Abstract
Adenocarcinoma of the pancreas has a poor prognosis. At present, no relevant personalized targets have been identified. Sequencing studies have implicated gene alterations of disruptor of telomeric silencing 1 like histone lysine methyltransferase (DOT1L) in pancreatic adenocarcinoma. DOT1L is part of the histone modification system and catalyzes methylation of H3K79, which is crucial in cell signaling and DNA damage repair. DOT1L is considered to be a target of therapy in mixed lineage leukemia gene-deficient leukemia cases and a potential target in breast carcinoma. The frequencies and importance of DOT1L copy-number variations and their specific correlation with protein expression in adenocarcinoma of the pancreas have yet to be investigated. In the present study, tissue microarrays of 230 resected pancreatic adenocarcinoma cases were constructed. The tumor tissue was analyzed using fluorescence in situ hybridization (FISH) and immunohistochemistry. In total, 10/225 carcinoma cases (4.4%) analyzed by immunohistochemistry demonstrated intense nuclear protein expression of DOT1L and in 9/224 tumors analyzed using FISH (4.0%), copy-number variations (CNV) were detectable. No DOT1L amplification was detected in the carcinoma cohort. To the best of our knowledge, the present study describes for the first time the frequency of CNV of DOT1L using the gold standard fluorescence in situ hybridization (FISH) and their specific correlation to the protein expression in adenocarcinomas of the pancreas. Although the positive cases by immunohistochemistry and copy-number variations by FISH were not congruent with each other, the data suggest a potential role for DOT1L in a small subset of pancreatic cancer cases. The significance of the two analysis methods concerning their druggability in pancreatic adenocarcinoma requires further studies.
Keywords: DOT1L, pancreatic adenocarcinoma, immunohistochemistry, fluorescence in situ hybridization, copy-number variations, personalized therapy
Introduction
Gene expression and regulation are based among other things on the posttranslational modification system of histone proteins (1). Disruptor of telomeric silencing 1 like histone lysine methyltransferase (DOT1L) is an evolutionarily conserved histone methyltransferase that, according to the current state of knowledge, exclusively catalyzes the methylation of H3K79 (2,3). In cancer, this methylation is involved in DNA damage repair and tumorigenesis. DOT1L was revealed to be crucial in leukemic transformation and is associated with poor prognosis in breast and colorectal cancer (4–7). Sequencing studies (www.cbioportal.org) demonstrate DOT1L alterations in ≥13% of pancreatic cancer cases. The frequencies of DOT1L copy-number variations determined using gold standard fluorescence in situ hybridization (FISH), and their specific correlation to protein expression, remain to be established.
At present, no relevant personalized treatable target exists in pancreatic adenocarcinoma, thus conventional chemotherapy and radiotherapy is still the standard systemic therapy (8–10). Nevertheless the prognosis of pancreatic adenocarcinoma remains poor, and surgery is the only curative option for patients. However, only 20% of adenocarcinomas are operably linked to a curative approach at the time of diagnosis (11,12). Targeted therapy strategies are urgently required for this severe form of carcinoma.
In the present study, DOT1L was evaluated as a potential therapeutic target in pancreatic adenocarcinoma. According to the results of genomic studies, immunohistochemical analysis and FISH were performed, hypothesizing that the identification of a subgroup of carcinoma cases may demonstrate a significant DOT1L copy-number variation or amplification.
Patients and methods
Patients and tumor samples
In the present retrospective study 230 patients with pancreatic adenocarcinoma who underwent surgical pancreatic resections at the University Hospital of Cologne (Cologne, Germany) between January 1999 and December 2014 were analyzed. For tissue microarray analysis (TMA), two tissue cores from various areas of each tumor were obtained and transferred into a TMA recipient block. TMA construction was performed as described previously (13,14). Patient characteristics are presented in Table I. In brief, tissue cylinders with a diameter of 1.2 mm each were punched from selected tumor tissue blocks using a homemade semi-automated precision instrument and inserted into empty recipient paraffin blocks. Four μm-thick sections of the resulting TMA blocks were transferred to an adhesive-coated slide system (Instrumedics Inc., Hackensack, NJ, USA). Consecutive sections were used for FISH and immunohistochemistry.
