Summary
Assembly of the tubulin-like cytoskeletal protein FtsZ into a ring structure establishes the location of the nascent division site in prokaryotes. Factors that modulate FtsZ assembly are essential for ensuring the precise spatial and temporal regulation of cytokinesis. We have identified ClpX, the substrate recognition subunit of the ClpXP protease, as an inhibitor of FtsZ assembly in Bacillus subtilis. Genetic data indicate that ClpX but not ClpP inhibits FtsZ-ring formation in vivo. In vitro, ClpX inhibits FtsZ assembly in a ClpP-independent manner through a mechanism that does not require ATP hydrolysis. Together our data support a model in which ClpX helps maintain the cytoplasmic pool of unassembled FtsZ that is required for the dynamic nature of the cytokinetic ring. ClpX is conserved throughout bacteria and has been shown to interact directly with FtsZ in Escherichia coli. Thus, we speculate that ClpX functions as a general regulator of FtsZ assembly and cell division in a wide variety of bacteria.
Introduction
In bacteria the precise spatial and temporal regulation of cytokinesis is dependent on the assembly dynamics of the essential cell division protein FtsZ (Romberg and Levin, 2003). FtsZ, a GTPase with a tertiary structure similar to tubulin, assembles into a ring structure (the Z ring) at mid-cell. The Z ring establishes the location of the division site and is responsible for recruiting other cell division proteins to the nascent septum (Romberg and Levin, 2003). GTP binding enhances FtsZ assembly (Mukherjee and Lutkenhaus, 1994) and dimerization leads to formation of a GTPase active site (Löwe and Amos, 1999; Scheffers et al., 2002). As with tubulin, GTP hydrolysis in vitro causes a conformational change in FtsZ polymer structure and rapid disassembly (Mukherjee and Lutkenhaus, 1998; Romberg et al., 2001). Photobleaching experiments indicate that the half-time for subunit replacement in the Escherichia coli Z ring is between 8 and 30 s (Stricker et al., 2002). The observation that FtsZ levels are equivalent in cells cultured at different growth rates suggests that Z-ring formation is primarily governed at the level of FtsZ assembly (Weart and Levin, 2003).
FtsZ is thought to exist in two states: (i) a cytoplasmic pool of monomers, small multimers and potentially small polymers and (ii) the membrane-associated polymers that constitute the Z ring. A precisely balanced network of regulatory factors act in concert to modulate FtsZ assembly to ensure that the Z ring forms in the proper position and is tightly co-ordinated with DNA replication, chromosome segregation and cell growth (Romberg and Levin, 2003). The majority of cell division genes identified to date were isolated in screens for cells with gross defects in cell division either as conditional mutants that blocked division altogether (e.g. ftsA, divIC) or mutants that divided aberrantly (e.g. the min genes). However, recent work has led to the identification of several genes that appear to function in more subtle ways to maintain the balance between assembled and unassembled FtsZ, thereby contributing to the dynamic nature of the cytokinetic ring. One of these, the well-conserved protein ZapA, was isolated as a multicopy suppressor of the filamentation associated with overexpression of the cell division inhibitor MinCD (Gueiros-Filho and Losick, 2002). In vitro, ZapA interacts directly with FtsZ and induces protofilaments to associate into large bundles (Gueiros-Filho and Losick, 2002). Although not required for division in wild-type cells, ZapA becomes essential for efficient division under conditions in which the cytoplasmic pool of unassembled FtsZ is reduced, in particular in the absence of the division inhibitor EzrA or in cells in which the intracellular concentration of FtsZ has been artificially lowered (Gueiros-Filho and Losick, 2002). Similarly, a null mutation in noc, a division inhibitor associated with the bacterial nucleoid, does not significantly affect cell division under normal conditions. However, when combined with a mutation in the division inhibitor minD, the resulting double mutant cells fail to properly assemble FtsZ at putative division sites resulting in filamentation (Wu and Errington, 2004).
In an effort to identify additional modulators of FtsZ assembly in Bacillus subtilis we isolated transposon insertions that suppressed the heat sensitivity of a conditional allele of ftsZ, ftsZts. We previously employed a similar approach to identify the division inhibitor EzrA (Levin et al., 1999). In this article we report the identification and characterization of ClpX as an inhibitor of FtsZ assembly. ClpX functions as the substrate recognition domain of the ClpXP protease. ClpX also functions in a ClpP-independent manner in which it can either prevent protein assembly/aggregation or remodel and disassemble pre-formed complexes/aggregates (Mhammedi-Alaoui et al., 1994; Levchenko et al., 1995; Wawrzynow et al., 1995). Our data indicate that ClpX inhibits Z-ring formation through a ClpP-independent pathway. Characterization of clpX null and overexpression phenotypes in combination with a variety of cell division mutants suggest that ClpX is important for maintaining the pool of unassembled FtsZ. Intriguingly, our biochemical data indicates that ClpX inhibition of FtsZ assembly does not require nucleotide hydrolysis on the part of ClpX suggesting that ClpX inhibits FtsZ assembly by sterically interfering with FtsZ assembly.
Results
A genetic screen for negative regulators of FtsZ-ring assembly
Fusing ftsZ to the gene encoding the green fluorescent protein (gfp) renders the resulting fusion protein heat-sensitive for function (the ftsZ–gfp allele and gene product are hereafter referred to as ftsZts and FtsZts respectively) (Levin et al., 1999). At the permissive temperature of 30°C most ftsZts cells have medial Z rings. In contrast, after 45 min at 45°C there are very few Z rings present. Not surprisingly, as Z-ring formation is an essential component of the division process, cells whose only source of FtsZ is the ftsZts allele are viable at 30°C but unable to form colonies at ≥45°C (Levin et al., 1999). The defect in FtsZ assembly most probably results from misfolding of the GFP moiety and potentially the entire fusion protein at higher temperatures and can be suppressed by increasing the intracellular concentration of FtsZts itself (Levin et al., 1999).
Previously, we identified ezrA, an inhibitor of FtsZ assembly, as a spontaneous suppressor of ftsZts heat sensitivity (Levin et al., 1999). In order to identify additional negative regulators of FtsZ assembly, we isolated extragenic Tn10 insertions that restored viability at 45°C to a heat sensitive ftsZts strain (PL874) (see Experimental procedures). After screening ~106 colonies, we identified seven that had stable suppressor mutations linked to a Tn10 transposon insertion. Linkage analysis indicated that two of these insertions were in ezrA and two others appeared to correct the ftsZts defect by truncating the gfp moiety on the fusion protein. Sequencing the DNA flanking the transposon in the remaining strains indicated that one suppressor was linked to a gene of unknown function and two were insertions in clpX. clpX encodes the 420 residue (~46 kDa) substrate recognition component of the ClpXP protease.
