Abstract
Background
Congenital deafness is one of the most distressing disorders affecting humanity and exhibits a high incidence worldwide. Most cases of congenital deafness in the Chinese population are caused by defects in a limited number of genes. A convenient and reliable method for detecting common deafness-related gene mutations in the Chinese population is required.
Methods
We developed a PCR-reverse dot blot (RDB) assay for screening 20 hotspot mutations of GJB2, GJB3, SLC26A4, and MT-RNR1, which are common non-syndromic hearing loss (NSHL)–associated genes in the Chinese population. The PCR-RDB assay consists of multiplex PCR amplifications of 10 fragments in the target sequence of the four above-mentioned genes in wild-type and mutant genomic DNA samples followed by hybridization to a test strip containing allele-specific oligonucleotide probes. We applied our method to a set of 225 neonates with deafness gene mutations and 30 normal neonates.
Results
The test was validated through direct sequencing in a blinded study with 100% concordance.
Conclusions
The results demonstrated that our reverse dot blot assay is a reliable and effective genetic screening method for identifying carriers and individuals with NSHL among the Chinese population.
Introduction
Hearing loss, which severely affects patients’ daily quality of life, is one of the most common sensory impairments, affecting approximately one to three newborns per every 1,000 live births[1, 2]. In China, there are approximately 0.8 million children (less than 7 years of age) with hearing impairments, and this proportion exhibits an annual increase of 30,000 children[3]. Hearing loss is etiologically heterogeneous, and it has been estimated that at least two thirds of the cases of childhood-onset hearing loss have genetic causes[4,5]. To date, more than 80 genes and more than a hundred genetic loci have been mapped and associated with hereditary hearing loss (http://hereditaryhearingloss.org/).
However, previous epidemiological studies have shown that nearly half of NSHL cases are related to the following genes with recurrent mutations in the Chinese population: GJB2 (OMIM: 121011), GJB3 (OMIM: 603324), SLC26A4 (OMIM: 605646) and the mitochondrial gene MT-RNR1 (OMIM: 561000) [6–8]. With respect to genetic factors of NSHL in China, GJB2 variants appear to be the most common (23.37%), followed by variants in the SLC26A4 (14.74%), MT-RNR1 (2.56%) and GJB3(1.97%) genes[9,10]. Mutations in the GJB2 gene are the most frequent causes of nonsyndromic autosomal recessive sensorineural hearing loss in most Asian populations, including Han Chinese [6]. Four common mutations (c.235delC, c.299_ 300delAT, c.176_ 191del16, and c.35delG) account for 88.0% of GJB2 mutant alleles in the Chinese population [11]. A SLC26A4 mutation is the second most common cause of deafness in China [12]. Mutations in the SLC26A4 gene are responsible for Pendred syndrome (PDS) and enlarged vestibular aqueduct syndrome (EVA), with hearing loss detected at birth or in early childhood [13,14]. In China, the most common mutation of SLC26A4 is IVS7-2 A>G, with an allele frequency ranging from 5.13% to 11.06% [10, 15]. The GJB3 gene, which is related to hereditary NSHL, was first cloned in the Chinese population, and mutations in GJB3 are associated with progressive hearing loss [7]. The most common mutations in GJB3 are c.538C>T and c.547G>A [16]. Although nuclear gene defects constitute the majority of cases of hereditary hearing loss, it has become clear that mutations in mtDNA can also cause NSHL. The best-studied mutations related to aminoglycoside susceptibility are m.1555A>G and m.1494T>C in the MT-RNR1 gene [17–19]. In a previous study, we showed that the carrier frequency of common mutations of the four above-mentioned genes among 9317 newborns was 3.84% [20], and another study conducted by Zhang showed a higher carrier frequency of 5.52% in northern China [21]. Congenital deafness accounts for the overwhelming majority of the population with prelingual deafness. Because it can affect language capacity, the timing of the detection of hearing impairment is very important. Therefore, early detection, diagnosis, and intervention are necessary for newborns who are susceptible to deafness. Deafness gene screening might identify the cause of deafness at the molecular level and is thus a reliable and effective method for identifying NSHL-associated gene mutation carriers.