Table I.
Clinicopathological characteristics of 230 pancreatic adenocarcinoma cases.
| Characteristic | No. of patients |
|---|---|
| Gender | |
| Male | 124 |
| Female | 106 |
| Age, years | |
| <50 | 51 |
| 50–60 | 70 |
| >70 | 99 |
| Tumor stage | |
| pT1 | 9 |
| pT2 | 25 |
| pT3 | 190 |
| pT4 | 6 |
| Lymph node metastasis | |
| N0 | 66 |
| N1 | 164 |
| Resection margin status | |
| R0 | 148 |
| R1 | 82 |
| Grading | |
| G1 | 3 |
| G2 | 152 |
| G3 | 75 |
Immunohistochemistry
Immunohistochemistry (IHC) was performed on TMA slides using the primary rabbit polyclonal antibody specific for DOT1L (#NB100-40845; dilution, 1:50; Novus Biologicals, Ltd., Cambridge, UK). Immunohistochemical staining was performed using the Ventana BenchMark stainer (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's protocol with on-slide controls of the appendix. As a secondary antibody the ready-to-use Ventana Detection kit (cat. no. 760–700; OptiView DAB IHC Detection kit; Ventana, Roche Diagnostics GmbH, Mannheim, Germany) for indirect biotin-free detection of primary rabbit antibody was used. DOTL1 exhibits a nuclear staining pattern. DOT1L staining was evaluated manually using light microscopy by two pathologists and discrepant results were resolved by consensus review.
FISH
FISH for evaluation of DOT1L gene copy numbers was performed with the DOT1L/CEN19 Dual Color Probe (Empire Genomics, Buffalo, NY, USA). Three-μm tissue sections on SuperFrost Plus slides were mounted by heating at 56°C, followed by semi-automated deparaffinization protease digestion washing steps (VP2000 processor system, Abbott Molecular, Wiesbaden, Germany). Protease digestion was performed using the ready-to-use FISH pretreatment kit (Vysis IntelliFISH Universal FFPE Tissue Pretreatment Protease; Abbott Molecular, Wiesbaden, Germany). Slides were hybridized at 37°C overnight with the FISH probe. The slides were stained with DAPI prior to analysis. Normal tissue including vessels, fibroblasts, or non-tumor lung tissue served as an internal positive control. Cases were only further evaluated if control tissue nuclei exhibited one or two clearly distinct signals of each color. Tumor tissue was scanned for amplification hot spots using ×63 objective (DM5500 fluorescent microscope; Leica Microsystems, Inc., Buffalo Grove, IL, USA). If the signals were homogeneously distributed, then random areas were used for counting the signals. Sixty tumor cells in three areas were evaluated by counting green DOT1L and orange centromere 19 (CEN19) signals. The reading strategy followed that of the cMET-FISH probe to evaluate different levels of amplification. Low-level amplification was defined as ≥4 DOT1L signals in ≥40% of cells. Intermediate-level amplification was defined as ≥5 DOT1L in ≥50% of cells. High-level amplification was defined as a DOT1L/CEN19 quotient of 2.0, ≥15 DOT1L signals in ≥10% of cells or an average DOT1L copy number of ≥6.
Procedures were followed as outlined in accordance with ethical standards formulated in the Helsinki Declaration 1975 (and revised in 1983) with pre-approval by the Ethics Committee at the University Hospital (Cologne, Germany; reference number: 09–232).
Results
Immunohistochemistry
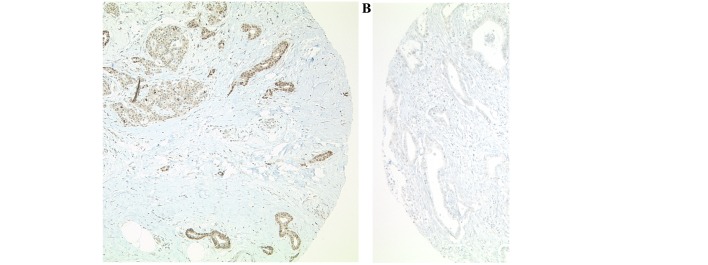
DOT1L immunohistochemistry was performed on TMA slides, including 230 pancreatic carcinomas. Tissue from the appendix vermiformis served as on-slide control. Of the 230 pancreatic carcinomas, 225 were evaluable. Ten carcinomas (4.4%) revealed strong nuclear staining (Fig. 1).