A deletion in clpX restores viability and FtsZts assembly at 45°C to cells bearing the ftsZts allele
The isolation of clpX::tn10 insertions in the above screen indicated that a null mutation in clpX restores function to the FtsZts protein at the restrictive temperature. To verify this, we proceeded to assess the ability of a previously characterized clpX deletion (clpX::spc; Liu et al., 1999) to suppress the heat-sensitive defect of FtsZts.
Our initial attempts to characterize the clpX::spc phenotype were complicated by the fact that B. subtilis clpX and clpP null mutants possess defects in a host of cellular processes including growth and genetic competence (Gerth et al., 1998; Msadek et al., 1998; Liu et al., 1999; Kruger et al., 2000). Most confounding was the fact that clpX mutant strains quickly acquire extragenic suppressor mutations that restore normal growth. Strains bearing these suppressor mutations rapidly outgrow the parent strain in liquid culture, making phenotypic analysis difficult (Zuber, 2004).
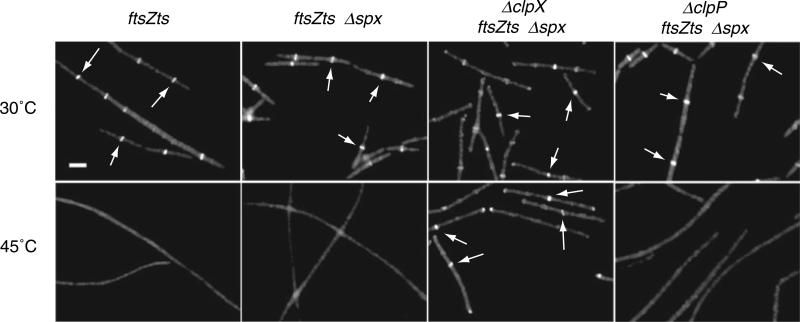
A null mutation in spx, encoding a global transcriptional regulator, relieves many of the negative phenotypes associated with clpX and clpP deletions including the severe defects in growth and competence (Nakano et al., 2001; 2003). We found that while an spx deletion (spx::neo) by itself was not sufficient to restore either viability or Z-ring formation at 45°C to ftsZts cells, an spx::neo clpX::spc double deletion restored ring formation and increased viability ~10 000-fold at the restrictive temperature (Table 1). Furthermore, these cells were proficient at assembling Z rings at the formerly restrictive temperature (Fig. 1). Thus, the clpX deletion restores FtsZts function at 45°C in a robust manner that is independent of the presence of Spx. Importantly, deletions in clpX and/or spx did not alter the intracellular concentration of either FtsZ or FtsZts as determined by quantitative immunoblot (data not shown). Based on these observations all subsequent analyses of the effect of clpX and clpP on FtsZ-ring assembly and cell division were conducted in the presence of the spx deletion.
Table 1.
A clpX null mutation suppresses the lethal ftsZts defect at 45°C.
| Strain | Plating efficiency (%) | Suppression (fold) |
|---|---|---|
| PL642 (ftsZts cm)a | 0.01 ± 0.011 | – |
| BW182 (Δspx ftsZts cm) | 0.01 ± 0.006 | 1 |
| BW189 (ΔclpX Δspx ftsZts cm) | 108 ± 11.93 | 10 800 |
| PL874 (ftsZts spc)a | 0.10 ± 0.131 | – |
| BW206 (ΔclpP Δspx ftsZts spc) | 0.30 ± 0.282 | 3 |
Reference strain.
Fig. 1. ClpX modulates FtsZts assembly in vivo.
A. A clpX null mutation restores Z-ring formation to ftsZts cells at 45°C. PL642 (ftsZts), BW182 (ftsZts spx::neo), BW189 (ftsZts spx::neo clpX::spc) and BW206 (ftsZts spx::neo clpP::cat) cells were cultured in LB at 30°C to mid-exponential phase (OD600 ~0.400) and then back-diluted into LB at 30°C or 45°C and grown for ~2.5 h. FtsZts (FtsZ–GFP) was visualized in fixed cells using fluorescence microscopy. Arrows indicate examples of medial Z rings. Scale bar = 2.5 μm.
ClpX inhibition of Z-ring formation is independent of ClpP
The best-characterized role for ClpX is that of the substrate recognition subunit of the ClpXP protease. The other principle component of this protease, ClpP, is the proteolytically active subunit (Gottesman et al., 1993; Wojtkowiak et al., 1993). ClpX also functions independently of ClpP to remodel and disassemble protein complexes (Mhammedi-Alaoui et al., 1994; Levchenko et al., 1995; Wawrzynow et al., 1995). Given these two disparate functions, it was important to assess whether ClpX was regulating FtsZts assembly by a ClpP-dependent or ClpP-independent mechanism.
We hypothesized that if ClpP is required for the modulation of FtsZts activity by ClpX, then a clpP deletion should suppress the FtsZts defect at high temperature to the same degree as the clpX deletion (Table 1, Fig. 1). However, if ClpX regulates FtsZts assembly in a manner that is independent of ClpP, then a clpP null mutation should not restore either Z-ring assembly or viability to ftsZts cells. Consistent with this latter possibility, we observed that an spx::neo clpP::cat double mutation did not restore either viability or Z-ring assembly to ftsZts cells at 45°C (Table 1, Fig. 1). These data argue that ClpX regulates Z-ring assembly in a manner that is independent of the presence of ClpP and that is therefore not related to ClpX's role in proteolysis.
Overexpression of ClpX inhibits cell division
To confirm that ClpX is a negative regulator of Z-ring formation in vivo, we assessed the effect of overexpressing ClpX within the cell, with the expectation that the overexpression of an FtsZ inhibitor would result in a block in FtsZ-ring formation. clpX was placed under the control of the IPTG-inducible Pspachy promoter (Quisel et al., 2001), the strongest inducible promoter available for B. subtilis, at the native clpX locus. The presence of inducer resulted in the accumulation of intracellular ClpX to concentrations twice that of wild-type cells (data not shown). This modest overexpression of ClpX led to a 20% increase in average cell length [from 4.87 μm (n = 272) to 5.87 μm (n = 300)], consistent with a delay in Z-ring assembly and cell division.
