Asymmetric cell shape depends on the ability of oriented layers of cellulose microfibrils to channel the nondirectional drive of turgor pressure. Tough microfibrils encircling the cell resist increases in girth and redirect the swelling force to a direction perpendicular to the alignment of the microfibrils. This transformation of an isotropic force into directional growth is crucial for setting up the main axis that allows an organism to project its photosynthetic and reproductive organs into the environment. But what controls the orientation of the microfibrils in the first place?
Cortical microtubules beneath the plasma membrane are thought to guide the directed extrusion of new microfibrils from cellulose-synthesizing machinery embedded in the plasma membrane. Newly laid cellulose microfibrils often are aligned above microtubules, which look like tracks for the cellulose-synthesizing enzymes. However, the molecular details of this hypothetical transmembrane system are scant, and there are now sufficient examples of the lack of coalignment to raise questions about the basic microtubule-microfibril paradigm. Before we can arrive at new models to explain phenomena as broad as “cell shape,” we need to answer more fundamental questions. Where are the microtubules formed? How do they move and link up to form a dynamic entity that covers the inner face of the plasma membrane? How can arrays form and reform so rapidly, and what controls their stability?
Several recent articles have provided a quantum leap in our understanding: one group of articles begins to show how the plant cell's unique cortical microtubule array is constructed, and the other shows that mutations in the microtubule's subunit protein—tubulin—affect the orientation of the entire microtubule array, resulting in changes in the direction of spiral growth.
MICROTUBULES BEGET MICROTUBULES
To understand how microtubules can be organized over large areas of plasma membrane, it is important to know where tubulin polymerization is initiated and how microtubules then self-organize into an ordered array. In animals, the microtubules are nucleated from a defined site—the centrosome—that consists of amorphous material gathered around conspicuous centrioles. The distal, growing (“plus”) ends of microtubules radiate from the centrosome, and the focused nucleating material concomitantly organizes the mi-crotubule array, a fact reflected in the term “microtubule-organizing center.” In acentriolar plant cells, nucleation and organization occur separately during interphase, and there has been considerable discussion regarding where the amorphous material is located. The nuclear surface seems to initiate microtubules for relatively brief periods during late G2 and transiently after cytokinesis, but studies on living cells have failed to find activity during interphase (Yuan et al., 1994; Granger and Cyr, 2001).
An important clue to understanding how the array is initiated has been provided by the finding that plant microtubules are severed from their nucleation sites by katanin. This ATPase is located in centrosomes in animals, where it is believed to cut microtubules free of their nucleation sites at the minus ends (McNally et al., 1996). Animal katanin is a heterodimer of a 60-kD subunit, p60, which exhibits ATPase activity required for microtubule-severing activity, and an 80-kD regulatory subunit. A homolog of the p60 subunit of katanin, AtKSS, has been identified in Arabidopsis (McClinton et al., 2001). Recombinant p60 is demonstrably active in vitro (Stoppin-Mellet et al., 2002), although no homolog of the 80-kD regulatory subunit has been detected in the Arabidopsis genome. The Arabidopsis katanin mutant fragile fiber (fra2) has a stunted growth habit and mechanically weakened cell walls with reduced cellulose (Burk et al., 2001), cor-relating with a reduction in cell length and a 50% reduction in height. Sequence com-parisons showed that FRA2 was similar to katanin, so it was renamed AtKTN1. In the mutant, the mitotic and cytokinetic microtubules appeared normal, but just after exit from cytokinesis, microtubule converging points remained at the nuclear envelope at the same time that microtubules appeared at the cortex in radiating clusters. Normally, microtubules radiating from the postcytokinetic nucleus are transient and do not overlap in time with the interphase array (Lambert and Lloyd, 1994). A gathering of minus ends at cortical nucleating sites also is antithetical to the formation of a dispersed array of parallel microtubules. In the mutant, conversion of these abnormal arrays to the evenly distributed parallel arrangement was delayed, and only limited expansion was seen during interphase. This was reported as well by Bichet et al. (2001) in the Arabidopsis mutant botero1, which is allelic to fra2. In the wild type, cortical microtubules in cells near the division zone in the meristem were loosely organized, but they became more highly aligned in transverse arrays the farther cells elongated away from the meristem. In bot1/fra2, this transition was not observed.
