Abstract
B cell-activating factor (BAFF) plays a dominant role in the B cell homeostasis. However, excessive BAFF promotes the development of autoreactive B-cells and several antibodies have been developed to block its activity. Bispecific antibodies with added functionality represent the next wave of biologics that may be more effective in the treatment of complex autoimmune disease. The single variable domain from the immunoglobulin new antigen receptor (VNAR) is one of the smallest antibody recognition units that could be combined with monospecific antibodies to develop bispecific agents. We isolated a panel of BAFF-binding VNARs with low nM potency from a semi-synthetic phage display library and examined their functional activity. The anti-BAFF VNARs blocked the binding of BAFF to all three of its receptors (BR3, TACI and BCMA) and the presence of the conserved DXL receptor motif found in the CDR3 regions suggests molecular mimicry as the mechanism of antagonism. One clone was formatted as an Fc fusion for functional testing and it was found to inhibit both mouse and human BAFF with equal potency ex vivo in a splenocyte proliferation assay. In mice, subchronic administration reduced the number of immature and transitional intermediates B cells and mature B cell subsets. These results indicate that VNAR single domain antibodies function as selective B-cell inhibitors and offer an alternative molecular format for targeting B-cell disorders.
Keywords: Anti-BAFF antibodies, Single domain antibodies, Variable domain of the immunoglobulin new antigen receptor (VNAR), B cell proliferation
1. Introduction
Monoclonal antibodies (mAbs) from their discovery in the 1970s have become essential tools for experimental research and diagnostics. The ability to generate high-affinity mAbs to virtually any foreign antigen made them an obvious class of agents for the pharmaceutical industry, and they quickly became key therapeutics for the treatment of human disease (Buss et al., 2012). Next-generation antibodies have been engineered to further increase potency, improve safety and add functionality and a variety of alternative antibody formats such as bispecifics are now moving into the clinic.
Two naturally occurring single chain antibodies, found in camelids and cartilaginous fish, bind their cognate antigen using a single variable domain (VHH and VNAR, respectively), in contrast to conventional antibodies whose binding domain is composed of the combined V domains of Heavy and Light chains (reviewed in Wesolowski et al., 2009). Due to their small size and high chemical and thermal stability, these single domain antibodies are easily reformatted into many different structures, allowing a “plug-and-play” approach to developing multifunctional therapeutic molecules. Although lagging behind VHH single domains in development, VNARs exhibit several unique properties that make them an attractive scaffold for developing novel therapeutics and hold great promise for the future.
VNARs primarily bind antigen via a single, long complementarity-determining region 3 (CDR3), which is oriented by non-canonical cysteine bridges (reviewed in Zielonka et al., 2015a,b). This distinctive feature facilitates access to cryptic epitopes in clefts and cavities, which are often sites of protein–protein interactions (Henderson et al., 2007). Interestingly, VNARs lack a CDR2 region but contain two additional hypervariable regions, HV2 and HV4. The HV2 region, which forms a belt-like structure at the bottom of the molecule, participates in antigen binding (Kovalenko et al., 2013) and can be engineered to independently bind a second target (Zielonka et al., 2015a,b). This versatility in antigen recognition, flexibility in formatting along with favorable biophysical properties offers a variety of novel options for antibody design.
High-affinity VNARs to a variety of target-specific antigens have been isolated from immune libraries (reviewed in Zielonka et al., 2015a,b). As an alternative strategy, we built semi-synthetic phage display libraries based on nurse shark (Ginglymostoma cirratum) VNAR sequence (Häsler and Rutkowski, 2016) and tested their ability to produce high-affinity leads against therapeutically-precedented targets. A library of type 2 VNARs (cysteine bridge between CDR3 and CDR1 loops) was first panned on human BAFF (also known as BLyS or TNFS13B) to compare to the characteristics of belimumab (LymphoStat B, Benlysta), which was isolated from a human single-chain antibody (scFv) phage library (Baker et al., 2003).
BAFF is a TNF superfamily ligand, crucial in the selection and survival of B cells for adaptive immunity. BAFF overproduction appears to play a role in a variety of autoimmune disorders and certain B cell malignancies and BAFF-blocking antibodies have been developed for their treatment, including belimumab, tabalumab, and blisibimod (peptibody) (for reviews see (Stohl, 2012; Stohl and Hilbert, 2012). While the approval of belimumab represents a major advance for mild to moderate Systemic Lupus Erythematosus (SLE), not all patients respond and the development of tabalumab was discontinued due to an insufficient level of efficacy despite evidence of BAFF inhibition (Morais et al., 2015).
Pro-survival signals in addition to BAFF mediate the positive selection of autoreactive B cells (Metzler et al., 2015) and robust efficacy may be difficult to achieve by targeting a single cytokine in such diverse and complex autoimmune diseases. A combination of cytokine antagonists may be more efficacious and could be tailored to particular diseases and patient subgroups. The goal of the study was to isolate VNARs to serve as building blocks for next generation BAFF antagonists.
2. Materials and methods
2.1. Isolation of BAFF-binding VNARs by phage display
A semi-synthetic library (1.6 × 1010 cfu) was constructed based on a Type 2 nurse shark VNAR (OsX-3) by integrating both a partial randomization of the CDR3 and a number of framework mutations. The vector used for both phage display and monomeric VNAR expression a modification of pSEX81 (Progen, Germany) in which a 6XHis tag, a FLAG tag, and an amber stop codon were inserted between the VNAR and the full-length PIII protein of the M13 phage.
Selection of BAFF-interacting VNARs, displayed as a fusion of the PIII protein on M13 bacteriophage, was performed as previously described (Griffiths et al., 1994). Briefly, human BAFF (ProSpec, Germany) was immobilized on Nunc Maxisorp 96 well plates and incubated with an excess of phage rescued from the OsX-3 library. After washing, the bound phages were eluted with triethylamine (100 mM) used to infect Escherichia coliEscherichia coli ER2738 after neutralization. The titer was estimated by the number of antibiotic-resistant colonies and the culture was infected with M13KO7 helper phage to produce phage for the next round of selection. Four rounds of selection were performed using increasingly stringent conditions consisting in progressively reducing the coated BAFF concentration at every round (50, 5, 2.5, and 1 μg/ml) and increasing the washes from 10 to 20.
