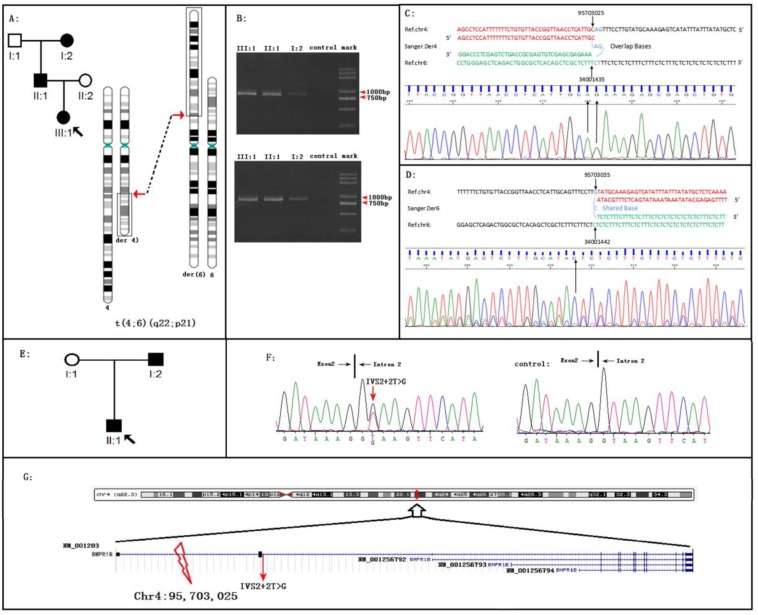
Figure 1. BMPR1B disruptions cause Pierre Robin Syndrome (PRS) in two unrelated families.
A. The PRS family 01 with the reciprocal translocation t(4;6)(q22;p22). B. Migration of amplified junction fragments on agarose gels. After whole genome sequencing, the junction fragments were amplified by the polymerase chain reaction (PCR). The PCR products of Der4 (upper panel) and Der6 (down panel) migrated in the 0.8% agarose gel. Two pairs of primers for Der4 (6-4F: AAGTCCCAGGGCATGAGGTATC and 6-4R: GCTTTGGTATCATATTAACCATTGC) and Der6 (4-6F: TGGTTGAATATAGGAGTCTGGAGT and 4-6R: AGGTCAATGGAGATAGTACAACCT) were used for PCR. The length of both predicted fragments is 800 bp. C. On the proband of family 01, Sanger sequencing of the fragment derived from DER 4 defined the breakpoints in the base-pair resolution: 4q22.3:95703025 fused to 6p21:34001435; AG was the shared sequence. D. On the proband of family 01, Sanger sequencing of the fragment derived from DER 6 defined the breakpoints in the base-pair resolution: 4q22.3:95703035 fused to 6p21.31:34001442; C was the shared sequence. E. The PRS family 02. F. Sequence chromatograms of the BMPR1B splicing mutation IVS2+2T>G. The mutation was occurred on I:2 and II:1 (left) but not on I:1 (right) in the family. G. The schematic of positions of two BMPR1B disruptions. Note: the human BMPR1B include four different transcripts, all two BMPR1B disruptions were occurred on the BMPR1B NM_001203 transcript.