Another implication of these studies is that the nucleation sites become scattered over the expanding cell's cortex; thus, it is important to find markers. γ-Tubulin is the foundation stone of microtubule nucleation, forming part of a ring-like complex that acts as the first turn for the addition of α/β-tubulin subunits to the growing tubule. In animal cells, it is located at the minus ends of microtubules. In plants, however, in addition to the nuclear surface, γ-tubulin is found all over the various microtubule arrays, not just at the minus ends (Panteris et al., 2000). This is consistent with the staggered arrangement of minus ends between parallel microtubules (Joshi and Palevitz, 1996), or γ-tubulin may function differently in plants (e.g., in microtubule stabilization). A recently discovered protein may prove to be a better minus-end marker. Erhardt et al. (2002) report that orthologs of Spc98p, an essential component of the γ-tubulin ring complex in animals, are present in rice and Arabidopsis. Like γ-tubulin, plant Spc98p is found at the nuclear surface but has a more restricted distribution on cortical microtubules than has γ-tubulin. This could mean that Spc98p is confined to nucleation sites, although its minus-end location remains to be confirmed. Therefore, core nucleating factors are present across phylogeny, regardless of whether those factors are gathered into discrete microtu-bule-nucleating bodies.
Drawing this information together, it appears that katanin is required first to release microtubules nucleated at the sur-face of the postcytokinetic nucleus and then to repeat this procedure once nucleating material has been transported to, or been activated at, the cortex. Wasteneys (2002) hypothesized that after being severed by katanin, the nucleating minus end of the cortical microtubule is transported to the plus end of the larger severed fragment by plus-end-directed kinesin. According to this “cut-and-grow” mecha-nism, the cortical array is self-regenerating.
CONVERTING DYNAMIC MICROTUBULES TO WHOLE-CELL ASSEMBLIES
Once formed, the cortical array gives the semblance of stability, but dynamic studies have shown that microtubules rapidly turn over tubulin subunits and are capable of reforming an array in a new direction (Yuan et al., 1994; Wymer and Lloyd, 1996; Granger and Cyr, 2001). It is assumed that the turnover of microtubules is caused by dynamic instability (i.e., stochastic switching between growth and catastrophic shrinkage of the plus end). In plant microtubules, with severed minus ends, it is possible that tubulin can be added to, or subtracted from, the minus end as well as the plus end in a process known as treadmilling. Factors that can stabilize and destabilize the microtubule ends will regulate these behaviors and are beginning to be described for plants (Lloyd and Hussey, 2001).
One factor recently shown to stabilize microtubules is MICROTUBULE ORGAN-IZATION1 (MOR1), which belongs to an evolutionarily conserved group of high molecular mass microtubule-associated proteins (MAPs) that includes human TOGp and Xenopus XMAP215. The temperature-sensitive mor1 mutant of Arabidopsis has cells that expand sideways instead of elongating, and as a result, the plants are extremely squat (Whittington et al., 2001). At the restrictive temperature, microtubules of the cortical array shortened and lost alignment. However, within minutes of reaching the permissive temperature, the microtubules lengthened and parallel order was restored. The effect of MOR1 on microtubule growth is consistent with what is known about its Xenopus relative, XMAP215, which stimulates microtubule polymerization (Vasquez et al., 1994). In animal cells, the stimulatory effect of XMAP215 is antagonized by the central motor kinesin, XKCM1 (Tournebize et al., 2000). According to Hussey and Hawkins (2001), the N-terminal repeat of MOR1 could negate the effect of a destabilizing kinesin, either by binding directly to it or by competing for the microtubule binding site. Since XMAP215 has recently been reported to anchor nascent minus ends of microtubule asters formed in vitro (Popov et al., 2002), a further possibility is that its plant homolog, MOR1, could itself have a direct effect on microtubule nucleation by helping to anchor growing ends at the cortex.