At present, different methods are used for the detection of NSHL-associated gene mutations, including classic polymerase chain reaction/restriction enzyme analysis (PCR-RFLP)[22], denaturing high-performance liquid chromatography (DHPLC) [23], matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) [24] and direct sequencing. Such methods present variable challenges; for example, the PCR-RFLP method can detect a limited number of mutations in a single run, making it insufficient for multi-allele detection. Other methods, however, require expensive equipment and personnel with specialized abilities, making them unsuitable for first-line screening in a clinic setting.
In this study, we aimed to develop a more efficient genetic diagnostic assay based on the PCR-RDB technique that could be used as a first-line screening tool for prevalent mutations in the Chinese population. We selected 20 variants of four genes (GJB2, GJB3, SLC26A4 and MT-RNR1) that are frequently found in the Chinese population and cause NSHL to varying degrees and with differing phenotypes. This platform will serve as a useful and cost-effective first-line screening tool for genetic NSHL in the Chinese population.
Methods
Subjects
We collected samples from neonates who were born in Dongguan Children’s Hospital between January 2015 and October 2015. All of the subjects had undergone traditional newborn hearing screening and deafness gene screening by MALDI-TOF mass spectrometry. A total of 225 neonates with at least one of the 20 variants of the GJB2, GJB3, SLC26A4 and MT-RNR1genes in homozygous, single-heterozygous, or compound-heterozygous forms were recruited. In addition, 30 normal control subjects who passed the traditional newborn hearing screening and did not have any of the 20 variants were also recruited. This study was approved and conducted in accordance with the protocols recommended by the Institutional Medical and Ethics Committee of Dongguan Children’s Hospital. Written informed consent was obtained from the parents or legal guardians of the subjects.
The positive controls used for the development of the assay were clinical blood samples with 18 common NSHL mutations, and the remaining two (c.167delT and c.589G>A) were obtained by site-directed mutagenesis (Shanghai Shanjing Biological Company).
Genomic DNA extraction
Peripheral whole-blood samples from all of the subjects were obtained in EDTA- containing tubes, and genomic DNA was extracted using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) according to the instructions provided by the manufacturer. A quantitative estimation of DNA expression was performed by spectrophotometry (Thermo-Fisher Nanodrop, DE, and USA). The genomic DNA products were stored in a freezer at -20°C.
Design of primers and probes
Ten sets of primers were designed for multiplex polymerase chain reactions (M-PCRs) to amplify the GJB2, GJB3, SLC26A4 and MT-RNR1 genes spanning the 20 mutation sites (Table 1). The 5’ ends of the primers were labeled with biotin. A total of 30 probes, including 10 normal and 20 mutant probes covering the mutation sites, were designed (Table 2). Ten of the normal probes were used as controls for 11 mutant probes, and these included 1226N for c.1226G>A and c.1229C>T. The remaining nine less-common mutations (c.167delT, c.281C>T, c.589G>A, IVS15+5G>A, c.547G>A, c.1975G>C, c.2027T>A, c.1174A>T, and c.2162C>T) had no positive controls. When positive result for any of these nine mutations was obtained, further verification was performed by Sanger sequencing.
Table 1. Multiplex PCR primers.