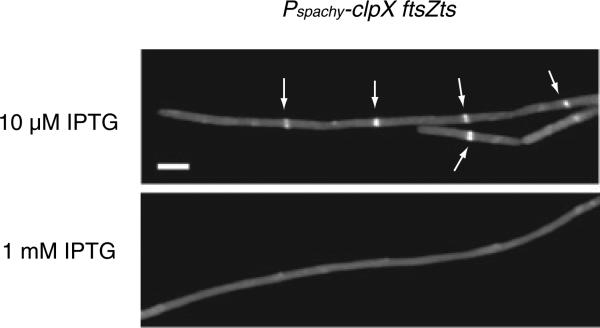
Although the limited overexpression we were able to achieve was insufficient to completely block assembly of wild-type FtsZ, we found that a twofold increase in ClpX levels had a dramatic effect on assembly of the defective FtsZ protein, FtsZts. Previous work indicates that the FtsZts protein is ‘sensitized’ to inhibitors of FtsZ assembly, presumably due to a mild defect in FtsZ polymerization even under permissive conditions (Haeusser et al., 2004). When Pspachy-clpX ftsZts cells were grown in the presence of low levels of inducer (10 μM IPTG was required to maintain basal ClpX expression to prevent the growth defects associated with a complete loss of ClpX), the cells were fully competent for FtsZts assembly and cell division (Fig. 2). However, full induction of ClpX by the addition of 1 mM IPTG, corresponding to a twofold increase in the intracellular concentration of ClpX, caused a severe block in FtsZts assembly and cell division that reduced viability ~1000-fold (Fig. 2). Overexpression of clpX did not affect the intracellular concentration of FtsZts (data not shown). Furthermore, as a control we overexpressed clpC from Pspachy at its native locus. We found that in contrast to ClpX, overexpression of ClpC did not alter FtsZts-ring formation indicating that inhibition of FtsZts assembly is specific to ClpX and not a general feature of this class of proteins (data not shown). These findings support a model in which ClpX acts as an inhibitor of Z-ring formation in vivo.
Fig. 2. ClpX overexpression blocks Z-ring assembly.
A. ClpX overexpression increases cell length. Cells encoding both an IPTG-inducible clpX allele and an ftsZts allele (BW243) were grown in LB at 37°C with 10 μM IPTG to maintain basal levels of ClpX expression. At mid-exponential phase (OD600 ~0.400) cultures were back-diluted into LB containing either 10 μM IPTG (grey bars) or 1 mM IPTG (black bars) and grown for ~2.5 h prior. FtsZts (FtsZ– GFP) was visualized in live cells using fluorescence microscopy. Arrows indicate Z rings in cells grown in LB with 10 μM IPTG. Scale bar = 2.5 μm.
ClpX helps maintain the balance between assembled and unassembled FtsZ
Recent work suggests that a host of regulatory factors act in concert to maintain the balance between assembled and unassembled FtsZ and, thus, ensure precise spatial and temporal regulation of Z-ring formation and cytokinesis (Romberg and Levin, 2003). This regulatory network appears to be robust: null mutations in a number of cell division genes have no discernible cell division phenotype unless combined with mutations in other genes known to affect FtsZ assembly dynamics (Gueiros-Filho and Losick, 2002; Wu and Errington, 2004). As further evidence of the importance of maintaining the balance between assembled and unassembled FtsZ, overexpression of an inhibitor of Z-ring formation can be compensated for either by the loss of another FtsZ inhibitor or by the overexpression of a protein that stabilizes the Z ring (Levin et al., 2001; Gueiros-Filho and Losick, 2002).
Our genetic data indicated that ClpX plays an important role in maintaining this balance between assembled and unassembled FtsZ. To confirm this possibility, we determined whether a null mutation in clpX, a putative inhibitor of FtsZ assembly, could compensate for the overexpression of another division inhibitor, MinCD. The MinCD complex is normally involved in preventing inappropriate Z-ring formation at the cell poles (Rothfield et al., 1999). However, 15-fold overexpression of MinCD results in a lethal block in FtsZ-ring formation and cell division in B. subtilis (Levin et al., 2001). This lethal overexpression of MinCD can be suppressed by a null mutation in a second inhibitor of Z-ring formation, EzrA (Levin et al., 2001). We therefore hypothesized that if ClpX was a bona fide inhibitor of wild-type FtsZ assembly, like EzrA, then a clpX null mutation should suppress the lethal phenotype associated with MinCD overexpression by restoring the proper balance to FtsZ assembly dynamics.
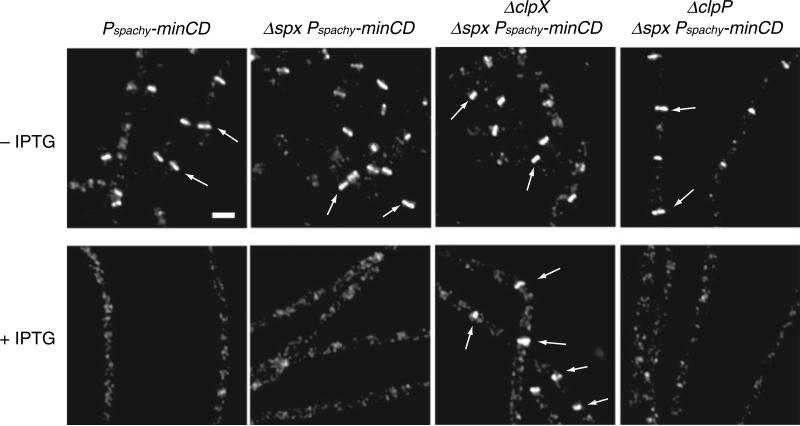
Z-ring formation was blocked and viability reduced ~5000-fold following the addition of IPTG to a strain encoding Pspachy-minCD. However, a clpX::spc spx::neo double mutation fully restored viability to Pspachy-minCD cells in the presence of inducer (Table 2). Moreover, immunofluorescence microscopy indicated that the presence of the clpX::spc spx::neo double mutation restored FtsZ assembly in cells overexpressing MinCD (Fig. 3). The spx::neo mutation alone had no effect on either viability or ring formation following MinCD overexpression (Fig. 3). In contrast to clpX, we found that the loss of clpP did not suppress the lethal block in Z-ring assembly caused by MinCD overexpression (Table 2, Fig. 3). The intracellular concentration of MinCD was not affected by the mutations in spx, clpX and/or clpP (data not shown).
Table 2.
A clpX null mutation suppresses the lethality of a ~15-fold MinCD overexpression.
| Strain | Plating efficiency (%) | Suppression (fold) |
|---|---|---|
| PL1145 (Pspachy-minCD)a | 0.02 ± 0.003 | – |
| BW287 (Δspx Pspachy-minCD) | 0.01 ± 0.003 | 0.5 |
| BW293 (ΔclpX Δspx Pspachy-minCD) | 101 ± 11.17 | 5050 |
| BW220 (ΔclpP Δspx Pspachy-minCD) | 0.05 ± 0.085 | 2.5 |
| BW299 (ΔclpC Pspachy-minCD) | 0.07 ± 0.014 | 3.5 |
Reference strain.
Fig. 3.
