individualized drug analysis
To analyze individual pathway aberrance induced by drugs, we introduced drug-set enrichment analysis (DSEA) [23], which quantifies the extent to which a set of drugs consistently dysregulates pathways.
Dataset
Gene expression data induced by 1309 drugs used in this study were obtained from a connectivity map (cMAP) [24] (http://www.broadinstitute.org/cmap/), including 7056 Affymetrix microarrays. After probes were mapped to gene symbols, gene expression values were normalized with quantile normalization [21].
Drug-induced pathways analysis
Gene-level statistics
Drug-induced gene-level statistics were calculated using the procedure in Pathway analysis.
Pathway level statistics
Drug-induced pathway-level statistics were calculated using DSEA, and the procedure was as follows:
We collected pathways in the KEGG database, removing those including no genes in drug-induce genes. A gene × drug matrix was converted to a pathway-oriented matrix. For each pathway i in the database and each matrix j, we computed a signed Enrichment Score ESij and a P-value using the Kolmogorov-Smirnov (KS) test [25]. The association is defined as
| (3) |
where F1 and F2 are the 2 empirical distribution functions corresponding to the ranks of the genes, and s is a function returning −1 or +1.
DSEA was assessed by comparing drug ranks as opposed to gene ranks. Each row i of the pathway i was sorted across the j=1… l drugs, generating a rank-based matrix R. Each element Rij in R represents the rank of drug j when sorting drugs according to their effect on pathway i.
Statistical analysis
Using the same method as the procedure detailed in Pathway analysis.
Thus, drug-induced pathways were obtained in 104 disease samples.
Comparative analysis of drug-induced pathways and disease pathways
To identify the drugs corresponding to disease, we performed a comparative analysis of drug and disease using the Pearson correlation coefficient [26]. The formula is
| (4) |
where X and Y are variables and N represents the variable value number. A score is +1 indicates a positive correlation and −1 indicates a negative correlation. A score is 0 indicates no correlation.
A parameter of rank score was introduced to assess Pearson correlation coefficients of a drug in all samples. The formula is
| (5) |
where ABS sum indicates the sum of the absolute value of Pearson correlation coefficients in all samples.
Results
Individualized disease-induced pathways analysis
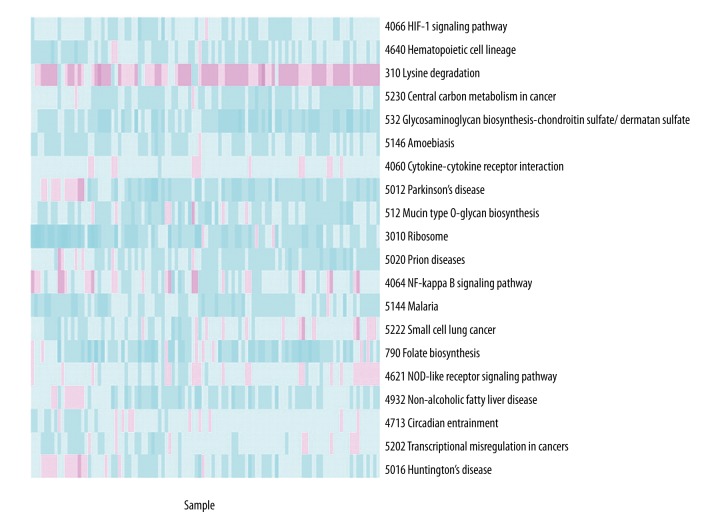
Pathway level statistics in individual pathway of all samples were obtained using iPAS. Normal distribution analysis was performed in pathway level statistics of disease samples after combining a disease case with all the normal samples. P-values of pathways in each disease sample were calculated. The pathways with P-values <0.05 were considered to be more prone to alteration. These pathways in an individual sample were recorded and presented in a heatmap, as shown in Figure 1, which intuitively reflects the frequency of altered pathways in the 104 disease samples. By ranking pathways with the distribution of pathways with P-values <0.05 in 104 samples, the top 20 pathways were identified and are listed in Table 1. The top-ranked pathway was “HIF-1 signaling pathway”, which was found in 93 samples.