| Gene | Mutation sites | Primer sequences | Size (bp) | PCR tube |
|---|---|---|---|---|
| GJB2 | c.35delG, c.167delT, c.176_191del16, c.235delC, c.299-300delAT |
HL1F:5’- AGAGCAAACCGCCCAGAGTAGAA -3’ HL1R: 5’-GAAGATGACCCGGAAGAAGATGCT -3’ |
420 | Tube I |
| SLC26A4 | c.281C>T |
HL4F: 5’- GGCTCCCCAAATACCGAGT -3’ HL4R: 5’- TGGTAGCTGGGGAGAAGTG -3’ |
150 | Tube I |
| c.589G>A |
HL5F: 5’- CAGCTAGAGTCCTGATTGCCA -3’ HL5R: 5’- GCCTTAATAAGTGGGGTCTTGC -3’ |
250 | Tube I | |
| IVS7-2A>G |
HL6F: 5’- CAGCATTATTTGGTTGACA -3’ HL6R: 5’- CCCTTGGGATGGATTTA -3’ |
304 | Tube I | |
| c.2162C>T, c.2168A>G |
HL10F: 5’- AATGCGGGTTCTTTGACGACA -3’ HL10R: 5’- AAATGGAACCTTGACCCTCTTGA -3’ |
115 | Tube I | |
| c.1174A>T, c.1226G>A, c.1229C>T |
HL7F: 5’- GGACCACCACGCAGAGTAG -3’ HL7R: 5’- TGCCATTCCTCGACTTGTTCTC -3’ |
287 | Tube II | |
| IVS15+5G>A |
HL8F: 5’- CAGTCCTATTTTCTATGGCAATGTC -3’ HL8R: 5’- TGCCCTACACAAAGGGAAGAGG -3’ |
177 | Tube II | |
| c.1975G>C, c.2027T>A |
HL9F: 5’- TGCTTACCAAGGAACAGTGTGT -3’ HL9R: 5’- GCCCATGTATTTGCCCTGTTG -3 |
334 | Tube II | |
| GJB3 | c.538C>T, c.547G>A |
HL2F: 5’- GCCCCCTGCCCCAACATCGTG -3’ HL2R: 5’- GTGGCAGCGGCAGGTGGAAGC -3’ |
200 | Tube II |
| MTRNR1 | m.1555A>G, m.1494C>T |
HL3F: 5’- TTAAGGGTCGAAGGTGGATTTAG -3’ HL3R: 5’- TGGTTTGGCTAAGGTTGTCTGGTA -3’ |
367 | Tube II |
Table 2. NSHL gene mutation detection probes.
| Name of probe | Mutation detection probe (5’→3’) | Name of probe | Normal probe (5’→ 3’) | Detected mutation sites |
|---|---|---|---|---|
| 35M | CGATCCTGGGGGTGTGA | 35N | GATCCTGGGGGGTGTGA | GJB2 c.35delG |
| 176M | CCTGCAGCCAGCTACGATCAC | 176N | CAGGCTGCAAGAACGTGTGCTAC | GJB2 c.176_191del16 |
| 235M | CTATGGGCCTGCAGCTG | 235N | CTATGGGCCCTGCAGCT | GJB2 c.235delC |
| 299M | CTACCGGAGACGAGAAGAAGA | 299N | CTACCGGAGACATGAGAAGAAG | GJB2c.299_300delAT |
| 538M | CTACATTGCCTGACCTACCG | 538N | CTACATTGCCCGACCTACC | GJB3 c.538C>T |
| 1494M | CCGTCACCCTTCTCAAGTATAC | 1497N | CCGTCACCCTCCTCAAGTAT | MT-RNR1 m.1494C>T |
| 1555M | TAGAGGAGGCAAGTCGTAACA | 1555N | GAGGAGACAAGTCGTAACATGG | MT-RNR1 m.1555A>G |
| IVS7-2M | TTATTTCGGACGATAATTGCT | IVS7-2N | GTTTTATTTCAGACGATAATTGCT | SLC26A4 IVS7-2A>G |
| 2168M | TGACGGTCCGTGATGCTA | 2168N | TGACGGTCCATGATGCTATAC | SLC26A4 c.2168A>G |
| 1226M | CACTGCTCTTTCCCACACG | 1226N/1229N | CTCTTTCCCGCACGGCC | SLC26A4 c.1226G>A |
| 1229M | GCTCTTTCCCGCATGGC | SLC26A4 c.1229C>T | ||
| 167M | GCAGCCAGCTACGATCAC | GJB2 c.167delT | ||
| 281M | CGGGAGTTAGTATTGGGC | SLC26A4 c.281C>T | ||
| 1174M | GATCAGCTACATCTTCTCAGGA | SLC26A4 c.1174A>T | ||
| 1975M | GCCTTCTGCTTGACTGTG | SLC26A4 c.1975G>C | ||
| 2162M | ACATTCTTTTTGATGGTCC | SLC26A4 c.2162C>T | ||
| 547M | ATTTTCTTCTTGGTAGGTCG | GJB3 c. 547G>A | ||
| 589M | CTGACTCTGCTGGTTAGAAT | SLC26A4 c.589G>A | ||
| IVS15+5M | AAGTCCACAGTAAATATTTTATCC | SLC26A4IVS15+5G>A | ||
| 2027M | AGTGAGATCACAGCGGGT | SLC26A4 c.2027T>A |
Multiplex PCR amplification
Multiplex PCR amplification was performed on an Mx3000p PCR machine (Stratagene, CA, USA). The reactions were conducted in a final volume of 25 μL containing 1×PCR buffer (MgCl2Plus), 1×Qiagen Hotstar buffer (Qiagen, Hilden, Germany), 0.2 mmol/L of each dNTP (Promega, CA,USA), 0.2 μmol/L of each of the twenty primers, 0.1 U/μL HotStarTaq DNA polymerase (Qiagen, Hilden, Germany) and 1~2 μg of genomic DNA. The amplificationswere performed in two tubes simultaneously (as detailed in Table 1). The PCR conditions were based on the manufacturer's instructions: pre-denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The products were subsequently visualized by electrophoresis on a 1.5% agarose gel.