A clpX null mutation restores Z-ring formation to cells overexpressing the FtsZ inhibitor MinCD. PL1145 (thrC::Pspachy-minCD), BW287 (thrC::Pspachy-minCD spx::neo), BW293 (thrC::Pspachy-minCD spx::neo clpX::spc) and BW220 (thrC::Pspachy-minCD spx::neo clpP::cat) cells were cultured in LB at 37°C to mid-exponential phase (OD600 ~0.400) and back-diluted into LB ± 1 mM IPTG and grown for ~2.5 h. FtsZ was visualized by immunofluorescence microscopy. Arrows indicate examples of FtsZ rings. Scale bar = 2.5 μm.
As further support of ClpX as an inhibitor of Z-ring formation, we found that twofold overexpression of clpX from the inducible Pspachy promoter at its native locus (see above) reduced minicell formation from 21.2% to 7.7% and polar Z-ring assembly from 43.4% to 24.2% in a minCD null mutant. This result indicates that the additional ClpX is able to compensate partially for the loss of MinCD and prevent aberrant polar Z-ring formation and septation.
These data support a model in which ClpX acts to modulate wild-type FtsZ assembly through a ClpP-independent mechanism. Moreover, these results indicate that ClpX, like EzrA, plays an important part in maintaining the balance between assembled and unassembled FtsZ within the cell.
ClpX inhibits FtsZ assembly in vitro
The genetic analyses described above establish that ClpX is an inhibitor of FtsZ assembly in vivo. To determine whether ClpX exerts its effects on FtsZ directly, we tested the ability of purified ClpX to inhibit assembly of purified wild-type FtsZ in vitro using a 90° angle light scattering assay (Mukherjee and Lutkenhaus, 1999).
All components in the reactions except GTP were mixed and incubated for 60 s to establish a baseline. FtsZ assembly was initiated by the addition of 1 mM GTP and allowed to continue for another 340 s. Reactions typically reached steady state ~180 s after the addition of GTP (Fig. 4A). All reactions were performed in the presence of ATP unless otherwise noted. ATP and its derivatives had no effect on either the kinetics or maxima of FtsZ assembly.
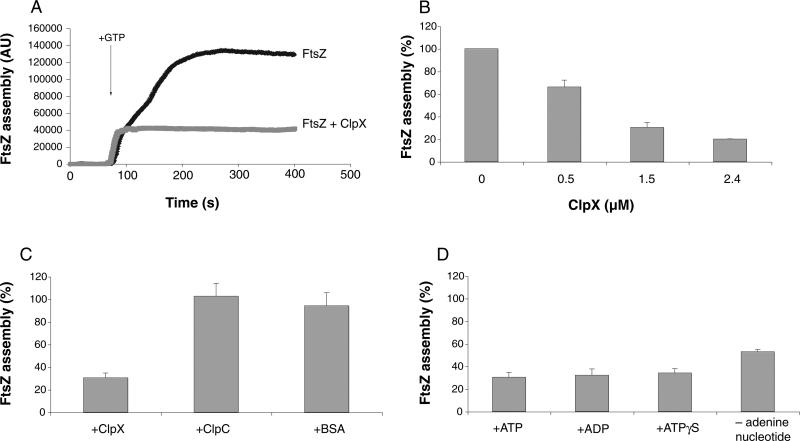
Fig. 4.
ClpX inhibits FtsZ assembly in vitro in a 90° angle light scattering assay. Unless indicated otherwise, all reactions are performed in the presence of 1 mM ATP with an FtsZ concentration of 2.5 μM. Assembly values in (B), (C) and (D) represent maximal levels of FtsZ assembly in the presence of ClpX relative to FtsZ assembly in the absence of ClpX. Error bars represent standard deviation.
A. Representative light scattering plot of FtsZ (black trace) and FtsZ + ClpX (grey trace). FtsZ assembly was initiated by the addition of 1 mM GTP.
B. Dose-dependent inhibition of FtsZ assembly by ClpX.
C. Inhibition of FtsZ assembly by ClpX is specific to ClpX. ClpX, ClpC and BSA were added to a final concentration of 1.5 μM.
D. ATP hydrolysis is not required for inhibition of FtsZ assembly by ClpX. ATP, ADP and ATPγS were added to 1 mM where appropriate.
To test the ability of purified ClpX to inhibit FtsZ assembly, we added increasing concentrations of ClpX to the light scattering reaction. FtsZ was kept at a constant concentration of 2.5 μM. ClpX inhibited FtsZ assembly in a concentration-dependent manner with near maximal inhibition at 1.5 μM ClpX, a ratio of 3 ClpX to 5 FtsZ (Fig. 4B). In light of these results and our difficulty obtaining pure and active ClpX at high concentration, all further experiments assessing the effect of ClpX on FtsZ assembly were performed at a 3:5 ratio of ClpX to FtsZ (concentrations of 1.5 μM ClpX and 2.5 μM FtsZ respectively). The in vivo ratio of ClpX to FtsZ is predicted to be between 1:1 and 2:1 (Feucht et al., 2001; Gerth et al., 2004).
We observed that the addition of 1.5 μM ClpX to 2.5 μM FtsZ resulted in a ~70% inhibition of FtsZ assembly (Fig. 4A and B). As a negative control, we assayed the ability of the ClpX-related chaperone ClpC to inhibit FtsZ assembly. ClpC, like ClpX, functions as a substrate recognition domain for ClpP in the ClpCP protease in B. subtilis (Msadek et al., 1994; Turgay et al., 1998). However, genetic data indicate that ClpC is not a negative regulator of FtsZ-ring assembly; a clpC null mutation does not suppress the lethal effects of a MinCD overexpression (Table 2). Correspondingly, we found that 1.5 μM ClpC had no effect on FtsZ assembly in vitro (Fig. 4C). As an additional negative control, we also tested the ability of bovine serum albumin (BSA) to inhibit FtsZ assembly and found that 1.5 μM BSA did not affect FtsZ assembly (Fig. 4C). Thus, the ability of ClpX to inhibit FtsZ assembly appears to be specific to ClpX.
ClpX inhibition of FtsZ assembly does not require ATP hydrolysis
ClpX is an ATP-binding protein and hydrolysis of ATP is required for many of ClpX's functions including both proteolysis and chaperone-like activities (Gottesman et al., 1993; Wojtkowiak et al., 1993; Kong and Dubnau, 1994; Mhammedi-Alaoui et al., 1994; Levchenko et al., 1995; Wawrzynow et al., 1995; Singh et al., 2000). We therefore tested the requirement for ATP hydrolysis in ClpX's ability to inhibit FtsZ assembly. Strikingly, we found that the level of inhibition by ClpX was virtually identical regardless of whether the reactions were performed in the presence of ATP, ADP or the non-hydrolysable ATP analogue ATPγS (Fig. 4D). Neither ADP, ATPγS nor GTP supports proteolysis of the known substrate Spx by ClpXP (data not shown), indicating that these nucleotides are not hydrolysed by ClpX to a significant degree. Therefore, these data suggest that nucleotide hydrolysis is dispensable for ClpX inhibition of FtsZ assembly. We also observed that ClpX was capable of inhibiting FtsZ assembly in the complete absence of adenine nucleotide, although at somewhat reduced levels when compared with reactions conducted in the presence of ATP, ADP or ATPγS (Fig. 4D). The decrease in inhibition is potentially due to changes in ClpX's oligomerization state resulting from the absence of adenine nucleotide (Wawrzynow et al., 1995; Grimaud et al., 1998).
