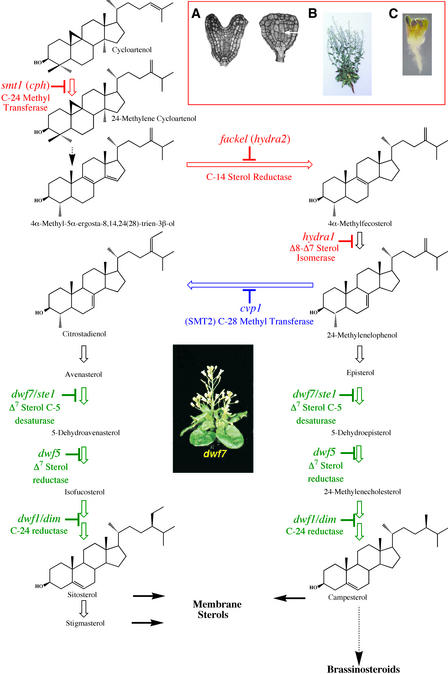
The molecular genetic, and biochemical analysis of sterol-deficient mutants in Arabidopsis strongly suggests an essential role for sterols in regulating multiple events in plant development, independent of their conversion to brassinosteroids (BRs). Embryogenesis, cell elongation, vascular differentiation, and hormone signaling all are affected by the alteration of sterol levels, but the molecular mechanism underlying these pleiotropic effects on development is not understood. Several models have been proposed in recent years, and they are discussed briefly below. A critical test of any of these models is whether they provide a clear explanation of why mutations that affect enzymes upstream of 24-methylenelophenol show embryonic defects but those downstream do not (Figure 1).
Figure 1.
Biosynthetic Pathways of Sterols Required for Normal Plant Development.
Cycloartenol, a polycyclic sterol intermediate unique to plants, is alkylated at C-24 by SMT1 to produce a triene in a series of steps (dashed arrow). FK, a C-14 sterol reductase, and HYD1, a Δ8-Δ7 isomerase, act on the triene to yield 24-methylenelophenol, a branch point in the pathway. Parallel pathways lead to the membrane sterols sitosterol and campesterol. SMT2 controls the ratio of the two sterols. Campesterol also serves as the progenitor of brassinosteroids, which are required for normal postembryonic development. Defects in embryogenesis are observed in mutants upstream of 24-methylenelophenol (in red) but not in downstream mutants (in blue or green). Mutants shown in green, such as dwf7/stel (pictured), show typical brassinosteroid-deficient phenotypes that can be rescued by brassinosteroid treatment.
(A) Morphological differences between wild-type (left) and fk (right) embryos at the heart stage. The arrow shows the lack of normal cell division and elongation in the central cells.
(B) An adult fk mutant shows some of the characteristics of brassinosteroid-deficient mutants, but it cannot be rescued by brassinosteroid treatment.
(C) The fk mutant often has multiple cotyledons.
Sterols are isoprenoid-derived lipids that have diverse and essential functions in all eukaryotes. Bulk sterols are integral components of the membrane lipid bilayer, where, in conjunction with phospholipids, they regulate membrane permeability and fluidity. In animals, cholesterol alone serves this structural function, whereas plant membranes consist of a variable mixture of several sterols, with sitosterol usually predominating (Hartmann, 1998). A typical plant membrane sterol profile is illustrated by light-grown 23-day-old pea seedlings, which contain ∼50% sitosterol, 25% stigmasterol, and <10% each campesterol, isofucosterol, and cholesterol in total membrane fractions (Nomura et al., 1999). As a consequence of their prominent role in maintaining membrane integrity and function, sterols also indirectly affect the activity of integral membrane proteins, including enzymes, ion channels, and signal transduction components. Moreover, animal membranes contain liquid-ordered assemblies of cholesterol and sphingolipid termed lipid rafts, which activate signal transduction by promoting a microenvironment in which the close contact of interacting proteins is enhanced (Simons and Toomre, 2000). Such structures might be expected to occur in plant membranes, but they have not been identified to date.