Reverse dot blot assay
The reverse dot blot (RDB) procedure performed in this study was based on that described by Lappin et al [25]. The carboxyl groups on the surface of the nylon membrane were negatively charged, and oligonucleotide probes were synthesized using a C6-amino-linker on the 5' end of the product, which was positively charged. The probes were fixed onto a Biodyne C nylon membrane (Pall Corporation) after the membrane was activated by incubating with 5% 1-ethyl 3-dimethyl aminopropyl carbodiimide (EDAC) for 30 minutes and then soaking with 0.1M NaOH for 5 minutes.
The target DNA was amplified with 5' biotinylated primers and hybridized to immobilized oligonucleotides on the membrane. All 30 above-mentioned probes were fixed to a Biodyne C membrane (Pall Corporation, NY, USA). The PCR products were heated to at least 95°C and then immediately cooled to 0°C. Each strip and 8mLof hybridization solution A solution consisting of 2×saline sodium citrate (SSC-3 mol/L NaCl and 0.3 mol/Lsodium citrate) with 0.1% SDS (pH 7.4) were pre-heated to 45°C. The denatured PCR products were then added, and the strips were incubated at 45°C for 2 h in a screw-top tube. The strips were subsequently washed with wash solution B (0.5×SSC and 0.1% SDS, pH 7.4) at 45°C for 10 min. Afterward, the strips were transferred to a hybridization solution A-diluted mixture containing 0.125 U/mL streptavidin-horseradish peroxidase conjugate (Roche, Mannheim, Germany) and incubated at room temperature for 30 min. Any excess conjugate was removed through two washes with solution A. The color-developing solution composed of 0.1 mg/mL tetramethylbenzidine dihydrochloride (TMB) substrate (Sigma T8768), 0.015‰ H2O2 and 0.1 mol/L sodium citrate (pH 5.4) were then added, and the color reaction was developed for 20 min. Blue dots indicated the positive detection. A positive control was performed in each experiment to ensure the reliability of the detection system. All of the samples were analyzed independently by DNA sequencing to confirm the accuracy of the assay.
Results
Establishment of the assay
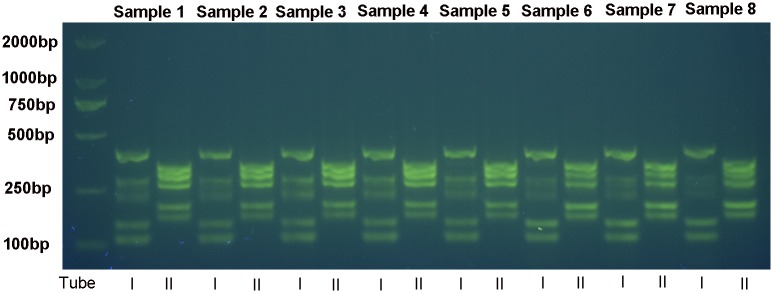
The assay was established using DNA samples from 18 subjects with mutations and two plasmid samples expressing the remaining two known mutations, which were introduced through site-directed mutagenesis. The successful electrophoretogram is shown in Fig 1. When analyzing the results, an absence of color development with all or the majority of the control probes served as a invalid results.
Fig 1. PCR products of eight samples obtained by electrophoresis in a 1.5% agarose gel stained with ethidium bromide.
I and II represents PCR products of tube I and II respectively.
The assay of the 20 positive control samples tested yielded the expected results (Fig 2 and S1 Fig). The normal DNA samples showed positive blue-colored spots with all of the control probes and negative white-colored spots with the mutation probes. Homozygous DNA samples showed blue-colored spots with the appropriate mutant probes and negative white-colored spots with the corresponding probes. Heterozygous and compound heterozygous DNA samples showed blue-colored spots with one and two mutant probes, respectively, and the corresponding white-colored control-probe spots.