Figure 1.
Immunohistochemistry of DOT1L in arrayed pancreatic adenocarcinomas. Nuclear staining for (A) DOT1L (positive) and (B) DOT1L negative tissues. Original magnification, ×100. DOT1L, disruptor of telomeric silencing 1 like histone lysine methyltransferase.
FISH
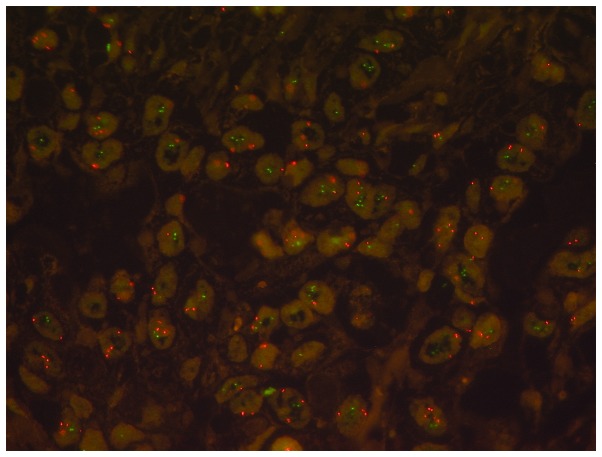
The DOT1L-FISH-signal pattern revealed an even distribution of the signals in the majority of cases. A total of 224 pancreatic adenocarcinomas were able to be analyzed using FISH; there were 9 cases with single tumor cells exhibiting microclusters of >5 DOT1L-signals accompanied by elevated CEN-signals, but no amplification according to the evaluation criteria (using common low-, intermediate- and high level amplification criteria of MET analyses) was detected (Table II). These elevated DOT1L signals were detected in cases where polysomy was already detected. Polysomy was slightly increased, ranging from 1.55–2.5 average CEN-signals/cell (Fig. 2).
Table II.
Fluorescence in situ hybridization analysis of copy number variations of DOT1L in 9 pancreatic adenocarcinoma cases.
| Patient | Green signals/60 cells | Orange signals/60 cells | Ratio green:orange | Average green signals | Average orange signals |
|---|---|---|---|---|---|
| 1 | 162 | 93 | 1.74 | 2.7 | 1.55 |
| 2 | 174 | 150 | 1.16 | 2.9 | 2.5 |
| 3 | 168 | 94 | 1.78 | 2.8 | 1.6 |
| 4 | 186 | 126 | 1.48 | 3.1 | 2.1 |
| 5 | 176 | 132 | 1.32 | 2.9 | 2.2 |
| 6 | 193 | 145 | 1.32 | 3.2 | 2.4 |
| 7 | 167 | 127 | 1.3 | 2.8 | 2.1 |
| 8 | 158 | 118 | 1.34 | 2.6 | 2.0 |
| 9 | 195 | 130 | 1.5 | 3.2 | 2.2 |
DOT1L signal is represented as green; CEN19 signal is represented as orange. DOT1L, disruptor of telomeric silencing 1 like histone lysine methyltransferase; CEN19, centromere 19.
Figure 2.
Fluorescence in situ hybridization analysis of DOT1L in arrayed pancreatic adenocarcinoma tissue. Copy number variations of DOT1L are shown as green and the signals of centromere CEN19 presented as orange. DOT1L, disruptor of telomeric silencing 1 like histone lysine methyltransferase; CEN19, centromere 19.
