Abstract
Background
MicroRNA (miRNA) control gene transcription by binding to and repressing the translation of messenger RNA (mRNA). Their role in the acute respiratory distress syndrome (ARDS) is undefined.
Methods
Blood leukocytes from 51 patients enrolled in a prior randomized trial of corticosteroids for ARDS were analyzed. After screening eight patients with microarrays for altered miRNA expression, 25 miRNAs were selected for further analysis using RT-PCR in all 51 patients.
Results
On day 0, the 51 patients had APACHE III score of 60.4 ± 17.7 and PaO2/FiO2 of 117 ± 49. 21 miRNA were expressed at increased levels in blood leukocytes at the onset of ARDS compared to healthy controls. These miRNA remained elevated at day 3 and increased further by day 7 (log2 fold change from 0.66 to 5.7 fold, p <0.05 compared to day 0). In a subgroup analysis (37 patients treated with corticosteroids and 14 treated with placebo), the interaction of miRNA expression over time and steroid administration was not significant suggesting that systemic corticosteroids had no effect on the miRNA detected in our study. In contrast, corticosteroids but not placebo decreased IL-6 and C-reactive protein at day 3 (p<0.001) demonstrating an early systemic anti-inflammatory response whereas both treatment arms had decreased values by day 7 (p<0.001).
Conclusions
Expression of miRNA are increased in blood leukocytes of patients with ARDS at day 0 and day 3 and rise further by day 7, when systemic inflammation is subsiding. These effects appear independent of the administration of steroids, suggesting different inflammatory modifying roles for each in the resolving phases of ARDS.
Keywords: acute respiratory distress syndrome, acute lung injury, microRNA, non-protein coding RNA, transcriptomics, corticosteroids
Introduction
The acute respiratory distress syndrome (ARDS) remains a substantial cause of morbidity and mortality in patients who develop hypoxemic respiratory failure following injury to the lung (1). Acute lung and systemic inflammatory responses occur throughout the course of the syndrome and the intensity of these responses is associated with higher mortality (2, 3). Decreasing these inflammatory responses by treating the underlying cause of injury when possible and limiting secondary injury from mechanical ventilation or nosocomial infection has improved survival (1). While anti-inflammatory therapy with corticosteroids has improved outcomes in ARDS it remains controversial due to dosing regimens and the timing and duration of therapy (4, 5). Further, the underlying cause may portend a different prognosis for outcome or differential response to treatments based on the etiology of ARDS (5).
Despite the disease-specific triggers of acute lung injury, it is assumed that there is commonality in the inflammatory pathways that culminate in diffuse alveolar damage (1). The global inflammatory state that occurs during ARDS is suggested by the similarities of genomic signatures found in alveolar macrophages and simultaneous peripheral blood samples (6). Several studies have analyzed transcriptomic signatures in blood leukocytes to provide insight into potential biomarkers and pathways associated with ARDS (6–9). Delineating the transcriptional pathways of inflammatory cells during these processes should enhance an understanding of the mechanisms that contribute to the initiation and resolution of inflammation and may be important in improving outcomes.
MicroRNA (miRNA) are short non-coding RNAs that negatively regulate gene expression by base pairing to specific messenger RNA (mRNA) targets and promoting their degradation or inhibiting their translation (10). A single miRNA may potentially target hundreds of mRNA. Similarly, a single mRNA species may have multiple matches for miRNA that may enhance their repression of gene expression (11). MiRNA regulate genes involved in both normal lung physiology as well as inflammatory diseases of the lung (12). Several studies in animal models of ARDS suggest that miRNA contribute to the pathogenesis of acute lung injury yet no information is available from human studies of ARDS (13).
Because of the intense immune activation present in peripheral blood leukocytes of patients with ARDS and the fundamental role miRNA play in the control of gene expression, we studied miRNA present in blood leukocytes of patients with ARDS during the first week of care. In a secondary analysis, we queried whether corticosteroid therapy had effects on miRNA expression during this period.