Figure 1.
A heatmap representing the distribution of the top 20 altered pathways in 104 samples. Red color indicates down-regulation and green color indicates up-regulation.
Table 1.
Pathways with P-values <0.05 and P-values <0.01.
| Pathway | Mean of statistics | SD of statistics | Mean of P-value | No. p005 | No. p001 |
|---|---|---|---|---|---|
| HIF-1 signaling pathway | −0.3262422 | 0.1485723 | 0.0388540 | 93 | 0 |
| Hematopoietic cell lineage | −0.3847170 | 0.1781843 | 0.0453297 | 93 | 0 |
| Lysine degradation | 0.3462189 | 0.2994140 | 0.0669152 | 81 | 67 |
| Central carbon metabolism in cancer | −0.3766408 | 0.1985446 | 0.0753532 | 80 | 0 |
| Glycosaminoglycan biosynthesis – chondroitin sulfate/dermatan sulfate | −0.6245317 | 0.2778061 | 0.0523940 | 79 | 0 |
| Amoebiasis | −0.3603284 | 0.1396767 | 0.0561224 | 77 | 0 |
| Cytokine-cytokine receptor interaction | −0.1711602 | 0.1304610 | 0.0833987 | 75 | 0 |
| Parkinson’s disease | −0.4871193 | 0.3378297 | 0.0771193 | 75 | 0 |
| Mucin type O-glycan biosynthesis | −0.3957148 | 0.2842250 | 0.0883046 | 73 | 1 |
| Ribosome | −0.6842832 | 0.3987781 | 0.0647567 | 72 | 0 |
| Prion diseases | −0.3939857 | 0.2532834 | 0.0900706 | 70 | 1 |
| NF-kappa B signaling pathway | −0.1501014 | 0.3221464 | 0.0728022 | 67 | 9 |
| Malaria | −0.4542561 | 0.2193519 | 0.0798666 | 66 | 0 |
| Small cell lung cancer | −0.2128115 | 0.2209183 | 0.1153846 | 62 | 2 |
| Folate biosynthesis | −0.5721935 | 0.3901549 | 0.1030220 | 61 | 0 |
| NOD-like receptor signaling pathway | −0.1747373 | 0.1945677 | 0.1456044 | 60 | 0 |
| Non-alcoholic fatty liver disease (NAFLD) | −0.3779647 | 0.2511017 | 0.1077316 | 60 | 0 |
| Circadian entrainment | −0.1950689 | 0.1449920 | 0.1197017 | 59 | 0 |
| Transcriptional misregulation in cancers | −0.2221240 | 0.1726676 | 0.1283359 | 58 | 0 |
| Huntington’s disease | −0.2635283 | 0.2501193 | 0.1342229 | 57 | 1 |
SD indicates standard deviation. No.p005 indicates the number of samples that the pathways with P-values <0.05 occurred. No.p001 indicates the number of samples that the pathways with P-values <0.01 occurred.
Individualized drug-induced pathways analysis
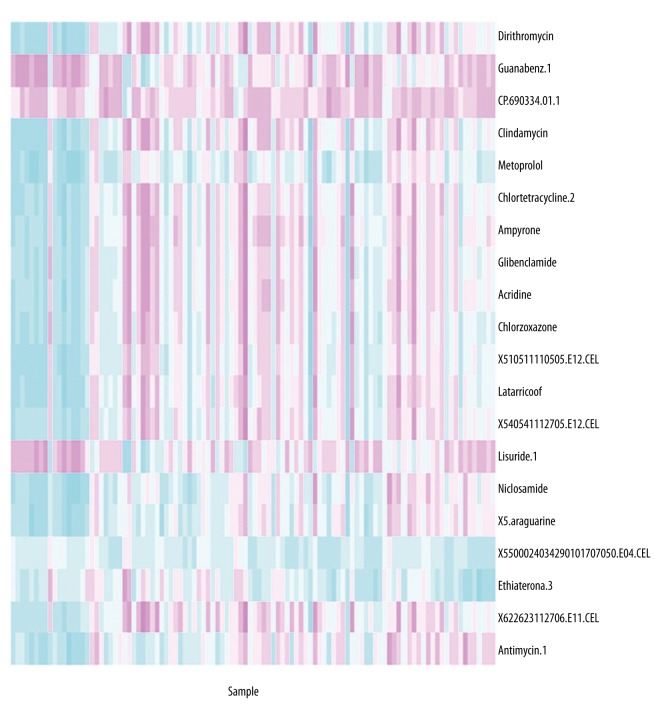
Each drug was analyzed to search for the induced pathways in the 104 samples. The drugs were more related to the disease if the induced pathways were the same or similar with disease-induced pathways. In this study, we presented the top 20 drugs most correlated with disease. The distribution of drugs in the 104 samples was presented in a heatmap, as shown in Figure 2.