Discussion
According to the analysis of sequencing data, certain cancer types exhibit DOT1L gene alterations. Concordant with the biology of MET, the present study hypothesized that a significant elevation of gene copy number of DOT1L may correlate with protein overexpression. The augmentation of DOT1L protein expression may further be associated with an increased response to a DOT1L-inhibitor therapy. DOT1L is a druggable target causing cell cycle arrest and leading to chemosensitivity in leukemia (15,16). The DOT1L inhibitor EPZ-5676 is under clinical investigation in a phase I study for mixed lineage leukemia (www.ClinicalTrials.gov; cat. no. NCT01684150). The pharmaco-physiological mechanism of DOTL1 inhibition using the drug EPZ-5676 in mixed lineage leukemia is not fully understood. The maintenance of the MLL-AF6 fusion protein requires H3K79 methylation (17). MLL fusion proteins recruit DOT1L, leading to aberrant DOT1L overexpression and H3K79 methylation (18,19). In breast carcinoma, DOT1L is associated with an aggressive phenotype and has also been revealed to be a potential therapeutic target (4,20,21).
Therefore, the present study immunohistochemically evaluated the frequency of pancreatic carcinoma cells that overexpress DOT1L, and how the protein expression correlates with the DOT1L gene copy number. No association between DOT1L gene copy number elevation and protein overexpression in tumor cells of the same cases were identified. Post-transcriptional modifications of DOT1L that inhibit appropriate protein expression may be one explanation. On the other hand, DOT1L protein overexpression (not dependent on an elevation of gene copy number) may be pharmaco-physiologically important if further studies are able to establish the expected association, in which more DOT1L protein be related to more intense methylation of H3K79. At present, the methylation of H3K79 is considered to accelerate the activity of various genes involved in cell cycle activity (22). Due to this perception, an inhibition of DOT1L in cancer may be important. Initial evidence suggests that the DOT1L inhibitor EPZ-5676 is effective in mixed lineage leukemia (MLL) (23,24).
According to sequence data, all DOT1L-altered pancreatic carcinoma harbor a KRAS mutation as well (The Cancer Genome Atlas data, www.cbioportal.org). This is not an unexpected finding, due to the fact that >90% of all pancreatic carcinomas have KRAS mutations (25). The present study speculates that CNVs of DOT1L in KRAS mutated tumors are likely to be an epiphenomenon. In accordance with this assumption, the KRAS mutation may knock out any therapeutically intended DOT1L inhibition. Furthermore, the distribution of CNVs or protein expression of DOT1L identified in the present study is highly heterogeneous. Tumor cells presenting with CNVs of DOT1L were intermixed with carcinoma cells with regular gene copy numbers. A similar result was observed in immunohistochemically positive tumors: Certain tumor cell groups were DOT1L positive and others were negative.
In conclusion, a subset of ~4% of pancreatic adenocarcinomas exhibit CNVs of DOT1L with heterogeneously-distributed single tumor cells showing microclusters of >5 DOT1L-signals (≤14 signals) or nuclear protein overexpression. None of the tumor samples meets the criteria of amplification. There is no correlation between CNVs and protein overexpression. The significance of possible druggability in pancreatic adenocarcinoma, particularly in the protein positive tumor subgroup, requires further study. However, the present study does not predict a successful therapeutic effect using DOT1L inhibitors in adenocarcinomas of the pancreas.