Fig 2. Layout of the probes in the nylon strip designed for the reverse dot blot assay and representative results of the genotyping of 20 NSHL-associated gene mutations using the reverse dot blot assay.
N denotes the wild-type probes,and M denotes the mutation probes. (A). Layout of the probes in the nylon strip; (B) c.167delT homozygous or heterozygous mutation; (C) c.2162C>T homozygous or heterozygous mutation; (D) m.1494C>T homoplasmic mitochondrial gene mutation; (E) IVS7-2A>G and c.2168A>Gheterozygous compound mutations; and (F) normal samples.
Validation of the assay
We applied our method to a group of 225 neonates who carried one of 20 deafness gene mutations and 30 normal neonates. All of the samples were subsequently examined by DNA sequencing, and 100% concordance was found between the two methodologies (S2 Fig).
Discussion
We developed a platform that can be easily performed with common equipment, has a low cost of less than US$3 and can be completed within 6 h. It is an ideal method for smaller clinics and laboratories with limited resources that could allow the front-line detection of NSHL genes without having to resort to direct DNA sequencing. Our results for 255 unrelated subjects showed 100% concordance with the results of independent direct sequencing.
At present, the MassARRAY system combined with an iPLEX assay, which is a high-throughput genotyping technology based on MALDI-TOF mass spectrometry, is the most common deafness gene-detecting method used in clinical settings[26–29]. This MassARRAY system can detect a large amount of variants in multiple samples simultaneously. Nevertheless, the MassARRAY system requires a very expensive platform for the analysis, which limits its use.
Genetic testing can potentially allow an accurate diagnosis of NSHL, and a diagnosis of the genetic etiology will aid the clinical management of patients, including the selection of the most appropriate treatment. Combined newborn hearing screeningand genetic screening of common deafness-related genes will benefit the early detection of infants at high risk of developing delayed hereditary sensorineural deafness or sensitivity to deafness-related drugsas well as the diagnosis of hearing loss in children. The screening of deafness-related genes might effectively prevent the occurrence and development of deafness. The molecular etiology, diagnosis, and effective genetic counseling of the probandare important for the prevention and treatment of deafness.
Because GJB2, GJB3, SLC26A4, and MT-RNR1 are the four most common causative genes of prelingual NSHL in the Chinese population and hotspot mutations of these four genes are included, our assay is expected to cover a substantial portion of prelingual severe-to-profound genetic NSHL in the Chinese population. The restricted targeting reduces the possibility of incidental findings but allows higher coverage at a lower cost compared with genome-wide approaches. This study shows that this simple and inexpensive method can be used for routine molecular diagnosis and potentially for large-scale genetic screening. However, it is possible that the RDB will be affected by the SNP near the mutation. Although this did not occur in the 225 positive samples, we will take test samples in the future study.
We propose that the PCR-RDB assay is a highly accurate, rapid and economical clinical assay that can be potentially used for neonatal hearing screening and personalized medicine. Children with the MT-RNR1mutation are recommended stay away from amino glycoside antibiotics to avoid the development of drug-induced hearing loss. Infants with homozygous or compound heterozygous forms of the same autosomal recessive gene may show the symptoms of deafness at birth, and clinical and audiology intervention should be performed as soon as possible. In infants with homozygous or heterozygous mutations in autosomal dominant genes, the mutations can cause deafness, However, some individuals mayexhibit normal hearing at birth and develop delayed deafness, and they should be guided regarding optimizing their listening environment to refrain from developing hearing impairment during their hearing development, e.g., avoidance of head trauma, and timely administration of treatment if their auditory threshold changes.
In conclusion, we developed a new method for the genetic screening of hearing loss and successfully detected hearing loss-related genetic mutations prevalent in the Chinese population in homozygousand heterozygous forms, as validated by Sanger sequencing. This assay is expected to serve as a primary screening tool for genetic hearing loss and could be helpful in gene-based and personalized hearing rehabilitation.