Figure 2.
A heatmap representing the distribution of top 20 drugs in 104 samples. Red color indicates down-regulation and green color indicates up-regulation.
Comparative analysis of drug and disease
We analyzed the correlations between drugs-induced pathways and disease-induced pathways in 104 samples using Pearson correlation analysis. The sum of Pearson correlation coefficients and rank scores of drugs in all samples were obtained. A small value of rank score indicated a closer correlation with disease. By ranking drugs by rank scores, the top 20 ranked drugs with the lowest rank scores were obtained and are presented in Table 2. A positive score indicates that the drug induces a similar mechanism with disease. The negative score indicates that the drug induces an adverse mechanism with disease. In total, there were 9 drugs with positive scores and 11 with negative scores. Positive scores indicate that the drug induces similar pathways with disease. Negative scores indicate that the drug induces adverse mechanisms with disease and provides possibilities for therapy.
Table 2.
Pearson Correlation analysis of 20 drugs in 104 disease samples.
| Drug | Sum | ABSsum | Rank score |
|---|---|---|---|
| CP.690334.01 | 19.599944 | 21.86954 | 0.1727514 |
| X5500024034290101707050.E04 | −19.307528 | 19.72240 | 0.2186308 |
| Methylbenzethonium.chloride | 15.509400 | 19.51497 | 0.2189624 |
| Proxymetacaine | −16.994467 | 17.08981 | 0.2350641 |
| Dirithromycin | −1.653874 | 22.41674 | 0.2448278 |
| Guanabenz | 14.260195 | 22.02237 | 0.2466435 |
| Primaquine | −14.400254 | 18.80216 | 0.2513455 |
| X5500024034290101707046.E10 | −14.732907 | 17.01374 | 0.2530762 |
| Mebendazole | −12.187068 | 17.40977 | 0.2550116 |
| Naproxen | 16.178064 | 16.40269 | 0.2555140 |
| Lidocaine | 14.575151 | 17.02255 | 0.2565994 |
| Chlorzoxazone | −4.121720 | 20.56024 | 0.2567613 |
| Nitrofural | −16.958575 | 19.39794 | 0.2567736 |
| Chenodeoxycholic.acid | 13.938889 | 16.12181 | 0.2573696 |
| Promethazine | −13.346200 | 17.62752 | 0.2585429 |
| Methotrexate | −2.564379 | 17.16506 | 0.2589465 |
| Clindamycin | −1.980943 | 21.75196 | 0.2599480 |
| Lisuride | 9.982621 | 20.29997 | 0.2604600 |
| Benzethonium.chloride | 14.071414 | 17.61407 | 0.2617151 |
| Ioxaglic.acid | 16.121434 | 16.87698 | 0.2621227 |
Sum indicates Pearson Correlation Coefficients in all samples. ABS is absolute value. Rank score indicates the rank of ABSsum.
Discussion
B cell chronic lymphocytic leukemia (B-CLL) is the most common adult leukemia in the Western world and remains an incurable disease. To search for therapeutic drugs, we introduced a new method by combining individual pathway analysis and DSEA.