In the absence of discrete microtubule-organizing centers, it is important to know how individual microtubules become organized into a whole-cell array. The crucial step in converting unitary microtubules to sheets of approximately parallel elements is cross-linking by bridges. MAP65 was discovered initially as a microtubule-bundling protein by Jiang and Sonobe (1993) and was found to decorate all four microtubule arrays (Chan et al., 1996). Biochemically isolated MAP65 from carrot cells was shown to induce brain microtubules to form parallel groups with the same 25- to 30-nm intermicrotubule spacing (Chan et al., 1999) observed in planta (Lancelle et al., 1986). Recently, Yasuhara et al. (2002) demonstrated that the MOR1 homolog, TMBP200, isolated from telophase tobacco cells also forms intermicrotubule bridges. However, these are 10 nm in length rather than the longer (>25 nm) bridges seen to cross-link microtubules during interphase (Lancelle et al., 1986).
One of the MAP65 genes, NtMAP65-1, has been cloned by Smertenko et al. (2000). Although it has no resemblance to the ubiquitous MAP2/4/tau group of animal MAPs, MAP65 does resemble PRC1, which was shown by Mollinari et al. (2002) to be a microtubule-bundling protein in animal cells. That study showed that PRC1 has functional domains that are regulated in a cell cycle–dependent manner. Applied to plant MAP65, this principle could explain the different functions of the bundling protein on the four microtubule arrays that form at different stages of the cycle. However, because there are nine MAP65 homo-logs in the Arabidopsis genome, it is likely that different isoforms have a cell cycle– and developmentally regulated pattern of expression. Immunostaining experiments support this notion. Although polyclonal antibodies to MAP65 fractions unselectively label all four microtubule arrays, including both sets of microtubules in the cytokinetic phragmoplast (Jiang and Sonobe, 1993; Chan et al., 1996), antibodies to recombinant NtMAP65-1a also stain all arrays but only the mid line of the phragmoplast where the two sets of plus ends meet. This finding suggests that different isoforms are targeted to different microtubule arrays and even to specific parts of the array. On SDS gels of cycling cells, MAP65 usually appears as three bands, but when the synthetic auxin is removed and all cells stop dividing and form interphase arrays, the pattern is simplified to one band (Barroso et al., 2000). Recent mass spectrometric evidence shows that this is a member of the most conserved subgroup of the multigene MAP65 family (J. Chan, G. Mao, and C. Lloyd, unpublished data). It will be interesting to determine if all nine isoforms have a specific spatiotemporal use. Therefore, although only a few components of the plant microtubule array were known a few years ago, there is now a fair amount of molecular detail regarding how microtubules polymerize and assemble into parallel groups. The next group of articles reviewed shows how microtubules influence the higher order process of spiral growth.
HELICAL MICROTUBULES AND SPIRAL GROWTH
The interphase microtubule array can appear very highly ordered, with microtubules sharing the same orientation over hundreds of micrometers. This degree of ordering means that cellular arrays can be classified easily as transverse, longitudinal, or oblique. In the oblique array, microtubules wind helically around the cell cortex, like the stripes on a barber's pole. The second set of articles throws surprising new light on the connection between this configuration and the spiral growth of plants. (A helix is a coil formed by winding a line around a uniform tube whose radius does not change [e.g., a Slinky, a spring, a corkscrew]. A spiral is a line that winds with a changing radius around a fixed point [e.g., a flat watch spring]. “Spiral growth” should more properly be described as helical growth.)
The three different configurations of the cortical array tend, with notable exceptions, to correlate with the growth status of the cells. In the root, elongating cells tend to have transverse arrays, and as they stop expanding, they tend to assume longitudinal patterns. In maize roots, the zones of transverse and longitudinal microtubules are separated by cells with left-handed oblique helices (Liang et al., 1996). Farther away from the root tip, beyond the zone of cells with longitudinal microtubules, is a fourth zone that also has oblique microtubules, except they are of the opposite helical sign. That entire fields of cells can have microtubule arrays angled in the same direction was confirmed in Arabidopsis. Clues to the genetic basis of this spatial regularity have been reported. Wild-type Arabidopsis grows straight, but two mutations, spiral1 and spiral2, grow with a right-handed twist (Furutani et al., 2000). Axial organs of the mutants grow spirally, and files of cells in the outer epidermis have a right-handed twist. This could be reversed to a left-handed twist by the addition of low amounts of a microtubule-depolymerizing herbicide, propyzamide, or microtubule-stabilizing taxol. Although it seems paradoxical, the microtubules in the right-hand-growing spiral mutants were arranged in left-hand helices (Figure 1).
Figure 1.