Each of the four input phage populations was tested for specificity to hBAFF by polyclonal phage ELISA. Briefly 1012 phages were incubated in 96 well plates coated at 1 μg/ml with hBAFF-Fc (Sino Biological, China), human transferrin receptor (Sino Biological), or HSA (Sigma, USA). After a 60-min incubation at room temperature and washing with 0.1% Tween-20 in PBS, the bound phage particles were detected using a specific anti-M13 antibody (GE Healthcare, UK). The DNA sequence of positive clones was determined using the specific oligonucleotide 5′-tcattaggcaccccaggctttacac-3′.
2.2. Screening for BAFF binding and blocking clones
Individual clones were grown in 96 deep-well plates in auto-induction medium (EMD Millipore, Germany) and the periplasmic fraction was extracted by osmotic shock as described in (Muller et al., 2012). For the binding ELISA, Maxisorp plates were coated at 1 μg/ml with either hBAFF-Fc or HSA and periplasmic fraction, in blocking buffer (2.5% non-fat dry milk and 0.1% Tween-20 in PBS) was exposed to the coated surface. After washing in PBS-0.1% Tween, bound VNARs were detected using a peroxidase-conjugated anti-FLAG antibody (Sigma) and absorbance at 450 nm was recorded. Cross-reactivity of selected clones to rhAPRIL (Sigma) was tested using an identical ELISA protocol. For the blocking ELISA, plates were coated at 1 μg/ml with extracellular domains of BR3, TACI, or BCMA (all from Peprotech, USA) and washed with blocking buffer. The pre-blocked periplasmic fraction with the addition of hBAFF-Fc (5 nM in the case of BR3 and 0.5 nM in the case of TACI and BCMA) was then exposed to the coated surface, and bound hBAFF was detected with a peroxidase-conjugated anti-human Fc (Sigma).
2.3. Expression and purification of VNAR proteins
Monomeric VNARs were purified from E. coli lysates by immobilized metal affinity chromatography on Ni-NTA resin (Qiagen, USA). The purified protein was concentrated on a Vivaspin 20 column (Sartorius, Germany) and endotoxin was removed using a Viva-Pure Q mini column (Sartorius). Selected VNARs were cloned and expressed in CHO cells as N-terminal fusions to either the human IgG1 or the mouse IgG2a Fc regions (Evitria AG, Switzerland). VNAR-Fc molecules were purified from the post-transfection media on protein A columns (Mab Select Sure, GE Healthcare) using an AKTA Express. Bound VNAR-Fc molecules were eluted with 0.1 M Glycine-HCl (pH 2.9) and neutralized with 1 M Tris (pH 8). The elution buffer was then exchanged with PBS using Vivaspin 20 (5 kDa) centrifugal concentrators and the protein estimated by absorbance at 280 nm. For in vivo studies, endotoxin was removed using Acti-Clean Etox resin (Sterogene, USA).
2.4. Biochemical assays
For epitope binning, 96-well Maxisorp plates were coated at 1 μg/ml with either hBAFF-Fc and washed with blocking buffer. Selected clones were grown in a 96 deep-well format in auto-induction medium and periplasmic extracts were pre-blocked in the presence of a competitor VNAR-Fc molecule at 2 μM final concentration. The extracts were then exposed to the coated surface and bound VNARs were detected using a peroxidase-conjugated anti-FLAG antibody (Sigma).
The biochemical EC50 of selected clones was determined by serially diluting purified monomeric VNARs in blocking buffer and binding to preblocked 96-well plates coated at 1 μg/ml with hBAFF-Fc. After washing in PBS-0.1% Tween, bound VNARs were detected using a peroxidase-conjugated anti-FLAG antibody (Sigma). The biochemical IC50 of selected clones on all three BAFF receptors was determined by serially diluting purified monomeric VNARs in blocking buffer containing 1 nM hBAFF-Fc in the case of BR3 or 0.5 nM in the case of TACI and BCMA. The pre-blocked proteins were then exposed to 96-well plates coated at 1 μg/ml with extra-cellular domains of BR3, TACI or BCMA. After washing in PBS-0.1% Tween, bound hBAFF was detected using a peroxidase-conjugated anti-human Fc. Absorbance at 450 nm was recorded and EC50/IC50 values were calculated by non-linear regression using GraphPad Prism®.
2.5. Murine splenocyte in vitro proliferation assay
Mouse splenocytes were obtained by dissociating spleens of C57BL/6 mice on a 70-μm cell strainer and lysing red blood cell in RBC buffer (Sigma). B cells were purified by depleting CD43-positive cells using magnetic microbeads (Miltenyi Biotec, USA) according to manufacturer’s instructions. The obtained B cells were subsequently stimulated with goat anti-mouse IgM antibody (Jackson ImmunoResearch, USA) at 10 μg/ml. Monomeric VNARs or VNAR-Fc fusion proteins were serially-diluted and pre-complexed with hBAFF-Fc at 5 ng/ml or mouse BAFF (R&D Systems, USA) at 0.5 ng/ml in RPMI 1640 supplemented with 10% FBS, for 30 min at 37 °C. Stimulated B cells were added to the pre-complexed proteins and further incubated for 72 h at 37 °C, 5% CO2. Cell proliferation was measured with WST-1 reagent (Sigma). IC50 values were calculated by non-linear regression as above.
2.6. Analysis of B cell populations in mouse spleen
Three groups of five 6–12 week old female C57BL/6 mice were injected intraperitoneally with 100 μg of purified VNAR-Fc fusion protein (or an equivalent volume of buffer) at days 0, 4, 8 and 12. On Day 15, blood samples were taken, serum was prepared, and mice were sacrificed. Spleens were extracted, weighed and splenocytes were prepared by dispersion on cell strainer followed by a Red Blood Cell Lysis step and cell count. Splenocytes were then stained and analyzed for total number of B-lymphocytes and subset frequency by FACS using the gating scheme based on (Scholz et al., 2008). BAFF serum levels were determined using a commercial ELISA kit (ab119580, AbCam, UK).