In this study, microarray data of 48 healthy samples and 104 chronic lymphocytic leukemia B cell samples underwent iPAS analysis. Personalized altered pathways were identified. With drug data from cMAP, DSEA was introduced to analyze drug mode in individual CLL patients. By comparing the heatmaps of Figures 1 and 2, we get an intuitive comparison between drugs and disease. Pearson correlation analysis was introduced to analyze the correlations between drugs-induced pathways and disease-induced pathways in 104 samples.
Among the 20 drugs, 9 drugs were in positive correlations with disease-induced pathways, indicating that the drugs induced similar mechanisms with disease, and 11 drugs showed negative relations with disease-induced pathways, which could be used for therapy.
Benzethonium chloride and its analog, methyl benzethonium chloride, were identified to be selectively toxic to human pharyngeal cancer cells but not to normal human or mouse cells [27]. They can induce apoptosis and caspase activation in tumor cells, delay the growth of xenograft tumors, and display in vivo additivity with local radiation therapy. It is proposed that benzethonium chloride should be highly effective in almost all leukemia [27]. However, Table 2 shows that methyl benzethonium chloride and benzethonium chloride are positively correlated with disease-induced pathways. The reason for this aberrance might be that the CLL B lymphocytes have been stimulated with an anti-IgM antibody, which influenced expression of genes and pathways.
Proxymetacaine showed a negative correlation with disease-induced pathways. Generally, it was used for cycloplegia because of the ease of administration and minimal adverse effects [28]. Leukemia is known to cause significant retinopathy [29], such as retinal lesions and retinal hemorrhages. Proxymetacaine was considered to relieve eye discomfort in CLL patients.
Mebendazole has been demonstrated to have cytotoxic anti-neoplastic potency in breast cancer [30], adrenocortical carcinoma [31] and several other cancers [32]. Its effect on cancer cells is rooted in the ability to promote suppression of migration/invasion (in vitro) metastasis (in vivo) and tumor growth rate (in vivo) [30]. In this study, we screened for drugs to treat CLL, and the clinical effect still needs further research to verify.
Guanabenz is a synthetic drug that can elevate eIF2α-p by inhibiting de-phosphorylation of eIF2α-p [33], similar to the effect of salubrinal. The phosphorylated eIF2α (eIF2α-p) inhibits eIF2α activity and translation initiation and was demonstrated to promote apoptosis [34]. It was reported that administration of salubrinal stimulates apoptosis of leukemic cells [35]. Thus, we hypothesized that guanabenz stimulates apoptosis of leukemic cells. However, Table 2 shows it is positively correlated with disease-induced pathways, similarly to methyl benzethonium chloride. The reason of this aberrance might be that the CLL B lymphocytes were stimulated with an anti-IgM antibody, which influenced expression of genes and pathways.
Primaquine is a drug for radical cure of Plasmodium vivax malaria, but its use is often hampered by safety concerns since it causes dose-dependent hemolysis in individuals who are deficient in glucose-6-phosphate dehydrogenase [36], and hemolysis one of the autoimmune complication that are common in CLL [37]. On the other hand, Soo et al. reported that primaquine increases imatinib clearance, bioavailability, and distribution pattern, which could improve the treatment of chronic myelogenous leukemia [38]. Therefore, we still need more research to verify its effect in CLL.
Naproxen is a potent nonsteroidal anti-inflammatory drug that inhibits both COX-1 and COX-2. It exhibits analgesic, anti-pyretic, and anti-inflammatory activities. It was revealed that naproxen induces significant apoptosis and inhibits Akt phosphorylation [39]. It has reported that Akt is constitutively active in various types of leukemia and is responsible for uncontrolled proliferation and resistance to apoptosis in leukemia cells, providing a potential therapeutic target in leukemia [40]. Thus, naproxen was suggested as a therapeutic drug for CLL treatment.
Our study has some limitations. In theory, the positively correlated drugs induce similar mechanism with disease, but in this study these drugs showed therapeutic effects for CLL. The reason may be that all the CLL B lymphocytes samples involved in the study have stimulated in vitro with an anti-IgM antibody, which activates the B cell receptor (BCR). Thus, it is better to collected samples without any treatment. In addition, the effects of drugs for CLL B lymphocytes have not been verified experimentally. The samples were not large enough. There are some other important drugs, such as the CD20 monoclonal antibodies, rituximab, ofatumumab and obinutuzumab, and the CD52 monoclonal antibody, alemtuzumab, which are currently being investigated in clinical trials and are not involved in the present study.