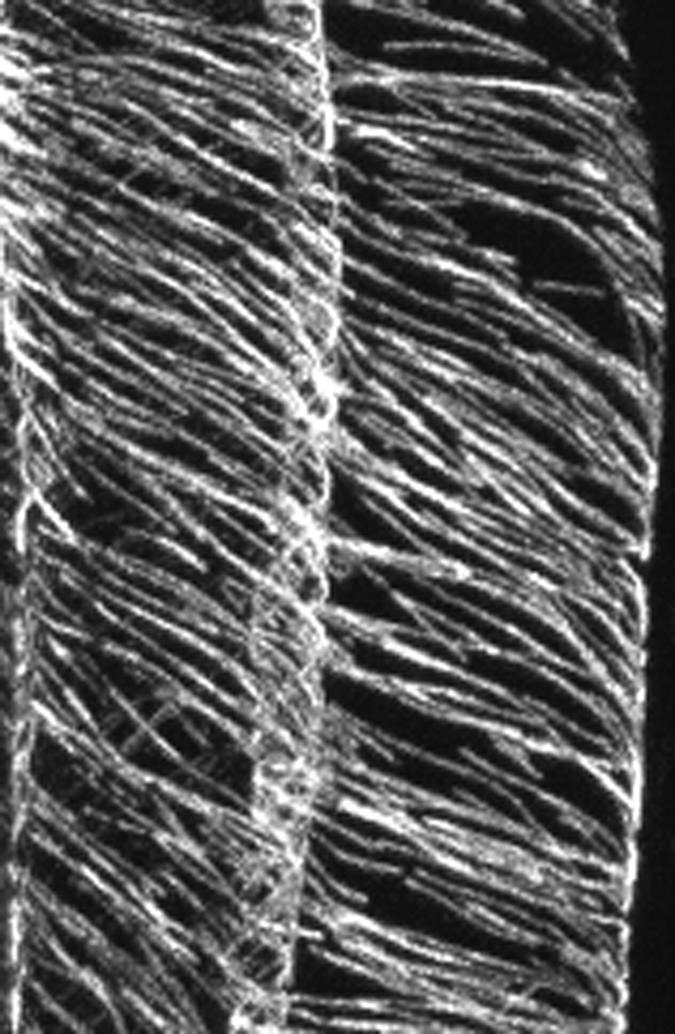
Microtubules in the spiral Mutant.
In the spiral mutant of Arabidopsis, the cortical microtubules of two epidermal cells wind around the cortex in left-handed S-shaped helices. Each single fluorescent strand is likely to represent a bundle of several microtubules. (Figure courtesy of Keiko Sugimoto.)
Subsequently, this group provided further molecular details regarding microtubular involvement. Thitamadee et al. (2002) generated suppressor mutants of spiral1 and spiral2, termed lefty1 and lefty2. As their name suggests, these mutants grow in a left-handed helix, contrary to spiral1 and spiral2. These suppressor mutants also reversed the helical sign of the cortical microtubule arrays in spiral, from a left-handed (S) to a right-handed (Z) helix. Intriguingly, these mutations result from single nucleotide changes in α-tubulin genes that affect the intradimer interface of the protein. One explanation suggested by this finding is that the helical sign of the microtubule array is a larger reflection of the chirality of the microtubules: the way in which the slight stagger between tubulin subunits in adjacent protofilaments imparts a helical structure to the microtubule itself. It is not known whether these mutant tubulins alter the basic microtubule lattice, thereby changing the helical sign in which the protein subunits appear to wind around the microtubule wall. If this proves correct, then imaginative explanations will be required for how the helical nature of the microtubule itself is negotiated into the helical sign of microtubules in an array. It is not easy to imagine how, but another explanation could involve microtubule dynamics, because the helical sign of the cortical microtubule array can be affected by agents that stabilize (such as taxol and MOR1) and destabilize (the herbicide propyzamide) microtubules (Furutani et al., 2000; Thitamadee et al., 2002; Wasteneys, 2002).
HOW DOES THE LEFT HAND KNOW WHAT THE RIGHT HAND DOETH?