ClpX does not unfold FtsZ
ClpX's principle enzymatic activity – its ability to unfold proteins – is dependent on the energy derived from ClpX's hydrolysis of ATP. The finding that ClpX does not require ATP hydrolysis to inhibit FtsZ assembly suggests either that ClpX is incapable of unfolding FtsZ or that the unfolding of FtsZ is not involved in ClpX regulation of FtsZ assembly. To examine this issue, we assessed the ability of the ClpXP protease to degrade FtsZ. ClpX-dependent unfolding is required for substrate proteolysis by ClpXP. Thus, the ability of ClpXP to degrade a given substrate is diagnostic of the ability of ClpX to unfold that substrate.
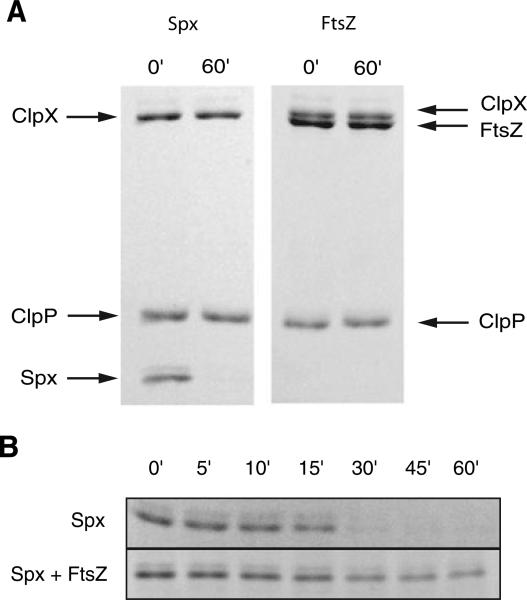
Proteolysis reactions were assembled on ice. ClpX and ClpP were added to a final concentration of 2.4 μM and 3.6 μM respectively. Either Spx or FtsZ was added to 0.1 μg μl−1. Reactions were initiated by the addition of 5 mM ATP and incubated at 37°C. Samples were removed at 0 and 60 min and analysed by SDS-PAGE. We found that ClpXP was unable to degrade FtsZ even under conditions that allowed the efficient degradation of a known ClpXP substrate, Spx (Fig. 5A). These data indicate that ClpX cannot unfold FtsZ and is consistent with the finding that deletions in clpX and clpP do not affect intracellular levels of FtsZ.
Fig. 5.
FtsZ interacts with, but is not degraded by, ClpXP.
A. ClpXP is able to degrade Spx but not FtsZ in vitro. Proteolysis reactions were performed at 37°C in the presence of 5 mM ATP and sampled at 0 and 60 min.
B. The presence of 10-fold mass excess of FtsZ relative to Spx inhibits the degradation of Spx by ClpXP. Proteolysis reactions were performed at 30°C and sampled at 0, 5, 10, 15, 30, 45 and 60 min. Samples in (A) and (B) were analysed by SDS-PAGE and stained with coomassie Brilliant Blue and representative gels are shown.
While ClpXP does not degrade FtsZ, FtsZ does inhibit the ability of ClpXP to degrade other substrates. We found that a 10-fold mass excess of FtsZ (1.0 μg μl−1) relative to the ClpXP substrate Spx (0.1 μg μl−1) strongly inhibited the proteolysis of Spx by ClpXP (Fig. 5B). In contrast, a 10-fold mass excess of BSA (1.0 μg μl−1) did not interfere with the degradation of Spx (data not shown). The simplest explanation for this finding is that the interaction between FtsZ and ClpX interferes with the docking between ClpX and either Spx or ClpP. FtsZ is unlikely to compete with ClpX for ATP as FtsZ does not possess an appreciable ATPase activity (data not shown).
ClpX does not affect FtsZ GTPase activity in vitro
Z-ring formation is thought to occur in two stages: (i) polymerization of FtsZ into single-stranded polymers or protofilaments and (ii) the assembly of FtsZ protofilaments into stable bundles via lateral interactions between polymers (Romberg and Levin, 2003). The light scattering assays discussed in the previous sections demonstrate that ClpX inhibits FtsZ assembly in vitro, but cannot distinguish which stage in FtsZ assembly is being affected. The GTPase active site is created by the interactions between the faces of two FtsZ subunits within FtsZ dimers or multimers (Erickson, 1998; Löwe and Amos, 1998; Sossong et al., 1999). Therefore, factors that block multimerization by sequestering free FtsZ monomers inhibit GTP hydrolysis (Trusca et al., 1998). To begin to elucidate which step in FtsZ assembly ClpX acts upon, we decided to examine whether ClpX alters FtsZ's intrinsic GTPase activity.
GTPase assays were performed under conditions analogous to those used for light scattering and all reactions contained 1 mM ATP. Components of the reactions were pre-assembled and assays were initiated by the addition of a mixture of unlabelled GTP and radioactively labelled [γ-32P]-GTP. Reactions were sampled every 5 min for 20 min, samples were separated by thin layer chromatography (TLC) and the generation of free radioactive phosphate was quantified by phosphorimaging coupled with densitometric analysis.
We found that the addition of 1.5 μM ClpX to a reaction containing 2.5 μM FtsZ, a condition that resulted in ~70% inhibition of FtsZ assembly (Fig. 4A and B), had no significant effect on FtsZ's GTPase activity (FtsZ: 0.15 ± 0.03 GTP FtsZ−1 min−1; FtsZ + ClpX: 0.18 ± 0.04 GTP FtsZ−1 min−1). ClpX on its own did not possess any detectable GTPase activity (data not shown). These data indicate that ClpX inhibits FtsZ assembly through a mechanism that does not affect FtsZ's intrinsic GTPase activity and argue against it functioning as a monomer sequestering protein.
Discussion
Using a battery of genetic and biochemical assays, we have established that B. subtilis ClpX is a potent inhibitor of Z-ring formation in vivo and FtsZ assembly in vitro. Our data indicate that ClpX inhibits FtsZ assembly in the absence of ClpP. ClpP-independent roles for ClpX have been described for E. coli ClpX and its substrates MuA from Phage Mu and Protein O from Phage Lambda (Figs 1 and 3, Tables 1 and 2). In these cases ClpX modulates the assembly/aggregation state of the relevant substrate complexes (Levchenko et al., 1995; Wawrzynow et al., 1995). ClpX is thought to use ATP hydrolysis to power the partial unfolding of target substrates within the complex which in turn destabilizes the substrate complex as a whole (Burton et al., 2001; Kenniston et al., 2003). This allows either the remodelling or disassembly of the substrate complex.