In addition to their structural role, sterols function as biosynthetic precursors of steroid hormones such as glucocorticoids, androgens, and estrogens in animals, ecdysteroids in insects, antheridiol and oogoniol in fungi, and BRs in plants. The role of animal steroids in the regulation of embryonic and postembryonic development along with adult homeostasis is well known (Beato et al., 1995). Like their animal steroid counterparts, BRs have been shown to regulate gene expression, stimulate cell division and differentiation, and modulate reproductive biology (reviewed by Clouse and Sasse, 1998). BRs also mediate growth responses unique to plants, including the promotion of cell elongation in the presence of a complex cell wall and influencing multiple developmental responses to darkness and light. The phenotype of BR-deficient and BR-insensitive mutants has been described extensively and includes extreme dwarfism, altered leaf morphology, reduced fertility or male sterility, delayed senescence, and altered vascular development, implicating BRs in all of these developmental processes (Clouse and Feldmann, 1999).
ARABIDOPSIS MUTANTS IN STEROL BIOSYNTHESIS
Plant sterols are derived from cycloartenol via a series of methylations, reductions, isomerizations, and desaturations (Figure 1). Because brassinolide, the most biologically active BR in many plants, is derived from campesterol, it would be expected that mutations leading to reductions in the levels of this sterol progenitor would result in BR deficiency and a mutant phenotype similar to BR-deficient dwarfs (Yokota, 1997). This has been shown for mutants affected in steps directly upstream from campesterol. The Arabidopsis mutants dwf7/ste1, dwf5, and dwf1/dim are blocked in each successive step in the conversion of episterol to campesterol. They each have mutant phenotypes similar to BR-deficient mutants downstream of campesterol, although not as severe. Moreover, the mutant phenotypes are rescued by treatment with brassinolide and biosynthetic intermediates in the BR-specific pathway (Klahre et al., 1998; Choe et al., 1999a, 1999b, 2000). Sitosterol differs from campesterol only in the degree of alkylation at C-24, and DWF7/STE1, DWF5, and DWF1/DIM also catalyze the conversion of avenasterol to sitosterol in a pathway parallel to the episterol-to-campesterol reactions (Figure 1). Thus, these mutants are deficient in both sitosterol and campesterol and might have consequent defects in membrane structure and function. However, the rescue of these mutants by BRs suggests that hormone deficiency rather than altered membrane integrity plays a greater role in these phenotypic alterations.
A BR-independent role for sterols in plant development was suggested ∼2 years ago when it was demonstrated that the Arabidopsis mutants sterol methyltransferase1 (smt1) and fackel (fk), both of which are affected in early steps of sterol biosynthesis, showed severe defects in embryogenesis that had not been reported previously in BR mutants and that could not be rescued by BR treatment (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000). SMT1 is a C-24 methyltransferase responsible for the conversion of cycloartenol to 24-methylenecycloartenol in the first step of sterol biosynthesis, whereas FK acts downstream of SMT1 with C-14 sterol reductase activity. Mutants affected in these steps have reduced sterol and BR levels and show some of the characteristics of BR mutants in adult plants, but they are affected uniquely at specific stages of embryo development. Both fk and smt1 show a lack of asymmetrical divisions in cells at the center of the globular-stage embryo, and although wild-type embryos progress to the heart stage, mutant embryos remain globular and disorganized (Figure 1). Multiple shoot apical meristems are initiated in the mutant, and developing seedlings often have more than two cotyledons, which appear to be attached directly to the root, with little development of the hypocotyls.
More recently, hydra2 (hyd2) has been identified as an allele of fk and cephalopod has been identified as an allele of smt1 (Schrick et al., 2002; Souter et al., 2002). Moreover, the hyd1 mutant blocks the Δ8-Δ7 isomerase step immediately downstream of fk. These new mutants share the same embryonic and seedling patterning defects of fk and smt1. Like fk, hyd2 shows severely aberrant vascular development, including duplicated vascular systems and discontinuities of vascular strands. Interestingly, this vascular abnormality was rescued partially when hyd mutants were crossed with auxin-resistant mutants such as axr1-12, suggesting that altered auxin signaling in hyd may be partially responsible for this phenotype (Souter et al., 2002). Further supporting this view, auxin-responsive promoters fused to reporter genes, such as DR5::GUS and ACS1::GUS, showed ectopic expression in the hyd mutant background. Root cell file patterning also is disrupted in the hyd mutants, including abnormal root hair formation. The root abnormalities were rescued by crossing hyd1 and hyd2 to the dominant ethylene-resistant mutant etr1-1, suggesting that hyd mutants also are defective in ethylene signaling (Souter et al., 2002).