Supporting information
(TIF)
(TIF)
Acknowledgments
This work was supported by the Dongguan Bureau of Science and Technology for the City Key Program of Science and Technology (Project No.2013108101018) and the Science and Technology Planning Project of Guangdong Province (Project No.2014A020213001). We also thank all of the neonates and their parents for their cooperation during this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Dongguan Bureau of Science and Technology for the City Key Program of Science and Technology (Project Number: 2013108101018) and the Science and Technology Planning Project of Guangdong Province (Project Number: 2014A020213001). We also thank all the neonates and their parents for their cooperation during this work.
References
- 1.Morton CC, Nance WE. Newborn hearing screening—a silent revolution. New England Journal of Medicine 2006;354:2151–64. 10.1056/NEJMra050700 [DOI] [PubMed] [Google Scholar]
- 2.Kral A, O'Donoghue GM. Profound deafness in childhood. New England Journal of Medicine 2010;363:1438–50. 10.1056/NEJMra0911225 [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Wang P, Han B, Ding Y, Pan L, Zou J, et al. Newborn hearing concurrent genetic screening for hearing impairment- a clinical practice in 58,397neonates in Tianjin,china. International Journal of Pediatric Otorhinolaryngology;77:1929–35. 10.1016/j.ijporl.2013.08.038 [DOI] [PubMed] [Google Scholar]
- 4.Petit C. Genes responsible for human hereditary deafness: Symphony of a thousand. Nat Genet 1996;14:385–91. 10.1038/ng1296-385 [DOI] [PubMed] [Google Scholar]
- 5.Willems PJ. Genetic causes of hearing loss. New England Journal of Medicine 2000;342:1101–9. 10.1056/NEJM200004133421506 [DOI] [PubMed] [Google Scholar]
- 6.Yuan Y, You Y, Huang D, Cui J, Wang Y, Wang Q, et al. Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in china. Journal of Translational Medicine 2009;7:79 10.1186/1479-5876-7-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia J-h, Liu C-y, Tang B-s, Pan Q, Huang L, Dai H-p, et al. Mutations in the gene encoding gap junction protein [beta]-3 associated with autosomal dominant hearing impairment. Nat Genet 1998;20:370–3. 10.1038/3845 [DOI] [PubMed] [Google Scholar]
- 8.Dai P, Yuan Y, Huang D, Zhu X, Yu F, Kang D, et al. Molecular etiology of hearing impairment in inner mongolia: Mutations in slc26a4 gene and relevant phenotype analysis. Journal of Translational Medicine 2008;6:74 10.1186/1479-5876-6-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Dai D, Chen Z, Cao X, Bu X, Wei Q, et al. Molecularscreening of patients with nonsyndromic hearing loss from Nanjing city of China. J Biomed Res 2011;25(5):309–18. 10.1016/S1674-8301(11)60042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Xiao Y, Bai X, Zhang F, Zhang D, Xu X,et al. GJB2, SLC26A4, and mitochondrial DNA12S rRNA hot-spots in 156 subjects with non-syndromic hearingloss in Tengzhou, China.Acta Otolaryngol 2016; 136(8):800–5. 10.3109/00016489.2016.1164893 [DOI] [PubMed] [Google Scholar]
- 11.Dai P, Yu F, Han B, Liu X, Wang G, Li Q, et al. GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med 2009; 14(7):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin F, Yuan Y, Deng X, Han W, Wang G, Zhao J, et al. Genetic mutations in nonsyndromic deafness patients of Chinese minority and Han ethnicities in Yunnan, China. J Transl Med 2013; 11:312 10.1186/1479-5876-11-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang QJ, Zhao YL, Rao SQ, Guo YF, Yuan H, Zong L, et al. A distinct spectrum of slc26a4 mutations in patients with enlarged vestibular aqueduct in china. Clinical Genetics 2007;72:245–54. 10.1111/j.1399-0004.2007.00862.x [DOI] [PubMed] [Google Scholar]
- 14.Yao G, Chen D, Wang H, Li S, Zhang J, Feng Z, et al. Novel mutations of SLC26A4 in chinese patients with nonsyndromic hearing loss. Acta Oto-Laryngologica 2013;133:833–41. 10.3109/00016489.2013.777160 [DOI] [PubMed] [Google Scholar]