We find that B. subtilis ClpX does not require ATP hydrolysis to block FtsZ assembly (Fig. 4D), suggesting that binding to FtsZ is sufficient to inhibit polymerization. A similar phenomenon has been observed with E. coli ClpX and its substrate Lambda Protein O. Protein O is susceptible to a heat-induced aggregation that is both preventable and reversible by ClpX (Wawrzynow et al., 1995). Wawrzynow et al. found that ClpX requires ATP hydrolysis to reverse Protein O aggregation but not to prevent it. Thus, there is precedence for ClpX regulating protein complex assembly in the absence of ATP hydrolysis as our data suggest occurs between B. subtilis ClpX and FtsZ.
ClpX oligomerizes into a hexameric ring in the presence of ATP, ATPγS and ADP and this oligimerization is important for ClpX's function as both a chaperone and subunit of the ClpXP protease. Interestingly, in the absence of exogenously added adenine nucleotide, we find that the inhibition of FtsZ assembly by ClpX is somewhat, but not completely, reduced. This result suggests that ClpX's effect on FtsZ is sensitive to the ClpX oligomerization state. Previous studies have suggested that ClpX remains partially assembled even in the absence of exogenously added nucleotide (Grimaud et al., 1998), possibly due to contaminating ADP that co-purifies with ClpX (Kim and Kim, 2003). As such, it is not immediately clear whether unassembled ClpX remains partially active against FtsZ or whether a fraction of the ClpX remains assembled and, therefore fully active in the absence of added adenine nucleotide. Uncertainty regarding the functional ClpX unit combined with limitations in our assays’ ability to distinguish between different oligomerization states of FtsZ (e.g. single-stranded protofilaments versus bundled polymers; see below) confounds our ability to interpret the stoichiometric ratios at which ClpX inhibits FtsZ assembly without additional experimentation.
Current models suggest that FtsZ assembly commences with the polymerization of FtsZ monomers into single-stranded polymers or protofilaments followed by the assembly of these polymers via lateral interactions into protofilament bundles. Based on analogy with factors governing cytoskeletal assembly in eukaryotes, there are many possible mechanisms for ClpX inhibition of FtsZ assembly (Desai and Mitchison, 1997; Pollard et al., 2000). One of the simplest models is that ClpX functions as a monomer-binding protein, sequestering free FtsZ monomers and preventing them from being added to growing polymers. This mode of action has precedence in the regulation of FtsZ assembly. The DNA damage-induced protein SulA inhibits Z-ring formation following DNA damage by sequestering FtsZ monomers (Trusca et al., 1998; Cordell et al., 2003). Because GTP hydrolysis is dependent on multimerization, SulA dramatically reduces GTPase activity when present at equimolar ratios by preventing interaction between FtsZ subunits (Trusca et al., 1998). However, in contrast to SulA, we found that ClpX has no observable effect on FtsZ's GTPase activity, even when assayed under conditions in which ClpX blocked FtsZ assembly by > 70%. Thus, ClpX is unlikely to bind to and sequester FtsZ monomers with high affinity.
It also seems unlikely that ClpX regulates FtsZ assembly by unfolding FtsZ subunits within the polymer and thereby disassembling pre-formed polymers. First, ATP hydrolysis is not required to block FtsZ assembly (Fig. 4D), suggesting a non-enzymatic mechanism of inhibition. Moreover, ClpXP is unable to proteolyse FtsZ (Fig. 5A), suggesting that ClpX is incapable of unfolding FtsZ subunits (Fig. 5A).
Although ClpXP does not degrade FtsZ, the observed competition between FtsZ and the ClpXP substrate Spx for access to the protease (Fig. 5B) argues that FtsZ is capable of docking physically with ClpX. Indeed, the C-terminus of FtsZ is rich in basic amino acids and as such resembles signal sequence motifs involved in interacting with ClpX (Flynn et al., 2003). Thus, the simplest model for the inhibition of FtsZ assembly by ClpX is that ClpX binds to FtsZ and sterically hinders FtsZ assembly. For example, ClpX might function to cap growing polymers of FtsZ rendering them too short to be incorporated efficiently into the FtsZ ring. Alternatively, ClpX may inhibit the lateral interactions between FtsZ protofilaments that lead to the formation of stable polymer bundles. Ultimately establishing the mechanism by which ClpX modulates FtsZ assembly will require extensive in vitro analysis of FtsZ assembly dynamics in the presence of the chaperone.
Bacterial cells maintain a reservoir of unassembled FtsZ throughout the cell cycle (Weart and Levin, 2003). Although FtsZ is present at concentrations 5- to 10-fold greater than the critical concentration required for assembly (Lu et al., 1998; Feucht et al., 2001), only 30% of available protein appears to be assembled into the Z ring at a given time (Stricker et al., 2002). The presence of a large pool of cytoplasmic FtsZ is presumably important for the maintenance and control of the highly dynamic FtsZ ring in vivo (Stricker et al., 2002). Our genetic data indicate that, like EzrA, ZapA and Noc, ClpX is important in maintaining the balance between assembled and unassembled FtsZ within the cell. Immunolocalization of ClpX indicates that it is distributed evenly throughout the cytoplasm (R.B. Weart, unpublished data), suggesting that ClpX impacts FtsZ assembly throughout the bulk of the cytosol.
ClpX is conserved among eubacteria and archaea and is therefore likely to be involved in the modulation of FtsZ assembly in organisms other than B. subtilis. Indeed, E. coli ClpX was recently shown to interact directly with FtsZ, suggesting that ClpX and/or ClpXP may play a role in regulating FtsZ assembly or stability in Gram-negative bacteria (Flynn et al., 2003). These data, together with our observation that ClpX is a potent inhibitor of FtsZ assembly in B. subtilis, suggest that ClpX's activity as a modulator of FtsZ assembly may be a widely conserved mechanism for regulating cell division in prokaryotes.
Experimental procedures
General methods
Cloning and genetic manipulations were performed as described previously (Sambrook et al., 1989; Harwood and Cutting, 1990). B. subtilis strains were grown in LB at either 37°C for strains bearing the wild-type ftsZ+ allele or 30°C for strains bearing the heat-sensitive ftsZts allele (Table 3). To assess viability, strains were grown to mid-exponential phase (OD600 ~0.400), serially diluted from 10−3 to 10−6, and equal volumes plated at the permissive (30°C for ftsZts strains; no IPTG for MinCD overexpression strains; 10 μM IPTG for ClpX overexpression strains) and restrictive (45°C for ftsZts strains; 1 mM IPTG for MinCD and ClpX overexpression strains) conditions. ‘Plating efficiency’ was calculated as the ratio of colonies at the restrictive versus permissive conditions. ‘Suppression’ was calculated as the ratio of the plating efficiency of an experimental strain versus the plating efficiency of a reference strain. Results are averages for either two (MinCD overexpression strains) or three (ftsZts strains) experiments and error bars represent standard deviation.