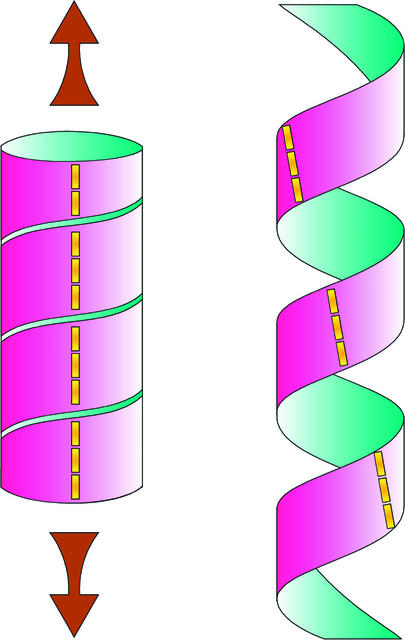
An intriguing finding from the spiral and lefty mutants is that the microtubular helices are of the opposite sign to the direction of spiral growth. How does this occur? Although there may be several layers of cellulose microfibrils in the cell wall, it is believed that the innermost layers, adjacent to the plasma membrane, exert the strongest influence on the direction of growth. When, as in the wild type, the microtubules (and presumably the cellulose microfibrils) are wrapped transversely around the cell, turgor pressure is converted to a directional force perpendicular to the restraining “hoops” (more likely a flat-pitched helix) of cellulose. Cellulose microfibrils lying side by side within a layer are held together by cross-linking glycans. To expand, these cross-links are cut, allowing the cellulose microfibrils to be teased apart under pressure and the cell to expand perpendicularly to them. According to Furutani et al. (2000), cell expansion perpendicular to a left-handed helix of cellulose microfibrils would result in a right-handed twist to the epidermis and of the inner tissues. Figure 2 shows a possible model for this process.
Figure 2.
Helices Unwind When They Are Stretched.
A cylinder, whose wall has a right-handed (Z) helical construction, unwinds in the opposite direction as it is stretched. Although the stretched helix maintains its right-handed construction (albeit with a steeper pitch), markers on its surface (vertical dashed line) are rotated to the left by the unwinding to form a contrary, left-handed S-shaped helix. (Figure courtesy of Tracey Cross.)
The observations of Furutani et al. (2000) and Thitamadee et al. (2002) suggest that microtubules are somehow involved in twisting growth and as such could be part of the mechanism for circumnutation, in which plants twine around growth supports. In 1961, Frei and Preston showed how the helicity of the cell wall translates into spiral growth. They took a strand of the filamentous marine alga Chaetomorpha and suspended it in a beaker of seawater. Tied to the lower end was a horizontal wire that provided an indicator showing the direction in which the filament twisted as it grew. This alga has a cell wall in which the cellulose is wrapped in separate concentric layers of a left-handed helix, which is flat (like a compressed spring), and a right-handed helix, which is steep (like a stretched spring). When seawater was replaced by distilled water (which would make the protoplast turgid), the filament twisted more rapidly in the same direction, and when placed in a hypertonic Suc solution (which would make the protoplast shrink), it twisted sharply in the reverse direction. It was concluded that spiral growth was induced by the stretching (and therefore twisting) of the helically arranged microfibrils in response to the wall stresses imposed by turgor pressure. Therefore, the twist is a reflection of the basically helical structure of the cell wall.
These elegant experiments on the cellulosic wall, conducted before the discovery of microtubules, were interpreted in terms of the multinet growth hypothesis, which stated that the angle of helically deposited wall lamellae is altered during subsequent cell expansion (Roelofsen and Houwink, 1953; Preston, 1982). More recent work on the filamentous freshwater alga Spirogyra confirmed that the angle adopted by cortical microtubules is changed similarly as cell size is modulated by changing the osmotic concentration of the medium to alter turgor pressure (Iwata, 1995; Iwata et al., 2001). In the experiments of Frei and Preston (1961), the angle of the cellulose helices changed as the cells elongated, with the flat (nearly transverse) helix steepening as it became stretched by growth. To do this, the helix “unwinds” and imparts a twist to the overall growth (Figure 2). However, files of cells in an Arabidopsis root are not equivalent to free filaments, because they are bound together to form a complex tissue. If individual cells are not free to rotate about their axes, then it can be imagined that the unwinding of cellulose helices oriented in the same direction around the epidermis could create a torsional force that would twist the entire growth axis. If, as shown in Figure 2, the stretching of a helix causes it to unwind in the opposite direction, this could account for the contrary signs of the microtubule helices and the spiral growth of the Arabidopsis mutants spiral and lefty.