Conclusions
In this study, we sought to find therapeutic drugs by combining the method of individual pathway aberrance analysis and drug-set enrichment analysis. Altered pathways in individual patients were identified and their relations with drug-induced pathways were analyzed by Pearson correlation analysis. The top 20 closely related drugs were obtained. Nine drugs were identified as being positively correlated with disease. Eleven were negatively correlated with disease, which suggests their usefulness for B-CLL therapy. Some drugs have been explored in earlier studies, and others showed poor research. There were 3 unknown drugs, which still need validated in a prospective study.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Sabattini E, Bacci F, Sagramoso C, Pileri SA. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica. 2010;102:83–87. [PubMed] [Google Scholar]
- 2.Redaelli A, Laskin BL, Stephens JM, et al. The clinical and epidemiological burden of chronic lymphocytic leukaemia. Eur J Cancer Care (Engl) 2004;13:279–87. doi: 10.1111/j.1365-2354.2004.00489.x. [DOI] [PubMed] [Google Scholar]
- 3.Hallek M, Fischer K, Fingerle-Rowson G, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: A randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–74. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 4.Ahn T, Lee E, Huh N, Park T. Personalized identification of altered pathways in cancer using accumulated normal tissue data. Bioinformatics. 2014;30:i422–29. doi: 10.1093/bioinformatics/btu449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guieze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126:445–53. doi: 10.1182/blood-2015-02-585042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juliusson G, Oscier DG, Fitchett M, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–24. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–16. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Rosati E, Sabatini R, Rampino G, et al. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood. 2009;113:856–65. doi: 10.1182/blood-2008-02-139725. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ten Hacken E, Burger JA. Microenvironment interactions and B-cell receptor signaling in Chronic Lymphocytic Leukemia: Implications for disease pathogenesis and treatment. Biochim Biophys Acta. 2016;1863:401–13. doi: 10.1016/j.bbamcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–57. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 12.Cuiru W, Ling L, Yunxia H, et al. [Effects of Bufei Capsule on pulmonary function and arterial blood gas of rat model of chronic obstructive pulmonary disease]. Lishizhen Medicine and Materia Medica Research. 2012;23:104–6. [in Chinese] [Google Scholar]
- 13.Wall S, Woyach JA. Chronic lymphocytic leukemia and other lymphoproliferative disorders. Clin Geriatr Med. 2016;32:175–89. doi: 10.1016/j.cger.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Hus I, Rolinski J. Current concepts in diagnosis and treatment of chronic lymphocytic leukemia. Contemp Oncol (Pozn) 2015;19:361–67. doi: 10.5114/wo.2015.55410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goede V, Fischer K, Engelke A, et al. Obinutuzumab as frontline treatment of chronic lymphocytic leukemia: Updated results of the CLL11 study. Leukemia. 2015;29:1602–4. doi: 10.1038/leu.2015.14. [DOI] [PubMed] [Google Scholar]
- 16.Forconi F, Fabbri A, Lenoci M, et al. Low-dose oral fludarabine plus cyclophosphamide in elderly patients with untreated and relapsed or refractory chronic lymphocytic Leukaemia. Hematol Oncol. 2008;26:247–51. doi: 10.1002/hon.868. [DOI] [PubMed] [Google Scholar]
- 17.Foon KA, Boyiadzis M, Land SR, et al. Chemoimmunotherapy with low-dose fludarabine and cyclophosphamide and high dose rituximab in previously untreated patients with chronic lymphocytic leukemia. J Clin Oncol. 2009;27:498–503. doi: 10.1200/JCO.2008.17.2619. [DOI] [PubMed] [Google Scholar]