Table 3.
Bacterial strains.
| Strains | Genotype | Reference |
|---|---|---|
| B. subtilis | ||
| PL642 | ftsZts cat | Levin et al. (1999) |
| PL874 | ftsZts spc | This work |
| PL990 | minCD::spc | Levin et al. (1998) |
| PL1145 | thrC::Pspachy-minCD erm | This work |
| PL1296 | ftsZts spc pHV1249 | This work |
| PL1610 | clpC::Pspachy-clpC | This work |
| PL1612 | clpC::Pspachy-clpC ftsZts spc | This work |
| BW182 | spx::neo ftsZts cat | This work |
| BW189 | clpX::spc spx::neo ftsZts cat | This work |
| BW206 | clpP::cat spx::neo ftsZts spc | This work |
| BW220 | clpP::cat spx::neo thrC::Pspachy-minCD erm | This work |
| BW239 | clpX::Pspachy-clpX | This work |
| BW243 | clpX::Pspachy-clpX ftsZts spc | This work |
| BW254 | minCD::spc clpX::Pspachy-clpX | This work |
| BW287 | spx::neo thrC::Pspachy-minCD erm | This work |
| BW293 | clpX::spc spx::neo thrC::Pspachy-minCD erm | This work |
| BW299 | clpC::spc thrC::Pspachy-minCD erm | This work |
| JRL672 | clpX::spc | Liu et al. (1999) |
| BD2243 | clpC::spc amyE::comG-lacZ | Kong and Dubnau (1994) |
| BD2590 | clpP::cat | Turgay et al. (1998) |
| ORB3834 | spx::neo | Nakano et al. (2001) |
| E. coli | ||
| PL1184 | ER2566 pBS58 pCXZ | Haeusser et al. (2004) |
| GZ1 | ER2566 pClpC | Nakano et al. (2002) |
| MMN509 | BL21(DE3) pLysS pMMN509 | Nakano et al. (2003) |
| PP61 | ER2566 pTYB1-clpP | D. Dubnau and P. Prepiak |
Isolation of suppressor mutations
Heat-sensitive strain PL1296 encoding ftsZts (ftsZ::ftsZ–gfp) and the transposon bearing plasmid pHV1249 (Petit et al., 1990) was taken from overnight culture and diluted 1:25 into fresh selective media (LB chloramphenicol 5 μg ml−1, spectinomycin 100 μg ml−1) at 30°C. When cell density reached OD600 ~0.25 the culture was diluted twofold into pre-warmed media at 51°C and cultured for 2 h to select for loss of the plasmid and chromosomal integration of the transposon. Colony-forming units were determined by serial dilution at 30°C. The remaining culture was frozen in 1 ml aliquots with 10% glycerol at −80°C. Frozen aliquots were thawed and plated onto selective medium at 45°C to identify those colonies bearing second site suppressor mutations that restored growth to ftsZts cells at the restrictive temperature. We obtained resistant colonies at a frequency of ~10−5. These colonies were passaged twice at 45°C to ensure that they retained their heat-resistant phenotype.
Linkage of suppressor mutations to the transposon was determined by transformation of chromosomal DNA into a fresh ftsZts background selecting for chloramphenicol resistance and screening for heat resistance. Further linkage analysis was used to eliminate transposon insertions in the previously identified cell division gene ezrA (Levin et al., 1999) and ftsZ. Inverse polymerase chain reaction (PCR) was employed to amplify DNA flanking the transposon in the three suppressor bearing strains that did not show linkage to ezrA or ftsZ. Sequencing and comparison of the B. subtilis database (http://genolist.pasteur.fr/SubtiList/) revealed two insertions in clpX and one in a gene of unknown function.
Microscopy
Fluorescence microscopy was performed using an Olympus BX51 microscope and a Hamamatsu OrcaERG camera. Images were processed using Openlabs version 3.5 (Improvision) and Adobe Photoshop CS version 8.0 (Adobe Systems).
Immunofluorescence microscopy
Cells were prepared as described previously (Levin, 2002), except that lysozyme treatment was extended to 10 min. FtsZ was detected using affinity-purified polyclonal rabbit anti-FtsZ sera (Levin and Losick, 1996) in combination with either Donkey anti-rabbit sera conjugated to Cy-3 (Jackson Immunoresearch) or goat anti-rabbit sera conjugated to Alexa 488 (Molecular Probes). Cell walls were visualized with wheat germ agglutinin (WGA) conjugated to either fluorescein or tetramethylrhodamine (Molecular Probes). Cell length measurements were performed on cells stained with WGA-tetramethylrhodamine using Openlabs version 3.5 (Improvision). Statistical analysis of cell-length distributions was performed using a chi-squared test for homogeneity with six degrees of freedom (df) and a significance (α) of 0.001.
GFP
Cells were fixed in glutaraldehyde/paraformaldehyde (Levin, 2002) and adhered to the slide with 1% poly-l-lysine (Sigma). Alternatively, live cells were visualized after adherence to an agarose pad (Levin, 2002).
Protein purification
FtsZ was purified as described (Haeusser et al., 2004) and stored at −80°C in 50 mM Mes pH 6.5, 2.5 mM Mg(CH3COO)2 and 1 mM EGTA. ClpX, ClpC and ClpP were purified using the Impact system (New England Biolabs). ClpX was subjected to further purification using Bio-Rad HighQ anion exchange column (Nakano et al., 2003). ClpX, ClpC and ClpP were dialysed into 20 mM Tris pH 7.5, 100 mM KCl, 10% glycerol, 1 mM dithiothreitol (DTT) and stored at −80°C. Spx-his was purified using a Ni-NTA column (Qiagen) followed by a Bio-Rad HighQ anion exchange column (Nakano et al., 2003). Spx-his was dialysed into 25 mM sodium phosphate pH 8.0, 100 mM KCl, 5 mM DTT, 1 mM TCEP, 5% glycerol and stored at −80°C.
90° angle light scattering
Light scattering assays were performed using a FluoroMax-2 fluorimeter. Assays were monitored at 310 nm with a slit width of 1 mm. Reaction components were at the following final concentrations: 25 mM Mes pH 6.5 and 5 mM Tris pH 7.5 to yield a final pH 6.5; 50 mM KCl; 0.25 mM DTT; 5% glycerol; 2.5 mM MgCl2. FtsZ was added to 2.5 μM. GTP was added to 1 mM. ATP, ADP or ATPγS was added, where appropriate, to 1 mM.