Antibodies used to stain splenocytes were: FITC-anti-mouse CD4 (553046), FITC-anti-mouse CD8 (553030), biotinylated anti-mouse CD23 (553138), PE-Cy5.5-anti-mouse CD21/35 (552957), PE-Cy7anti-mouse IgM (552867) and PE-Texas Red-Streptavidin (562284) from BD Biosciences (USA); and APC-anti-mouse AA4 (17.5892.81) and APCCy5.5 anti-mouse B220 (45.0452.80) from e-Bioscience (USA). For FACS analyses, forward and side scatter were used to identify lymphocytes and singlet events. Viable cells were identified by dead cell exclusion and non-B cells were excluded by staining for CD4 and CD8. B cells were then gated according to expression of B220 (CD45R) and AA4.1 (CD93). Cell surface expression of IgM and CD23 were examined on the B220 + AA4.1+ cells, whilst IgM and CD21/35 were visualised on the B220 + AA4.1- subset. The total number of cells in each mature B cell subset (namely T1, T2, T3, FO, MZ) was determined. An unpaired two-sample Student’s t-test with a confidence interval of 95% was used to determine significant differences between the means.
2.7. Structural modeling
The structure of the VNAR A07 clone was modeled based on homologous sequences of a nurse shark IgNAR with known structure containing the same cysteine bond arrangement recovered from an NCBI BLAST search (accession # 2i26). The clone sequence was threaded onto the template structure using Swiss-Model software (Biasini et al., 2014). The VNAR-BAFF complex was modeled based on the assumption that the DXL motif shared between multiple BAFF binding VNAR clones and the BAFF receptor has a similar binding conformation in the two proteins (BAFF–BAFF receptor complex structure protein data bank accession 4v46). The backbone structure of the VNAR CDR3 region corresponding to the sequence YDPLT was adjusted to fit that of the BAFF receptor binding loop FDLLV by optimally grafting the backbone of the BAFF receptor loop onto the CDR3.
3. Results
3.1. Selection of BAFF-binding VNARs
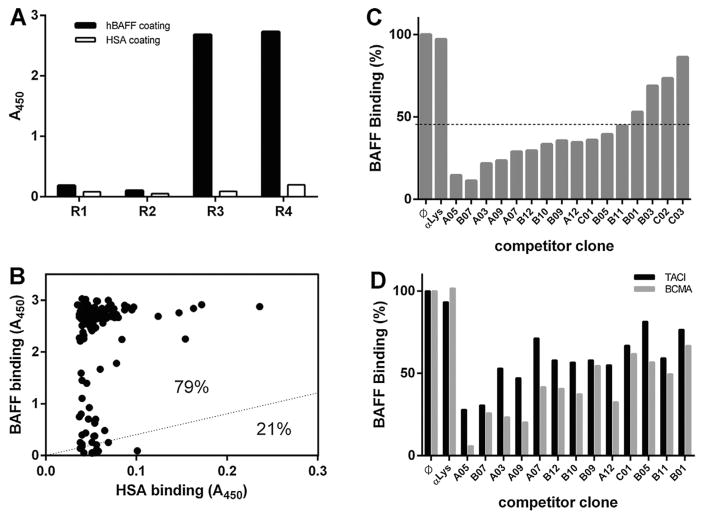
VNARs specific for human BAFF (hBAFF) were isolated from a VNAR semi-synthetic phage display library by four rounds of selection and amplification on immobilized recombinant hBAFF. To select for the highest affinity clones, the stringency of selection was increased at each round by decreasing the hBAFF coating density and increasing the number of washing steps. The enrichment of BAFF-binding clones during the selection procedure was first evaluated by polyclonal phage ELISA. The inputs of each selection rounds were exposed to immobilized recombinant hBAFF or human serum albumin (HSA), and bound phage were detected with an anti-M13 antibody. A significant increase in binding phage was observed from round 3, specifically on the hBAFF-coated surface (Fig. 1A), revealing an efficient selection of BAFF-specific clones.
Fig. 1. Identification of VNARs blocking the interaction between BAFF and its receptors.
A. Polyclonal phage ELISA. Phage inputs of each panning round were exposed to hBAFF- or HSA-coated surfaces before being detected with an anti-M13 antibody. A significant increase in the signal of phage binding specifically to hBAFF was observed from round 3. B. Independent clones (randomly picked from round 3 and 4) were expressed as a periplasmic fraction before being exposed to a surface coated with either hBAFF or HSA. Bound VNAR was detected using an anti-FLAG antibody. 79% of the tested clones displayed a binding to hBAFF at least four times higher than their respective binding to HSA. C. The ability of selected clones to block the interaction between hBAFF and BR3 was tested in an ELISA format. As compared to a negative control VNAR anti-Lysosyme (αLys), thirteen of the sixteen tested clones appeared to block the interaction between BAFF and BR3 (line = 45% inhibition threshold). D. Same as C. As compared to a negative control VNAR anti-lysosyme (αLys), the thirteen clones blocking BAFF/BR3 interaction appeared to also block the interaction between BAFF and both TACI and BCMA
Selection outputs were then evaluated in a monoclonal format by ELISA. Clones (186 in total) were randomly selected from rounds 3 and 4 and soluble monomeric VNARs were produced in the periplasm of E. coli. Their ability to bind hBAFF specifically was tested by exposing their respective periplasmic fractions to hBAFF-and HSA-coated surfaces. After detecting bound VNAR, a threshold of BAFF-binding signal to HSA-binding signal higher than four was used to define positive clones (Fig. 1B). Almost 80% of the tested clones bound hBAFF specifically using this criterion. Positive clones were then sequenced and sorted into groups based on sequence identity. The output of the selection contained sixteen unique hBAFF-binding VNARs.