In the current issue of The Plant Cell, Carland et al. (pages 2045–2058) reveal a mutation that affects yet another step in sterol biosynthesis. The cotyledon vascular pattern1 (cvp1) mutant shows a discontinuous, poorly axialized venation pattern in cotyledons. Like the downstream mutants dwf7, dwf5, and dwf1, cvp1 does not exhibit the altered embryogenesis of fk, hyd1, and smt1, but unlike the downstream mutants, its altered phenotype is not rescued by BR treatment. A role for BRs in vascular differentiation is apparent from examination of the BR-deficient mutant constitutive photomorphogenesis and dwarfism, which exhibits unequal division of the cambium, producing extranumerary phloem cell files at the expense of xylem cells (Szekeres et al., 1996). The sterol- and BR-deficient mutant dwf7 shows the same increase in phloem versus xylem cells, and the number of vascular bundles is reduced from eight in the wild type to six in the mutant. Furthermore, the spacing between vascular bundles is irregular, and two vascular bundles can be joined without a separating layer of parenchyma cells (Choe et al., 1999b). However, the unique vascular patterning defects observed in cvp1 and hyd/fk mutants clearly indicate a BR-independent role for sterols in vascular differentiation.
CVP1 encodes SMT2, which acts at a branch point in sterol biosynthesis, directing 24-methylenelophenol toward sitosterol and away from campesterol and downstream BRs (Figure 1). The ratio of campesterol to sitosterol has been shown to modulate Arabidopsis growth. Plants overexpressing SMT2 have increased sitosterol/campesterol ratios and exhibit reduced stature and growth that can be rescued by BR treatment. Conversely, plants underexpressing SMT2 have high campesterol levels at the expense of sitosterol and show reduced growth, increased branching, and low fertility that cannot be restored by BR treatment (Schaeffer et al., 2001). As expected, cvp1 mutants have increased campesterol and reduced sitosterol, and their growth defects cannot be rescued by BR (Carland et al., 2002).
EFFECT OF ALTERED STEROL COMPOSITION ON MEMBRANE AND CELL WALL STRUCTURE AND FUNCTION
One obvious explanation for the defective growth phenotypes in sterol-deficient mutants is altered membrane structure and function. The smt1 mutant is hypersensitive to calcium ions and was proposed to result from altered membrane permeability in the mutant (Diener et al., 2000). Membrane trafficking of regulatory proteins also could be altered by the unusual sterol composition of the membranes. However, Souter et al. (2002) showed by immunolocalization that the predicted components of the auxin efflux carrier system, PIN1 and PIN2, are localized normally in hyd1 and hyd2/fk mutants. Gross structural abnormalities in membranes caused by sitosterol and campesterol deficiencies would be expected to affect cell elongation and division and thus diverse aspects of development.
However, it is difficult to reconcile the specific phenotypes observed in many of the sterol-deficient mutants with global defects in membrane structure. For example, basic cell division is not affected globally in fk, because mutant callus culture cells proliferate and cell expansion defects in the embryo are not observed until the globular stage, by which time ∼60 cells already have undergone division (Schrick et al., 2000). Furthermore, as seen in Table 1, downstream mutants such as dwf1/dim are affected as severely in sterol composition as fk and smt1, yet they do not exhibit embryogenic defects and can be rescued by BR treatment. One possible explanation for the normal embryogenesis in downstream mutants is that redundant genes for dwf7, dwf5, and dwf1 are expressed specifically during embryogenesis. However, the rescue of growth defects by BR in these mutants suggests that altered bulk sterol composition (i.e., sterols downstream of 24-methylenelophenol) does not prevent cell expansion in response to BR. It is possible that 24-methylenelophenol or an undetected upstream sterol acts in concert with BR to promote elongation, but both smt1 and fk hypocotyls respond to exogenous BR treatment by elongating to some extent (Diener et al., 2000; Schrick et al., 2000).
Table 1.