- 15.Du W, Cheng J, Ding H, Jiang Z, Guo Y, Yuan H. A rapid method for simultaneous multi-gene mutation screening in children with nonsyndromic hearingloss. Genomics 2014;104(4):264–70. 10.1016/j.ygeno.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 16.Wu H, Feng Y, Jiang L, Pan Q, Liu Y, Liu C, et al. Application of a New Genetic Deafness Microarray for Detecting Mutations inthe Deaf in China. PLoS One 2016;11(3):e0151909 10.1371/journal.pone.0151909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lerer I, Sagi M, Malamud E, Levi H, Raas-Rothschild A, Abeliovich D. Contributionof connexin 26 mutations to nonsyndromic deafness in Ashkenazi patientsand the variable phenotypic effect of the mutation 167delT. Am J Med Genet 2000, 95(1):53–56. [DOI] [PubMed] [Google Scholar]
- 18.Gasparini P, Rabionet R, Barbujani G, Melchionda S, Petersen M, Brondum-Nielsen K, et al. High carrierfrequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur J Hum Genet 2000,8(1):19–23. 10.1038/sj.ejhg.5200406 [DOI] [PubMed] [Google Scholar]
- 19.Kudo T, Ikeda K, Kure S, Matsubara Y, Oshima T, Watanabe K, et al. : Novel mutations in the connexin 26 gene (GJB2) responsible for childhood deafness in the Japanese population. Am J Med Genet 2000, 90(2):141–145. [DOI] [PubMed] [Google Scholar]
- 20.Peng Q, Huang S, Liang Y, Ma Y, Li S, Yang L, et al. Concurrent Genetic and Standard Screening for Hearing Impairment in 9317 Southern Chinese Newborns. Genet Test Mol Biomarkers 2016, 20(10):603–608. 10.1089/gtmb.2016.0055 [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Wang P, Han B, Ding Y, Pan L, Zou J, et al. Newborn hearing concurrentgenetic screening for hearing impairment-a clinicalpractice in 58,397neonates in Tianjin, China. Int J Pediatr Otorhinolaryngol 2013, 77(12):1929–35. 10.1016/j.ijporl.2013.08.038 [DOI] [PubMed] [Google Scholar]
- 22.Pandya A, Xia X, Radnaabazar J, Batsuuri J, Dangaansuren B, Fischel-Ghodsian N, Nance WE. Mutation in the mitochondrial 12S rRNA gene in two families from Mongolia with matrilineal aminoglycoside ototoxicity. Journal of Medical Genetics 1997;34(2):169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin D, Goldstein JA, Mhatre AN, Lustig LR, Pfister M, Lalwani AK. Assessment of denaturing high-performance liquid chromatography (DHPLC) in screening for mutations in connexin 26 (GJB2). Hum Mutat 2001;18(1):42–51. 10.1002/humu.1148 [DOI] [PubMed] [Google Scholar]
- 24.Huang S, Xu Y, Liu X, Zhou M, Wu X, Jia Y. Molecular newborn screening of four genetic diseases in guizhou province of south china. Gene 2016;591(1):119–22. 10.1016/j.gene.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 25.Lappin S, Cahlik J, Gold B. Robot printing of reverse dot blot arrays for human mutation detection. J Mol Diagn 2001;3(4):178–88. 10.1016/S1525-1578(10)60670-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alves RM, da Silva Costa SM, do Amôr Divino Miranda PM, Ramos PZ, Marconi TG, Santos Oliveira G, et al. Analysis of mitochondrial alterations in Brazilian patients with sensorineural hearing loss using MALDI-TOF mass spectrometry. BMC Med Genet. 2016;17(1):41 10.1186/s12881-016-0303-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, Xu Y, Liu X, Zhou M, Wu X, Jia Y.Molecular newborn screening of four genetic diseases in Guizhou Province of South China.Gene. 2016. 591(1):119–22. 10.1016/j.gene.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 28.Yao GD, Li SX, Chen DL, Feng HQ, Zhao SB, Liu YJ, et al. Combination of hearing screening and genetic screening for deafness-susceptibility genes innewborns. Exp Ther Med. 2014. 7(1):218–222. 10.3892/etm.2013.1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Wang P, Han B, Ding Y, Pan L, Zou J, et al. Newborn hearing concurrent genetic screening for hearing impairment-a clinical practice in 58,397 neonates in Tianjin, China. Int J Pediatr Otorhinolaryngol. 2013. 77(12):1929–1935. 10.1016/j.ijporl.2013.08.038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.