3.2. Identification of VNARs blocking BAFF/BAFF receptors interaction
The ability of the 16 selected hBAFF-binding clones to block the interaction between BAFF and its main receptor BR3 was tested by a “blocking ELISA”. Soluble monomeric VNARs were produced in the periplasmic fraction and pre-incubated with recombinant hBAFF before being exposed to a BR3-coated surface. The interaction was then measured by specifically detecting the amount of BAFF bound to the plate. By using a threshold at 45% of binding inhibition, 13 of the 16 binding clones blocked the interaction between BAFF and BR3 (Fig. 1C). The ability of these 13 VNARs to block the interaction between BAFF and its other two receptors (TACI and BCMA) was then tested using an identical experimental setup on surface coated with either TACI or BCMA. The thirteen BR3 blockers also blocked the interaction between BAFF and TACI or BCMA, although to a lesser extent (Fig. 1D).
3.3. BAFF specificity and epitope binning
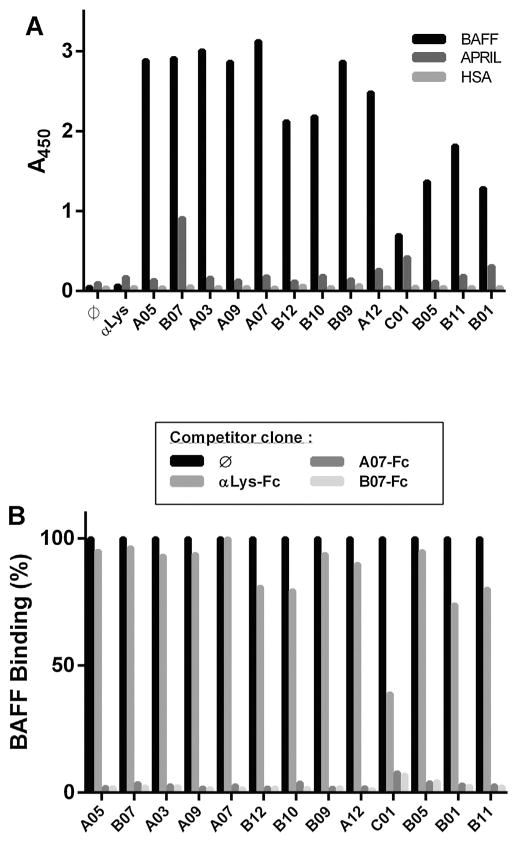
In order to further study specificity, VNAR clones that blocked the interaction between BAFF and its receptors were tested for their ability to bind to BAFF’s closest-related TNF superfamily ligand APRIL. With the exception of B07, which showed a slight binding to APRIL, every blocking clone was BAFF-specific (Fig. 2A). The blocking clones were then grouped into different categories based on their epitope using an epitope-binning protocol. The binding of monomeric VNARs, expressed in the periplasmic fraction, was competed with an excess of VNAR-Fc fusion molecules of two BAFF-specific clones (A07 and B07) and one irrelevant VNAR specific to hen egg lysozyme (αLys) (Dooley et al., 2003). The binding of every clone of the panel specific for BAFF was efficiently competed by both the A07-Fc and B07-Fc (Fig. 2B) and not by the irrelevant VNAR-Fc. All the blocking clones of the selected panel therefore target the same, or at least overlapping, epitope on the BAFF protein.
Fig. 2. Specificity of the interaction and epitope binning.
A. The 13 blocking clones were expressed as a periplasmic fraction and their binding to hBAFF, APRIL, and HSA was analyzed by ELISA. B. The binding of the 13 blocking clones (expressed as a periplasmic fraction) to hBAFF was competed by an excess (2 μM) of two an anti-BAFF VNAR expressed as a human Fc fusion. A07 and B07 Fc fusions were used in this assay together with the αLys-Fc negative control.
3.4. Biochemical characterization: binding and blocking potencies
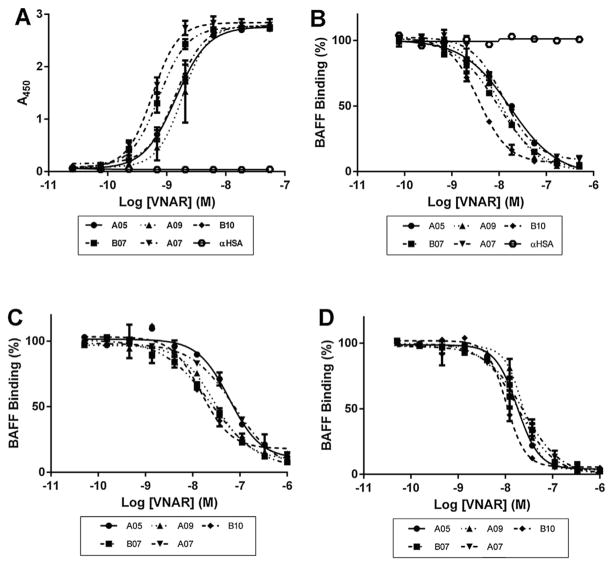
The 13 selected blocking clones were produced at larger scale, IMAC purified from the periplasmic fraction, and their binding and inhibitory potency was investigated. ELISA binding curves were generated by applying increasing concentrations of purified VNARs to immobilized hBAFF (Fig. 3A). After curve fitting, the calculated EC50s – ranging from 0.5 to 17 nM (Table 1) – revealed that the selected clones have a very high affinity. The blocking potency of the selected panel of clones was then measured by ELISA. As described earlier, the binding of recombinant hBAFF to immobilized BR3 was competed by purified monomeric VNARs. The calculated IC50s (Fig. 3B, Table I), which ranged from 2.8 to 148 nM, revealed the high blocking capacity of the selected clones. The clones were then ranked based on both their EC50 and IC50 values to select a restricted panel of five lead molecules to be progressed for cellular assays. The ability of these five clones to block the interaction between BAFF and its other two receptors TACI (Fig. 3C) and BCMA (Fig. 3D) was also quantified using a similar ELISA protocol. The IC50 of the five selected leads were lower on BCMA than on TACI (10–24 nM and 14–83 nM) (Table 1).
Fig. 3. Biochemical characterization.