Endogenous Sterol Levels in Wild-Type and Sterol Biosynthetic Mutant Plants
| Mutant/Wild-Type Ratio
|
|||||||
|---|---|---|---|---|---|---|---|
| Sterol | smt1-1 | fk-X224 | fk-J79 | hyd2 | hyd1 | dim/dwf1 | cvp1-3 |
| 24-Methylenecholesterol | 2.50 | U/T | –a | – | – | 30.7/T | 25.2 |
| Campesterol | 1.31 | T/13.1 | 0.51 | 0.00 | 0.12 | 0.09 | 3.59 |
| Isofucosterol | 1.86 | U/T | – | – | – | 53.0 | 1.06 |
| Sitosterol | 0.57 | 0.77 | 0.50 | 0.04 | 0.02 | 0.06 | 0.29 |
| Stigmasterol | – | 1.18 | – | 3.22 | 1.82 | 0.31 | 0.51 |
| Cholesterol | 5.42 | 1.87 | – | – | – | 0.43 | 3.53 |
Data derived from Diener et al. (2000), Schrick et al. (2000), Jang et al. (2000), Souter et al. (2002), Klahre et al. (1998), and Carland et al. (2002). T, trace; U, undetected.
–, no data.
Alterations in membrane structure attributable to abnormal sterol levels also might affect membrane-bound signal transduction elements. The altered auxin and ethylene signaling exhibited by hyd mutants could be explained by such effects (Souter et al., 2002). Also, the severe cvp1-3 allele has some of the features of BR mutants (Carland et al., 2002), which might arise from the faulty function of the membrane-bound BRI1 receptor kinase involved in BR perception (Friedrichsen and Chory, 2001). Another possible effect of altered sterol content is on lipid raft assembly (Simons and Toomre, 2000). The lipid raft concept is intriguing, but it remains highly speculative until it can be shown that (1) plants contain lipid rafts in their membranes, (2) the structure of these rafts is affected by altering the campesterol and sitosterol ratios, and (3) signaling molecules associated with the smt1, fk, hyd1, and cvp1 mutants occur in these rafts.
The synthesis and orientation of cellulose microfibrils is an essential process in cell elongation and division. Recently, Peng et al. (2002) showed that sitosterol-β-glucoside is a primer for cellulose synthesis in plants. Thus, plants deficient in sitosterol might be deficient in cellulose biosynthesis, and this was proposed to be at least a partial cause of the phenotype in sterol-deficient mutants such as fk. However, if cellulose biosynthesis cannot occur normally at reduced sitosterol levels, it is difficult to explain the rescue of dwf1/dim to the wild type by BR treatment, because this mutant is more severely sitosterol deficient than smt1 or fk (Table 1).
POSSIBLE SIGNALING ROLES FOR PLANT STEROLS
Recent studies in animal systems have shown that cholesterol can serve as a signaling molecule in its own right, without conversion to steroid hormones (Farese and Herz, 1998; Edwards and Ericsson, 1999). Such a role also can be postulated for plant sterols. The 60–amino acid homeodomain is a sequence-specific DNA binding segment that is highly conserved among eukaryotic transcription factors involved in regulating the spatial control of cell differentiation and pattern formation (Gehring et al., 1990). The homeodomain–Leu zipper (HD-ZIP) family of putative plant transcription factors possesses a unique arrangement of an N-terminal homeodomain followed by a Leu zipper. A subset of the HD-ZIP family, which is involved in the morphogenesis of a variety of plant organs, also contains a C-terminal region with sequence similarity to the mammalian sterol/lipid binding domain termed START (steroidogenic acute regulatory protein–related lipid transfer) (Ponting and Aravind, 1999).
Steroidogenic acute regulatory protein plays an important role in the initial stages of steroid hormone biosynthesis by transferring cholesterol from the outer mitochondrial membrane to a series of enzymes on the inner membrane, where the sterol is converted to pregnenolone (Kallen et al., 1998). However, the appearance of a START domain in plant HD-ZIP proteins suggests a possible role for sterol binding in regulating the activity of these important modulators of early development. Arabidopsis HD-ZIP proteins that contain a START domain include ATHB-8, which is expressed specifically in procambial cells of embryos and developing organs (Sessa et al., 1998); INTERFAS-CICULAR FIBERLESS1/REVOLUTA, which spatially regulates fiber differentiation in Arabidopsis (Zhong and Ye, 1999; Ratcliffe et al., 2000); GLABRA2, which is required for the regulation of trichome development and root hair formation (Masucci et al., 1996); and PHABULOSA (PHB) and the related PHAVOLUTA (PHV), which are involved in the perception of radial positional information in the leaf primordium, leading to the differentiation of adaxial versus abaxial surfaces (McConnell et al., 2001).