Unless noted, all components except GTP were mixed in a quartz cuvette (Starna Cells) and a baseline was established for 60 s. Reactions were initiated by the addition of GTP and allowed to continue for an additional 340 s. Reactions containing FtsZ and either ClpX, ClpC or BSA were preceded by a reference reaction that contained FtsZ alone. Relative assembly was calculated by dividing the maxima for the ClpX-, ClpC- or BSA-containing reactions by the maxima for the FtsZ-only reference. Assembly values reflect an average of either two (ATPγS-containing reactions) or three (all other reactions) experiments and error bars reflect the standard deviations.
Proteolysis assay
Reactions were assembled on ice. Reaction components were added to the following concentrations: 4.6 mM Mes pH 6.5 and 14 mM Tris pH 7.5 and 2.7 mM sodium phosphate pH 8.0 to yield a final pH 7.5; 80.8 mM KCl; 0.38 mM DTT; 7.54% glycerol; 0.23 mM Mg(CH3COO)2, 5 mM MgCl2, 0.09 mM EGTA. ClpX and ClpP were added to final concentrations of 2.4 μM and 3.6 μM respectively. Spx, FtsZ or BSA was added to final concentrations of 1 μg μl−1. Reactions were initiated by the addition of 5 mM ATP and incubated at 37°C. Samples were taken at 0 and 60 min. Substrate competition assays were performed as above except that FtsZ was added to a final concentration of 10 μg μl−1, reactions were incubated at 30°C and samples were taken at 0, 5, 10, 15, 30, 45 and 60 min. All samples were analysed by SDS-PAGE in conjunction with Kodak 1D version 3.5.3 after staining with coomassie Brilliant Blue.
GTPase assay
Reaction components were at the same final concentrations as described for the 90° angle light scattering assays except that Tris pH 7.5 was added to 10 mM yielding a final pH of ~6.5. All reaction components were assembled in the absence of GTP and reactions were initiated by the addition of a mixture of unlabelled GTP (1 mM final concentration) and radioactively labelled [γ-32P]-GTP (0.5 μCi). Reactions were sampled at 5, 10, 15 and 20 min and samples were quenched using an equal volume of 8 M urea, 20 mM Tris pH 7.5 and 20 mM EDTA. Samples were run on PEI-cellulose TLC plates (JT Baker) and the release of free phosphate was quantified using a Bio-Rad molecular Imager FX phosphorimager in conjunction with Quantity One version 4.2.1 (Bio-Rad). GTP hydrolysis rates represent an average of three experiments and error bars reflect the standard deviation.
Acknowledgements
We extend our thanks to Nir Keren and the Pakrasi laboratory for the use of and assistance with their fluorimeter. We are grateful to Jeffrey Errington and Tania Baker for their generous provision of antisera and to Michiko Nakano, David Dubnau, Peter Prepiak and Alan Grossman for the gift of several bacterial strains. We are indebted to Peter Prepiak and Michiko Nakano for helpful advice in purifying proteins, the Kranz lab and Doug Chalker for their assistance with the GTPase assays and Laura Romberg for her advice regarding the 90° angle light scattering assay. We appreciate Daniel Haeusser's assistance in purifying FtsZ and are also grateful to members the Levin Laboratory for helpful discussions. Finally, we would like to thank Laura Romberg and Peter Chivers for insightful comments on this manuscript. This work was supported in part by NIH Public Health Service Grants GM45898 to P.Z. and GM64671 to P.A.L.
References
- Burton BM, Williams TL, Baker TA. ClpX-mediated remodeling of mu transpososomes: selective unfolding of subunits destabilizes the entire complex. Mol Cell. 2001;8:449–454. doi: 10.1016/s1097-2765(01)00307-0. [DOI] [PubMed] [Google Scholar]
- Cordell SC, Robinson EJ, Lowe J. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc Natl Acad Sci USA. 2003;100:7889–7894. doi: 10.1073/pnas.1330742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cel Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Atomic structures of tubulin and FtsZ. Trends Cell Biol. 1998;8:133–137. doi: 10.1016/s0962-8924(98)01237-9. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Gerth U, Kruger E, Derre I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- Gerth U, Kirstein J, Mostertz J, Waldminghaus T, Miethke M, Kock H, Hecker M. Fine-tuning in regulation of Clp protein content in Bacillus subtilis. J Bacteriol. 2004;186:179–191. doi: 10.1128/JB.186.1.179-191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Clark WP, de Crecy-Lagard V, Maurizi MR. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol. 2004;52:801–814. doi: 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. John Wiley and Sons; Chichester: 1990. [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kong L, Dubnau D. Regulation of competence-specific gene expression by Mec-mediated protein–protein interaction in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:5793–5797. doi: 10.1073/pnas.91.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger E, Witt E, Ohlmeier S, Hanschke R, Hecker M. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol. 2000;182:3259–3265. doi: 10.1128/jb.182.11.3259-3265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Luo L, Baker TA. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- Levin PA. Sansonetti PJ, Zychlinsky A, editors. Light microscopy techniques for bacterial cell biology. In Molecular Cellular Microbiology. London: Academic Press. 2002:115–132. [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Levin PA, Shim JJ, Grossman AD. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Schwartz RL, Grossman AD. Polymer stability plays an important role in the positional regulation of FtsZ. J Bacteriol. 2001;183:5449–5452. doi: 10.1128/JB.183.18.5449-5452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cosby WM, Zuber P. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima – quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Mhammedi-Alaoui A, Pato M, Gama MJ, Toussaint A. A new component of bacteriophage Mu replica-tive transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc Natl Acad Sci USA. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msadek T, Dartois V, Kunst F, Herbaud ML, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol Microbiol. 2001;42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Zheng G, Nakano MM, Zuber P. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J Bacteriol. 2002;184:3664–3670. doi: 10.1128/JB.184.13.3664-3670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci USA. 2003;100:4233–4238. doi: 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MA, Bruand C, Janniere L, Ehrlich SD. Tn10-derived transposons active in Bacillus subtilis. J Bacteriol. 1990;172:6736–6740. doi: 10.1128/jb.172.12.6736-6740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol. 2001;183:6573–6578. doi: 10.1128/JB.183.22.6573-6578.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Romberg L, Simon M, Erickson HP. Polymerization of FtsZ, a bacterial homolog of tubulin: is assembly cooperative? J Biol Chem. 2001;4:11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Scheffers DJ, de Wit JG, den Blaauwen T, Dries-sen AJ. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry. 2002;41:521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossong TM, Jr, Brigham-Burke MR, Hensley P, Pearce KH., Jr Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38:14843–14850. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, et al. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Zuber P. Spx–RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]