- 18.Fischer K, Cramer P, Busch R, et al. Bendamustine in combination with rituximab for previously untreated patients with chronic lymphocytic leukemia: A multicenter phase II trial of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol. 2012;30:3209–16. doi: 10.1200/JCO.2011.39.2688. [DOI] [PubMed] [Google Scholar]
- 19.Vallat L, Kemper CA, Jung N, et al. Reverse-engineering the genetic circuitry of a cancer cell with predicted intervention in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2013;110:459–64. doi: 10.1073/pnas.1211130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;171:783–90. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano F, Sirci F, Carrella D, di Bernardo D. Drug-set enrichment analysis: A novel tool to investigate drug mode of action. Bioinformatics. 2016;32:235–41. doi: 10.1093/bioinformatics/btv536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler J, Parmryd I. Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A. 2010;77:733–42. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 27.Yip KW, Mao X, Au PY, et al. Benzethonium chloride: A novel anticancer agent identified by using a cell-based small-molecule screen. Clin Cancer Res. 2006;12:5557–69. doi: 10.1158/1078-0432.CCR-06-0536. [DOI] [PubMed] [Google Scholar]
- 28.Sutherland MS, Young JD. Does instilling proxymetacaine before cyclopentolate significantly reduce stinging? The implications of paediatric cycloplegia. Br J Ophthalmol. 2001;85:244–45. doi: 10.1136/bjo.85.2.238g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo V, Scott IU, Querques G, et al. Orbital and ocular manifestations of acute childhood leukemia: clinical and statistical analysis of 180 patients. Eur J Ophthalmol. 2008;18:619–23. doi: 10.1177/112067210801800420. [DOI] [PubMed] [Google Scholar]
- 30.Coyne CP, Jones T, Bear R. Gemcitabine-(C4-amide)-[anti-HER2/neu] anti-neoplastic cytotoxicity in dual combination with mebendazole against chemotherapeutic-resistant mammary adenocarcinoma. J Clin Exp Oncol. 2013;2(2) pii: 1000109. [PMC free article] [PubMed] [Google Scholar]
- 31.Martarelli D, Pompei P, Baldi C, Mazzoni G. Mebendazole inhibits growth of human adrenocortical carcinoma cell lines implanted in nude mice. Cancer Chemother Pharmacol. 2008;61:809–17. doi: 10.1007/s00280-007-0538-0. [DOI] [PubMed] [Google Scholar]
- 32.Bai RY, Staedtke V, Aprhys CM, et al. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neuro Oncol. 2011;13:974–82. doi: 10.1093/neuonc/nor077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 34.Hamamura K, Yokota H. Stress to endoplasmic reticulum of mouse osteoblasts induces apoptosis and transcriptional activation for bone remodeling. FEBS Lett. 2007;581:1769–74. doi: 10.1016/j.febslet.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koizumi M, Tanjung NG, Chen A, et al. Administration of salubrinal enhances radiation-induced cell death of SW1353 chondrosarcoma cells. Anticancer Res. 2012;32:3667–73. [PubMed] [Google Scholar]
- 36.White NJ. Primaquine to prevent transmission of falciparum malaria. Lancet Infect Dis. 2013;13:175–81. doi: 10.1016/S1473-3099(12)70198-6. [DOI] [PubMed] [Google Scholar]
- 37.Hamblin TJ. Autoimmune complications of chronic lymphocytic leukemia. Semin Oncol. 2006;33:230–39. doi: 10.1053/j.seminoncol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Soo GW, Law JH, Kan E, et al. Differential effects of ketoconazole and primaquine on the pharmacokinetics and tissue distribution of imatinib in mice. Anticancer Drugs. 2010;21:695–703. [PubMed] [Google Scholar]
- 39.Kim MS, Kim JE, Lim do Y, et al. Naproxen induces cell-cycle arrest and apoptosis in human urinary bladder cancer cell lines and chemically induced cancers by targeting PI3K. Cancer Prev Res (Phila) 2014;7:236–45. doi: 10.1158/1940-6207.CAPR-13-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shehata M, Schnabl S, Demirtas D, et al. Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood. 2010;116:2513–21. doi: 10.1182/blood-2009-10-248054. [DOI] [PubMed] [Google Scholar]