The functional importance of the plant START domain is demonstrated by the fact that all five dominant phb mutations as well as the four known phv lesions lie within this region. A model for the role of PHB in the development of leaf polarity has been proposed in which the protein is initially distributed equally throughout the young, unpolarized leaf primordium. A ligand (possibly a sterol?) is distributed unequally within the leaf primordium as development proceeds, activating PHB in a position-dependent manner (McConnell et al., 2001). Such a model also could provide an explanation for why sterol biosynthetic mutants upstream of 24-methy-lenelophenol show embryonic defects but those downstream do not. In this situation, 24-methylenelophenol or its derivative would bind to the regulatory domain of a transcription factor that controls stage-specific events in embryogenesis. These events would be disrupted in smt1, fk, and hyd1, which fail to produce sufficient 24-methylenelophenol levels, whereas dwf7, dwf5, and dwf1 have normal 24-methylenelophenol concentrations, and thus normal embryogenesis, but they remain BR deficient as a result of insufficient campesterol production. Thus, downstream mutants can be rescued by BR treatment, whereas upstream mutants cannot. Interestingly, exogenous application of bulk sterols, such as sitosterol and campesterol, fails to rescue smt1, fk, and hyd1 (Diener et al. 2000; Jang et al., 2000; Souter et al., 2002). Because sterols are less hydrophilic than BRs, this could be an uptake problem, but metabolic studies with Arabidopsis seedlings grown in liquid culture show that campesterol is taken up and converted to campestanol efficiently (Noguchi et al., 1999). A potentially informative experiment, which has not yet been reported, would involve determining if 24-methylenelophenol application rescues upstream mutants.
Such a model also could account for the specific vascular defects in cvp1, particularly in view of the known effects of INTERFASCICULAR FIBERLESS1 and ATHB-8 on vascular development (Sessa et al., 1998; Zhong and Ye, 1999). However, because cvp1 accumulates high levels of 24-methylenelophenol, an alternative sterol, perhaps originating from citrostadienol, would need to be postulated as the putative ligand. CVP1 is expressed throughout the embryo (Carland et al., 2002), yet mutations that affect this enzyme function do not lead to the type of embryonic defects observed in smt1, fk, and hyd1, consistent with a possible role of 24-methylenelophenol deficiency in the distinctive embryonic phenotype of upstream mutants.
Because mutants in the currently characterized HD-ZIP family members with START domains do not have embryonic defects identical to those of smt1, fk, and hyd1, an uncharacterized member of this family or an unrelated regulatory protein with specific sterol binding properties would need to be postulated as part of this model. Moreover, it has not been shown by direct experimentation with labeled lipids that plant START domains actually bind sterols. Specificity of binding also would be essential for this model, and it should be noted that the mammalian steroidogenic acute regulatory protein has affinity for both its physiological substrate cholesterol and the plant sterol sitosterol (Kallen et al., 1998).
A second scenario for plant sterol signaling in embryogenesis can be derived by analogy from mammalian systems, in which cholesterol has been shown to play an essential role in embryonic development, independent of its membrane structural and steroid precursor functions (Farese and Herz, 1998). Hedgehog proteins participate in embryonic pattern formation by a process of proteolytic cleavage followed by direct covalent attachment of cholesterol to the N-terminal peptide. This sterol-protein ligand then binds to a cell surface receptor, which activates a signaling pathway essential for embryogenesis. Moreover, a number of free oxysterols, such as 27-hydoxycholesterol, have been found to serve as ligands for nuclear receptors, transcriptionally activating the expression of specific genes (Edwards and Ericsson, 1999). Although plants apparently do not have members of the superfamily of nuclear receptors, it is well known that the BRI1 receptor kinase is part of a complex that binds brassinolide (Friedrichsen and Chory, 2001). Of the hundreds of predicted plant receptor kinases in Arabidopsis, it is conceivable that one or more might function as a receptor for specific sterols, leading to a signal transduction cascade that activates specific events in embryogenesis. Again, as in the HD-ZIP scenario, 24-methylenelophenol or its derivative might serve as the ligand for such a receptor.