CONCLUDING REMARKS
The importance of the articles reviewed here is that we are now beginning to understand how microtubules build the arrays necessary for an organized cell wall to support axial growth. Because cortical microtubules do not always parallel the nascent cellulose microfibrils, it is clear that there is not an obligatory one-to-one relationship. The loosening of this stricture, from time to time, has encouraged the thought that microtubules are not necessary for organized cellulose deposition, that cellulose patterns are self-organizing, and that this feeds back across the membrane to the alignment of microtubules. Although we are no nearer to understanding the circumstances under which microtubule-microfibril parallelism can be uncoupled, the elongation mutants mor1 and bot1/fra2 reaffirm the basic importance of microtubules for proper wall organization. In the previous issue of The Plant Cell, Burk and Ye (2002) used a combination of field emission scanning electron microscopy (to study the wall) and immunofluorescence (to study the microtubules). They were able to demonstrate that the disorganized microtubules of the katanin mutant fra2 correlated with the loss of oriented cellulose deposition in primary and secondary cell walls and with the formation of abnormal bundles of cellulose. The use of mutants as genetic tools is more selective than the use of antimicrotubule herbicides, which are challenged as having unpredicted side effects, and these results were suggested to support a direct role for microtubules in controlling wall organization (Burk and Ye, 2002).
Another important outcome from the spiral studies is that the cell wall and its cytoskeletal scaffolding are seen to be expressions of a basically helical structure. This has important implications for any model that tries to unify microtubules and the synthetic machinery for cellulose. By showing images of both the cell wall and the microtubular cytoskeleton, Burk and Ye (2002) have bridged two areas that often are examined separately. Early electron microscopy studies concentrated on the overall patterns of cellulose microfibrils in different lamellae and on how these changed with growth, but relatively little such work is performed today. Rather than focusing too closely on who does what in the microtubule-microfibril debate, it might be timely—as Burk and Ye (2002) have shown—to use combinations of whole-cell methods to study these higher order phenomena.
References
- Barroso, C., Chan, J., Allan, V., Doonan, J., Hussey, P., and Lloyd, C.W. (2000). Two kinesin-related proteins associated with the cold stable cytoskeleton of carrot cells: Characterization of a novel kinesin, DcKRP120-2. Plant J. 24, 859–868. [DOI] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Hofte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.-H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule severing protein. Plant Cell 14, in press. [DOI] [PMC free article] [PubMed]
- Chan, J., Jensen, C.G., Jensen, L.C.W., Bush, M., and Lloyd, C.W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA 96, 14931–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Rutten, T., and Lloyd, C.W. (1996). Isolation of microtubule-associated proteins from carrot cytoskeletons: A 120 kDa MAP decorates all four microtubule arrays and the nucleus. Plant J. 10, 251–259. [DOI] [PubMed] [Google Scholar]
- Erhardt, M., Stoppin-Mellet, V., Campagne, S., Canaday, J., Mutterer, J., Fabian, T., Sauter, M., Muller, T., Peter, C., Lambert, A.-M., and Schmit, A.C. (2002). The plant Spc98p homologue colocalizes with γ-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 115, 2423–2431. [DOI] [PubMed] [Google Scholar]
- Frei, E., and Preston, R.D. (1961). Cell wall organization and wall growth in the filamentous green algae Cladophora and Chaetomorpha. II. Spiral growth and spiral structure. Proc. R. Soc. B 155, 55–81. [Google Scholar]
- Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., Thitamadee, S., Shikanai, T., and Hashimoto, T. (2000). The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Granger, C.L., and Cyr, R.J. (2001). Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP-MBD. Protoplasma 216, 201–214. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., and Hawkins, T.J. (2001). Plant microtubule-associated proteins: The HEAT is off in temperature-sensitive mor1. Trends Plant Sci. 6, 389–392. [DOI] [PubMed] [Google Scholar]
- Iwata, K. (1995). Regulation of the orientation of cortical microtubules in Spirogyra cells. J. Plant Res. 108, 531–534. [DOI] [PubMed] [Google Scholar]
- Iwata, K., Tazawa, M., and Itoh, I. (2001). Turgor pressure regulation and the orientation of cortical microtubules in Spirogyra cells. Plant Cell Physiol. 42, 594–598. [DOI] [PubMed] [Google Scholar]
- Jiang, C.-J., and Sonobe, S. (1993). Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J. Cell Sci. 105, 891–901. [DOI] [PubMed] [Google Scholar]
- Joshi, H.C., and Palevitz, B.A. (1996). γ-Tubulin and microtubule organization in plants. Trends Cell Biol. 6, 41–44. [DOI] [PubMed] [Google Scholar]
- Lambert, A.-M., and Lloyd, C.W. (1994). The higher plant microtubule cycle. In Microtubules, J.S. Hyams and C.W. Lloyd, eds (New York: Alan R. Liss), pp. 327–341.