A. The binding of the 13 blocking clones was characterized by titrating the purified VNAR monomers on hBAFF by ELISA. Five representative anti-BAFF VNARs are shown together with a negative control VNAR anti-HSA (αHSA). B. The blocking of the interaction between hBAFF and BR3 was quantified by adding increasing amounts of purified monomeric VNAR to a BAFF solution and exposing it to a BR3-coated surface. C. Same as (B) with TACI, but without αHSA. D. Same as (C) with BCMA.
Table 1.
Biochemical properties of the selected clones.
| CLONE | EC50 on hBAFF (nM) | IC50 for BR3 (nM) | IC50 for TACI (nM) | IC50 for BCMA (nM) | CELLULAR IC50 AS MONOMER on hBAFF (nM) | CELLULAR IC50 AS Fc-FUSION on hBAFF (nM) | CELLULAR IC50 AS Fc FUSION on mBAFF (nM) |
|---|---|---|---|---|---|---|---|
| A07 | 0.5 | 6.8 | 82.7 | 24.5 | 58 | 2.47 | 1.95 |
| B07 | 0.8 | 6.8 | 24 | 18.9 | 60 | 3.78 | 604 |
| A05 | 1.5 | 13.8 | 57.8 | 17.4 | 135 | 4.14 | n.d. |
| B10 | 1 | 2 | 14.1 | 10.5 | ≈ 100 | 5.53 | n.d. |
| A09 | 4.9 | 9.7 | 27.2 | 23.6 | 215 | n.d. | n.d. |
| C01 | 2.13 | 3.85 | n.d. | n.d. | n.d. | n.d. | n.d. |
| A12 | 4.57 | 49.16 | n.d. | n.d. | n.d. | n.d. | n.d. |
| B11 | 2.49 | 6.85 | n.d. | n.d. | n.d. | n.d. | n.d. |
| B12 | 2.7 | 9.32 | n.d. | n.d. | n.d. | n.d. | n.d. |
| B09 | 4.07 | 36.77 | n.d. | n.d. | n.d. | n.d. | n.d. |
| B01 | 7.4 | 91.2 | n.d. | n.d. | n.d. | n.d. | n.d. |
| A03 | 17.16 | 12.33 | n.d. | n.d. | n.d. | n.d. | n.d. |
| B05 | 11.82 | 148.2 | n.d. | n.d. | n.d. | n.d. | n.d. |
3.5. Inhibition of B cell proliferation and species cross-reactivity
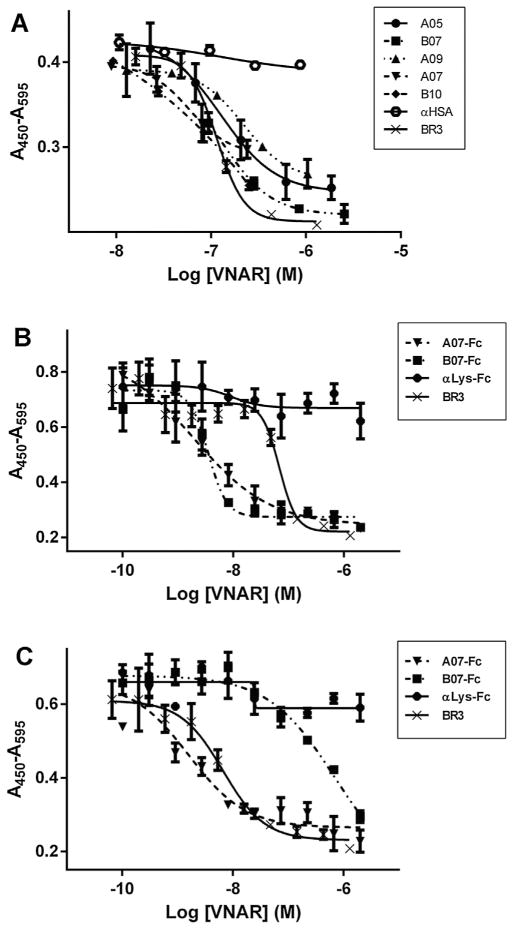
In order to test the ability of the selected VNARs to inhibit BAFF biological activity, the five lead molecules were tested in a mouse splenocyte proliferation assay. Mouse primary splenic B cells were stimulated with hBAFF in the presence or in the absence of increasing concentrations of antagonist monomeric VNAR. Cell proliferation was estimated 72 h later and a cellular IC50 was determined for the five molecules. The values ranged from 60 to 200 nM (Table 1), which was in the same range as the BR3 extracellular domain, which displayed an IC50 of 121 nM in this assay (Fig. 4A). A07 and B07 showed the highest level of inhibition of B cell proliferation, with EC50s of 58 nM and 60 nM, respectively.
Fig. 4. Effect of anti-BAFF VNARs on mouse splenocytes proliferation.
A. Mouse splenocytes were stimulated with hBAFF in the presence or in the absence of increasing concentrations of antagonist monomeric VNAR. The five tested clones inhibited hBAFF-induced proliferation in a similar range as recombinant BR3 (IC50 = 121 nM), while an irrelevant VNAR (αHSA) did not have any significant effect. B. Same as (A) with two anti-BAFF VNARs formatted as an N-terminus human Fc fusion, and one irrelevant VNAR-Fc (αLys-Fc). Both anti-BAFF VNAR-Fc molecules inhibited hBAFF-induced proliferation more efficiently than recombinant BR3 (IC50 = 88 nM), while the irrelevant VNAR-Fc did not have any significant effect. C. Same as (B) with splenocytes stimulated with recombinant mouse BAFF (mBAFF). Both anti-BAFF VNAR-Fc molecules inhibited mBAFF-induced proliferation. A07-Fc in the same range as recombinant BR3 (IC50 = 7 nM), B07 at a higher concentration. The irrelevant VNAR-Fc did not show any significant effect.