CONCLUSION
The identification of Arabidopsis mutants that affect seven steps in the biosynthetic pathway from cycloartenol to campesterol and sitosterol has greatly advanced our understanding of the multiple developmental processes affected by the alteration of sterol composition in plants. Numer-ous testable models have been advanced to account for the diversity of pathways affected by sterol defects, and it is likely, as with cholesterol, that both structural membrane functions and specific signaling events can be attributed to specific plant sterols.
Acknowledgments
Thanks to J.-C. Jang, S. Choe, and K. Feldmann for providing photographs of sterol mutants. Work in the author's laboratory is supported by the National Science Foundation (Integrative Plant Biology), the U.S. Department of Agriculture National Research Initiative (Plant Growth and Development), and the North Carolina Agricultural Research Service.
References
- Beato, M., Herrlich, P., and Schutz, G. (1995). Steroid hormone receptors: Many actors in search of a plot. Cell 83, 851–857. [DOI] [PubMed] [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14, 2045.–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Gregory, B.D., Ross, A.S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. a). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C.P., Gregory, B.D., Ross, A.S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. b). The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to bras-sinosteroid biosynthesis. Plant Cell 11, 207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Tanaka, A., Noguchi, T., Fujioka, S., Takatsuto, S., Ross, A.S., Tax, F.E., Yoshida, S., and Feldmann, K.A. (2000). Lesions in the sterol delta reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 21, 431–443. [DOI] [PubMed] [Google Scholar]
- Clouse, S., and Feldmann, K. (1999). Molecular genetics of brassinosteroid action. In Brassinosteroids: Steroidal Plant Hormones, A. Sakurai, T. Yokota, and S. Clouse, eds (Tokyo: Springer), pp. 163–190.
- Clouse, S., and Sasse, J. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., Li, H., Zhou, W., Whoriskey, W.J., Nes, W.D., and Fink, G.R. (2000). Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, P.A., and Ericsson, J. (1999). Sterols and isoprenoids: Signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 68, 157–185. [DOI] [PubMed] [Google Scholar]
- Farese, R.V., Jr., and Herz, J. (1998). Cholesterol metabolism and embryogenesis. Trends Genet. 14, 115–120. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D., and Chory, J. (2001). Steroid signaling in plants: From the cell surface to the nucleus. Bioessays 23, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Gehring, W.J., Muller, M., Affolter, M., Percival-Smith, A., Billeter, M., Qian, Y.Q., Otting, G., and Wuthrich, K. (1990). The structure of the homeodomain and its functional implications. Trends Genet. 6, 323–329. [DOI] [PubMed] [Google Scholar]
- Hartmann, M.-A. (1998). Plant sterols and the membrane environment. Trends Plant Sci. 3, 170–175. [Google Scholar]
- Jang, J.C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kallen, C.B., Billheimer, J.T., Summers, S.A., Stayrook, S.E., Lewis, M., and Strauss, J.F., III (1998). Steroidogenic acute regulatory protein (StAR) is a sterol transfer protein. J. Biol. Chem. 273, 26285–26288. [DOI] [PubMed] [Google Scholar]
- Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., Yoshida, S., and Chua, N.-H. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Takatsuto, S., Sakurai, A. Yoshida, S., Li, J., and Chory, J. (1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120, 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Kitasaka, Y., Takatsuto, S., Reid, J.B., Fukami, M., and Yokota, T. (1999). Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 119, 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L., Kawagoe, Y., Hogan, P., and Delmer, D. (2002). Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295, 147–150. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P., and Aravind, L. (1999). START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24, 130–132. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Riechmann, J.L., and Zhang, J.Z. (2000). INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. Plant Cell 12, 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, A., Bronner, R., Benveniste, P., and Schaller, H. (2001). The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 25, 605–615. [DOI] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., Schmidt, J., and Jurgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Martin, G., Bellini, C., Kuhnt, C., Schmidt, J., and Jurgens, G. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31, 61–73. [DOI] [PubMed] [Google Scholar]
- Sessa, G., Steindler, C., Morelli, G., and Ruberti, I. (1998). The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38, 609–622. [DOI] [PubMed] [Google Scholar]
- Simons, K., and Toomre, D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D., and Lindsey, K. (2002). hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14, 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Yokota, T. (1997). The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 2, 137–143. [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]