- Lancelle, S.A., Callaham, D.A., and Hepler, P.K. (1986). A method for rapid freeze fixation of plant cells. Protoplasma 131, 153–165. [Google Scholar]
- Liang, B.M., Dennings, A.M., Sharp, R.E., and Baskin, T.I. (1996). Consistent handedness of microtubule helical arrays in maize and Arabidopsis primary roots. Protoplasma 190, 8–15. [Google Scholar]
- Lloyd, C., and Hussey, P. (2001). Microtubule-associated proteins in plants: Why we need a MAP. Nat. Rev. Mol. Cell Biol. 2, 40–47. [DOI] [PubMed] [Google Scholar]
- McClinton, R.S., Chandler, J.S., and Callis, J. (2001). cDNA isolation, characterization, and protein intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma 216, 181–190. [DOI] [PubMed] [Google Scholar]
- McNally, F.J., Okawa, K., Iwamatsu, A., and Vale, R.D. (1996). Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J. Cell Sci. 109, 561–567. [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman, J.-P., Jiang, W., Hunter, T., and Margolis, R.L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteris, E., Apostolakos, P., Graf, R., and Galatis, B. (2000). Gamma-tubulin colocalizes with microtubule arrays and tubulin paracrystals in dividing vegetative cells of higher plants. Protoplasma 210, 179–187. [Google Scholar]
- Popov, A.V., Severin, F., and Karsenti, E. (2002). XMAP215 is required for the microtubule-nucleating activity of centrosomes. Curr. Biol. 12, 1326–1330. [DOI] [PubMed] [Google Scholar]
- Preston, R.D. (1982). The case for multinet growth in growing walls of plant cells. Planta 155, 356–363. [DOI] [PubMed] [Google Scholar]
- Roelofsen, P.A., and Houwink, A.L. (1953). Architecture and growth of the primary wall in some plant hairs and in the Phycomyces spo-rangiosphore. Acta Bot. Neerl. 2, 218–225. [Google Scholar]
- Smertenko, A., Saleh, N., Igarashi, H., Mori, H., Hauser-Hahn, I., Jiang, C.J., Sonobe, S., Lloyd, C.W., and Hussey, P.J. (2000). A new class of microtubule-associated proteins in plants. Nat. Cell Biol. 2, 750–753. [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet, V., Gaillard, J., and Vantard, M. (2002). Functional evidence for in vitro microtubule severing by the plant katanin homolog. Biochem. J. 365, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee, S., Tuchihara, K., and Hashimoto, T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417, 193–196. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., Popov, A., Kinoshita, K., Ashford, A.J., Rybina, S., Pozniakovsky, A., Mayer, T.U., Walczak, C.E., Karsenti, E., and Hyman, A.A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Vasquez, R.J., Gard, D.L., and Cassimeris, L. (1994). XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 127, 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115, 1345–1354. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Wymer, C., and Lloyd, C. (1996). Dynamic microtubules: Implications for cell wall patterns. Trends Plant Sci. 1, 222–228. [Google Scholar]
- Yasuhara, H., Muraoka, M., Shogaki, H., Mori, H., and Sonobe, S. (2002). TBMP200, a microtubule bundling polypeptide isolated from telophase tobacco BY-2 cells is a MOR1 homologue. Plant Cell Physiol. 43, 595–603. [DOI] [PubMed] [Google Scholar]
- Yuan, M., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc. Natl. Acad. Sci. USA 91, 6050–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]