To address the human/mouse cross-reactivity of the lead molecules, and to select the best molecule for testing in a mouse model, the two molecules showing the highest level of inhibition in B cell proliferation (A07 and B07) were expressed and purified as a human IgG1 Fc fusion molecule, together with a negative control (anti-lysozyme) VNAR. These molecules were then tested for their ability to block the proliferation of primary mouse splenic B cells stimulated with either mouse (Fig. 4B) or human (Fig. 4C) recombinant BAFF. Both lead molecules cross-reacted with human and mouse BAFF. A07 having a significantly lower IC50 on mouse BAFF (1.95 nM) than B07 (604 nM), it was selected for subsequent in vivo studies.
3.6. Effect of anti-BAFF VNAR-Fc on mouse splenic B cell subsets in vivo
In order to study the effect of anti-BAFF VNAR-Fc molecules on B cell development in vivo, 12-week-old female C57BL/6 wild type mice were injected intraperitoneally four times over a period of 12 days with 100 μg of VNAR mIgG2a Fc fusion. Three different treatments were administered: A07-Fc as BAFF-specific, αLys-Fc as a negative control, and PBS-only as mock injection. On day 15, spleens were extracted, splenocytes were prepared and the different B cell subpopulations were analyzed by FACS as described in (Scholz et al., 2008).
Neither of the treatments induced any difference in the splenic or body weights. The viable splenocyte number, however, was lower in the A07-Fc-treated group (not shown). Analysis of the total cell numbers within individual mature B cell subpopulations showed that treatment with A07-Fc resulted in a general 40–60% decrease in all B cell subsets (Fig. 5A–E). The difference was mature follicular (FO) and marginal zone (MZ) B cells statistically signifi-cant (p < 0.05) as compared to either the 5A7-Fc or mock treatment groups despite the sample samples size (N = 5). In contrast, serum mBAFF was not decreased in the A07-Fc-treated mice and actually exhibited a modest but significant increase relative to both control groups (Fig. 5F). Although the assay does not discriminate between bound versus free BAFF, the increase could result from the longer half-life of antibody-antigen complexes in the circulation.
Fig. 5. Effect of anti-BAFF VNARs on B cell development in vivo.
C57BL/6 mice were injected intraperitoneally with 100 μg of either anti BAFF VNAR-Fc fusion protein (A07-Fc), negative control VNAR-Fc (αLys-Fc), or an equivalent volume of vehicle (mock) four times over a period of 15 days after which spleens were extracted and splenocytes prepared. Total numbers of Follicular (FO) (A) Marginal Zone and Marginal Zone Precursors (MZ/MZP) (B) and Transitional 1, 2, and 3 (T1, T2, T3) (C–E) B cells were determined by FACS analysis. The serum level of endogenous BAFF was determined by ELISA (F).
3.7. Molecular mimicry of anti-BAFF VNARs
As previously shown in Fig. 1C–D, the vast majority of the BAFF-specific clones also blocked the interaction between BAFF and its three receptors. Sequence analysis revealed that 85% of the clones contain an identical short motif in CDR3, composed of an aspartic acid separated from a leucine by one residue (Asp-X-Leu or DXL) (Fig. 6A). Interestingly, in most of the clones this motif was also located just one residue upstream of the single cysteine present in the typical Type II VNAR CDR3, suggesting that the orientation of this DXL motif is important for the binding of the VNAR to BAFF.
Fig. 6. Molecular mimicry of anti-BAFF VNARs.
A. CDR1 and CDR3 sequences of the 13 blocking clones. DXL motifs are highlighted in the CDR3. B. Molecular modeling of the anti-BAFF clone A07 in complex with a BAFF trimer. The BAFF trimer is shown space filled in different shades of grey and the modeled VNAR is shown as a ribbon diagram with the CDR1 in green and the CDR3 in orange. The residues of the DXL are shown in red. C. Superimposition of the A07CDR3 and a BAFF receptor interacting loop (mini BR3 peptide) binding to a BAFF monomer. mini BR3 is shown as a blue ribbon with the DXL motif shown as a stick diagram illustrating its close alignment with the DXL motif of the VNAR.
Such a DXL motif has been found previously in other BAFF-binding antibodies like belimumab, which harbors one in its CDR1 and one in its CDR3 (Edwards et al., 2003; Gordon et al., 2003; Gordon et al., 2010). This motif is also present in all three BAFF receptors, and structures of BAFF-BR3, BAFF-BCMA and BAFF-bhpBR3 complexes showed that DXL accounts for the majority of the ligand-receptor interaction (Kim et al., 2003; Liu et al., 2003; Gordon et al., 2003).
The CDR3 region of the anti-BAFF VNARs we identified seems to mimic the BAFF binding site of the receptors. We therefore modeled the interaction of the lead VNAR A07 with a BAFF monomer based on published crystal structures. The A07CDR3 protrudes out and targets a shallow pocket on the surface of BAFF, which is complemented by the hydrophobic side chain of the leucine residue in the DXL motif (Fig. 6B). We then superimposed the CDR3 of A07 and the portion of the BR3 ectodomain responsible for BAFF binding (previously denoted miniBR3 (Kayagaki et al., 2002; Gordon et al., 2003) on a BAFF monomer (Fig. 6C), and both of them fit the cavity in the same way. This may explain the efficiency of the selected VNAR clones in antagonizing BAFF biological activity.
4. Discussion
Screening of a semi-synthetic VNAR phage display library allowed the identification of a panel of binders to BAFF that antagonize its interaction with all three BAFF receptors (BR3, TACI and BCMA). The majority of the VNARs isolated directly from the library have low nanomolar EC50s for BAFF and the main lead molecule A07 has an EC50 of 0.5 nM. The average CDR3 length of the best 11 BAFF hits is 18+/− 3 amino acids, suggesting that there was a selection bias towards longer CDR3s. Moreover, most of the CDR3s contain a particular structural motif (DXL) and its position was conserved relative to the cysteine residue. The DXL motif is found in all three BAFF receptors and is essential for binding a pocket created at the monomer–monomer interface in the BAFF trimer (Kim et al., 2003). Structural modeling with A07 suggests that VNARs inhibit BAFF activity by receptor mimicry.