References
- 1.Wang X, Chen CW, Armstrong SA. The role of DOT1L in the maintenance of leukemia gene expression. Curr Opin Genet Dev. 2016;36:68–72. doi: 10.1016/j.gde.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Lacoste N, Utley RT, Hunter JM, Poirier GG, Côte J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 3.Farooq Z, Banday S, Pandita TK, Altaf M. The many faces of histone H3K79 methylation. Mutat Res Rev Mutat Res. 2016;768:46–52. doi: 10.1016/j.mrrev.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JY, Kong G. DOT1L: A new therapeutic target for aggressive breast cancer. Oncotarget. 2015;6:30451–30452. doi: 10.18632/oncotarget.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oktyabri D, Ishimura A, Tange S, Terashima M, Suzuki T. DOT1L histone methyltransferase regulates the expression of BCAT1 and is involved in sphere formation and cell migration of breast cancer cell lines. Biochimie. 2016;123:20–31. doi: 10.1016/j.biochi.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Caparello C, Meijer LL, Garajova I, Falcone A, Le Large TY, Funel N, Kazemier G, Peters GJ, Vasile E, Giovannetti E. FOLFIRINOX and translational studies: Towards personalized therapy in pancreatic cancer. World J Gastroenterol. 2016;22:6987–7005. doi: 10.3748/wjg.v22.i31.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaori K, Bassi C, Biankin A, Brunner TB, Cataldo I, Campbell F, Cunningham D, Falconi M, Frampton AE, Furuse J, et al. IAP/EPC study group on the clinical managements of pancreatic cancer: International Association of Pancreatology (IAP)/European Pancreatic Club (EPC) consensus review of guidelines for the treatment of pancreatic cancer. Pancreatology. 2016;16:14–27. doi: 10.1016/j.pan.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Gall TM, Tsakok M, Wasan H, Jiao LR. Pancreatic cancer: Current management and treatment strategies. Postgrad Med J. 2015;91:601–607. doi: 10.1136/postgradmedj-2014-133222. [DOI] [PubMed] [Google Scholar]
- 11.Wagner M, Redaelli C, Lietz M, Seiler CA, Friess H, Büchler MW. Curative resection is the single most important factor determining outcome in patients with pancreatic adenocarcinoma. Br J Surg. 2004;91:586–594. doi: 10.1002/bjs.4484. [DOI] [PubMed] [Google Scholar]
- 12.Badger SA, Brant JL, Jones C, McClements J, Loughrey MB, Taylor MA, Diamond T, McKie LD. The role of surgery for pancreatic cancer: A 12-year review of patient outcome. Ulster Med J. 2010;79:70–75. [PMC free article] [PubMed] [Google Scholar]
- 13.Simon R, Mirlacher M, Sauter G. Tissue microarrays. Methods Mol Med. 2005;114:257–268. doi: 10.1385/1-59259-923-0:257. [DOI] [PubMed] [Google Scholar]
- 14.Helbig D, Ihle MA, Pütz K, Tantcheva-Poor I, Mauch C, Büttner R, Quaas A. Oncogene and therapeutic target analyses in atypical fibroxanthomas and pleomorphic dermal sarcomas. Oncotarget. 2016;7:21763–21774. doi: 10.18632/oncotarget.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Deng L, Song Y, Redell M. DOT1L inhibition sensitizes MLL-rearranged AML to chemotherapy. PLoS One. 2014;9:e98270. doi: 10.1371/journal.pone.0098270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein EM, Tallman MS. Mixed lineage rearranged leukaemia: Pathogenesis and targeting DOT1L. Curr Opin Hematol. 2015;22:92–96. doi: 10.1097/MOH.0000000000000123. [DOI] [PubMed] [Google Scholar]
- 17.Deshpande AJ, Chen L, Fazio M, Sinha AU, Bernt KM, Banka D, Dias S, Chang J, Olhava EJ, Daigle SR, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013;121:2533–2541. doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Wong M, Polly P, Liu T. The histone methyltransferase DOT1L: Regulatory functions and a cancer therapy target. Am J Cancer Res. 2015;5:2823–2837. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Deng L, Chen F, Yao Y, Wu B, Wei L, Mo Q, Song Y. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget. 2014;5:10665–10677. doi: 10.18632/oncotarget.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ, Lee CH, Oh SH, Song YS, Kim HS, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun. 2015;6:7821. doi: 10.1038/ncomms8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim W, Choi M, Kim JE. The histone methyltransferase Dot1/DOT1L as a critical regulator of the cell cycle. Cell Cycle. 2014;13:726–738. doi: 10.4161/cc.28104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaus CR, Iwanowicz D, Johnston D, Campbell CA, Smith JJ, Moyer MP, Copeland RA, Olhava EJ, Scott MP, Pollock RM, et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. J Pharmacol Exp Ther. 2014;350:646–656. doi: 10.1124/jpet.114.214577. [DOI] [PubMed] [Google Scholar]
- 25.Morris JP, IV, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]