Clone A07 was formatted as an Fc fusion for functional testing and it was found to inhibit both mouse and human BAFF with equal potency ex vivo in a splenocyte proliferation assay. In mice, subchronic administration reduced the number of immature and transitional intermediates B cells by 50–60% and 40% mature B cell subsets, as expected for a selective B-cell inhibitor. While its in vivo efficacy cannot be compared directly to belimumab (which only binds hBAFF), in a challenge model with hBAFF, belimumab decreased the number of splenic B cells by the same order of magnitude that was observed with A07-Fc. This represents only the second reported demonstration of a VNAR with in vivo effi-cacy against a therapeutic target since their discovery over 20 years ago (Greenberg et al., 1995). The first report showed that an anti-TNFα VNAR reduced lethality in a mouse model of endotoxic shock (Bojalil et al., 2013).
Major hurdles to the isolation of VNAR suitable for development include even more challenging animal husbandry for generating immunized libraries than with camelids and the limited diversity provide by semi-synthetic libraries described to date (Nuttall et al., 2003) (Liu et al., 2007). These libraries used a single framework with randomized CDR3s of only 3 different lengths. Extensive sequencing of naturally occurring VNARs in the nurse shark revealed a wide range in CDR3 length and additional diversity in the framework. These features were incorporated into our VNAR library, which contain CDR3 regions ranging from 11 to 18 residues within 10 framework variants. The level of representation of the unique structural motif found in our anti-BAFF VNARs, suggests that the size, diversity and complexity of the library will enabled the rapid and efficient isolation of potent hits to a variety of other antigens. Smaller VNAR-based therapeutics would likely provide more tissue-penetrant molecules or could be used to produce novel bis-pecifics antibodies with added functionality.
A potential application of anti-BAFF VNARs is to enhance efficacy or improve tissue targeting of existing treatments for autoim-mune diseases and B cell malignancies. For example, smaller VNAR-Fc antibodies would have better access to B cells in ectopic lymphoid-like structures, which often develop at sites of inflamma-tion (Pitzalis et al., 2014). Alternatively, a BAFF antagonist (VNAR, belimumab or peptibody) could be linked to a VNAR carrier for CNS transport (Rutkowski et al., 2015) for the treatment of cerebral lupus or multiple sclerosis. Bispecific approaches could also be used to either enhance selectivity or broaden the activity against particular B cell subsets. BAFF antibodies selectively target autoreactive B cells over non-self-reactive B cells might be enhanced if combined with antagonists to other pro-survival signals such as CD40 or toll-like receptors (Metzler et al., 2015). Conversely, an anti-BAFF VNAR fused to an existing CD20 antibody such as rituximab would produce a more complete B cell depletion, which was required for significant therapeutic benefit in mouse models of SLE (Lin et al., 2015) and might prevent post-rituximab flares in SLE patients characterized by elevated circulating BAFF (Ehrenstein and Wing, 2016). Similarly, a BAFF/CD20 bispecific may counteract the resistance to CD20 antibody therapy facilitated by BAFF that occurs in patients with chronic lymphocytic leukemia (Wild et al., 2015). These examples highlight the potential for VNARs in specifically tailoring next generation of biologics for a particular therapeutic use.
Acknowledgments
The authors are grateful to Yatindra Tirunagari for purification and characterization of recombinant proteins.
Footnotes
Conflict of interest
JH, FSW and JLR were employed by Ossianix, Inc. at the time of completion of this work. MFF received research support from Ossianix and GW was a paid consultant in molecular modeling and bioinformatics.
References
- Baker KP, Edwards BM, Main SH, Choi GH, Wager RE, Halpern WG, Lappin PB, Riccobene T, Abramian D, Sekut L, Sturm B, Poortman C, Minter RR, Dobson CL, Williams E, Carmen S, Smith R, Roschke V, Hilbert DM, Vaughan TJ, Albert VR. Generation and characterization of LymphoStat-B, a human monoclonal antibody that antagonizes the bioactivities of B lymphocyte stimulator. Arthritis Rheum. 2003;48(11):3253–3265. doi: 10.1002/art.11299. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojalil R, Mata-Gonzalez MT, Sanchez-Munoz F, Yee Y, Argueta I, Bolanos L, Amezcua-Guerra LM, Camacho-Villegas TA, Sanchez-Castrejon E, Garcia-Ubbelohde WJ, Licea-Navarro AF, Marquez-Velasco R, Paniagua-Solis JF. Anti-tumor necrosis factor VNAR single domains reduce lethality and regulate underlying inflammatory response in a murine model of endotoxic shock. BMC Immunol. 2013;14:17. doi: 10.1186/1471-2172-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss NA, Henderson SJ, McFarlane M, Shenton JM, de Haan L. Monoclonal antibody therapeutics: history and future. Curr Opin Pharmacol. 2012;12(5):615–622. doi: 10.1016/j.coph.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Dooley H, Flajnik MF, Porter AJ. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol Immunol. 2003;40(1):25–33. doi: 10.1016/s0161-5890(03)00084-1. [DOI] [PubMed] [Google Scholar]
- Edwards BM, Barash SC, Main SH, Choi GH, Minter R, Ullrich S, Williams E, Du Fou L, Wilton J, Albert VR, Ruben SM, Vaughan TJ. The remarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol. 2003;334(1):103–118. doi: 10.1016/j.jmb.2003.09.054. [DOI] [PubMed] [Google Scholar]
- Ehrenstein MR, Wing C. The BAFFling effects of rituximab in lupus: danger ahead? Nat Rev Rheumatol. 2016 doi: 10.1038/nrrheum.2016.18. [DOI] [PubMed] [Google Scholar]
- Gordon NC, Pan B, Hymowitz SG, Yin J, Kelley RF, Cochran AG, Yan M, Dixit VM, Fairbrother WJ, Starovasnik MA. BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site. Biochemistry. 2003;42(20):5977–5983. doi: 10.1021/bi034017g. [DOI] [PubMed] [Google Scholar]
- Gordon NC, Lien S, Johnson J, Wallweber HJ, Tran T, Currell B, Mathieu M, Quan C, Starovasnik MA, Hymowitz SG, Kelley RF. Multiple novel classes of APRIL-specific receptor-blocking peptides isolated by phage display. J Mol Biol. 2010;396(1):166–177. doi: 10.1016/j.jmb.2009.11.041. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374(6518):168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13(14):3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler JR, Rutkowski JL. WO2015200883 A3 Semi-synthetic nurse shark VNAR libraries for making and using selective binding compounds WO2015200883 A3. 2016
- Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, Batchelor AH, Gupta A, Bai T, Murphy VJ, Anders RF, Foley M, Nuttall SD. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15(11):1452–1466. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Yan M, Seshasayee D, Wang H, Lee W, French DM, Grewal IS, Cochran AG, Gordon NC, Yin J, Starovasnik MA, Dixit VM. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2. Immunity. 2002;17(4):515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, Yoo OJ, Lee H, Lee JO. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat Struct Biol. 2003;10(5):342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Olland A, Piche-Nicholas N, Godbole A, King D, Svenson K, Calabro V, Muller MR, Barelle CJ, Somers W, Gill DS, Mosyak L, Tchistiakova L. Atypical antigen recognition mode of a shark immunoglobulin new antigen receptor (IgNAR) variable domain characterized by humanization and structural analysis. J Biol Chem. 2013;288(24):17408–17419. doi: 10.1074/jbc.M112.435289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Seshasayee D, Lee WP, Caplazi P, McVay S, Suto E, Nguyen A, Lin Z, Sun Y, DeForge L, Balazs M, Martin F, Zarrin AA. Dual B cell immunotherapy is superior to individual anti-CD20 depletion or BAFF blockade in murine models of spontaneous or accelerated lupus. Arthritis Rheumatol. 2015;67(1):215–224. doi: 10.1002/art.38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hong X, Kappler J, Jiang L, Zhang R, Xu L, Pan CH, Martin WE, Murphy RC, Shu HB, Dai S, Zhang G. Ligand-receptor binding revealed by the TNF family member TALL-1. Nature. 2003;423(6935):49–56. doi: 10.1038/nature01543. [DOI] [PubMed] [Google Scholar]
- Liu JL, Anderson GP, Goldman ER. Isolation of anti-toxin single domain antibodies from a semi-synthetic spiny dogfish shark display library. BMC Biotechnol. 2007;7:78. doi: 10.1186/1472-6750-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler G, Kolhatkar NS, Rawlings DJ. BCR and co-receptor crosstalk facilitate the positive selection of self-reactive transitional B cells. Curr Opin Immunol. 2015;37:46–53. doi: 10.1016/j.coi.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais SA, Vilas-Boas A, Isenberg DA. B-cell survival factors in autoimmune rheumatic disorders. Ther Adv Musculoskelet Dis. 2015;7(4):122–151. doi: 10.1177/1759720X15586782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MR, O’Dwyer R, Kovaleva M, Rudkin F, Dooley H, Barelle CJ. Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol Biol. 2012;907:177–194. doi: 10.1007/978-1-61779-974-7_9. [DOI] [PubMed] [Google Scholar]
- Nuttall SD, Krishnan UV, Doughty L, Pearson K, Ryan MT, Hoogenraad NJ, Hattarki M, Carmichael JA, Irving RA, Hudson PJ. Isolation and characterization of an IgNAR variable domain specific for the human mitochondrial translocase receptor Tom70. Eur J Biochem. 2003;270(17):3543–3554. doi: 10.1046/j.1432-1033.2003.03737.x. [DOI] [PubMed] [Google Scholar]
- Pitzalis C, Jones GW, Bombardieri M, Jones SA. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat Rev Immunol. 2014;14(7):447–462. doi: 10.1038/nri3700. [DOI] [PubMed] [Google Scholar]
- Rutkowski JL, Tirunagari Y, Häsler J. Transferrin receptor-mediated delivery of BACE1 therapeutic antibodies to the brain. World Preclinical Congress: Blood-brain Barrier; Boston, MA. 2015. [Google Scholar]
- Scholz JL, Crowley JE, Tomayko MM, Steinel N, O’Neill PJ, Quinn WJ, 3rd, Goenka R, Miller JP, Cho YH, Long V, Ward C, Migone TS, Shlomchik MJ, Cancro MP. BLyS inhibition eliminates primary B cells but leaves natural and acquired humoral immunity intact. Proc Natl Acad Sci U S A. 2008;105(40):15517–15522. doi: 10.1073/pnas.0807841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl W, Hilbert DM. The discovery and development of belimumab: the anti-BLyS-lupus connection. Nat Biotechnol. 2012;30(1):69–77. doi: 10.1038/nbt.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl W. Biologic differences between various inhibitors of the BLyS/BAFF pathway: should we expect differences between belimumab and other inhibitors in development? Curr Rheumatol Rep. 2012;14(4):303–309. doi: 10.1007/s11926-012-0254-6. [DOI] [PubMed] [Google Scholar]
- Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, Schwarz N, Adriouch S, Boyer O, Seman M, Licea A, Serreze DV, Goldbaum FA, Haag F, Koch-Nolte F. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009;198(3):157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J, Schmiedel BJ, Maurer A, Raab S, Prokop L, Stevanovic S, Dorfel D, Schneider P, Salih HR. Neutralization of (NK-cell-derived) B-cell activating factor by Belimumab restores sensitivity of chronic lymphoid leukemia cells to direct and Rituximab-induced NK lysis. Leukemia. 2015;29(8):1676–1683. doi: 10.1038/leu.2015.50. [DOI] [PubMed] [Google Scholar]
- Zielonka S, Empting M, Grzeschik J, Konning D, Barelle CJ, Kolmar H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs. 2015a;7(1):15–25. doi: 10.4161/19420862.2015.989032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka S, Empting M, Konning D, Grzeschik J, Krah S, Becker S, Dickgiesser S, Kolmar H. The shark strikes twice: hypervariable loop 2 of shark IgNAR antibody variable domains and its potential to function as an autonomous paratope. Mar Biotechnol (NY) 2015b;17(4):386–392. doi: 10.1007/s10126-015-9642-z. [DOI] [PubMed] [Google Scholar]