Materials and Methods
Samples of archived peripheral blood leukocytes were analyzed from patients with ARDS enrolled in a multi-center trial conducted by one of the authors (GM) (14). The randomized, double blind, placebo-controlled trial compared placebo and standard supportive therapy (usual care) to standard therapy plus early low-dose methylprednisolone infusion (1 mg/kg/day for 14 days and then tapered over 2 wks) using a 1:2 randomization schedule in intubated patients enrolled within 72 hours of ARDS onset. Fifty-one patients (37 treated with corticosteroids and 14 with standard care alone) of the 91 patients enrolled in the original study had blood leukocyte samples available for analysis within 24 hours of enrollment. Blood leukocyte samples from six healthy subjects (median age 55, range 29–61) were obtained as a reference of miRNA expression in healthy persons.
All patients or legally authorized representatives consented to sample collection. The institutional review board of each participating centers approved the protocol for blood sample collection (14). The laboratory studies were performed by the Critical Care Medicine Department at the Clinical Center, National Institutes of Health with approval by the Office for Human Subjects Research, Clinical Center, National Institutes of Health.
Isolation of peripheral blood cells and RNA
Blood leukocytes were isolated by centrifugation of 30 ml of heparinized whole blood at 500 g for 10 min. The buffy coat was obtained by removing the upper layer of plasma. The cells were then incubated with lysing buffer (10mM Tris HCl with 0.15M NH4Cl, pH 7.5) to remove erythrocytes and washed with phosphate buffered saline pH 7.4 (PBS). Following re-suspension in 1 ml of PBS, the cells were stored in tissue culture medium (RPMI 1640) with 15% dimethylsulfoxide and 20% heat-inactivated fetal bovine serum at −80°C (all Invitrogen). Thawed blood leukocytes were washed 3 times with 10 ml PBS and then lysed (Qiazol, Qiagen, Valencia, CA). Total RNA, including miRNA was isolated (miRNeasy Mini Kit) as per manufacturer’s instructions.
miRNA microarray
In order to screen miRNA for further study, we assessed their differential expression in a pilot study of eight patients (four received placebo/usual care and the other four received corticosteroids/usual care) from the 51 patient cohort using a microarray that measures 1733 human mature miRNA (GeneChip miRNA 3.0 Array, Affymetrix, Santa Clara, CA). We analyzed the microarray data from these eight patients at day 0 (prior to receiving usual care or corticosteroids) and at day 3 of treatment. Total RNA (500 ng including miRNA) was labeled with Poly (A) tail and biotinylated tag (Flashtag Biotin HSR Kit, Affymetrix) as per the manufacturer’s instructions.
Microarray analysis
Signal intensity values were assessed using the miRNA Arrays-RMA+DABG procedure in the Expression Console™ (Affymetrix). Probe set identifications were annotated using miRNA-3.0 (release date 2011) resulting in 1733 human mature miRNA probe sets. Probe sets were selected if at least half of the chips had present call values on day 0 or day 3 in either the placebo/usual care or corticosteroid/usual care group, resulting in a total of 768 probe sets identified for further analysis. Intensities based on Robust Multichip Average (RMA)-summarized probe sets without background adjustment were preprocessed using Bioconductor (http://www.bioconductor.org). Linear models were used to test various contrasts of interest between day 0 and day 3 peripheral blood samples on the filtered list of 768 probe sets using R limma package (15, 16). False discovery rates (FDR) were calculated using the Benjamini-Hochberg method (17) and the top-ranked genes were selected by FDR cut-off of 0.05.
Selection of miRNA species from microarray analysis for analysis by RT-PCR
In an exploratory analysis, we used the following criteria for the contrasts of interest to identify 25 miRNA species for further analysis with RT-PCR: 1) 16 miRNA were selected based on between-subject comparisons and an FDR <0.05. and 2) 9 miRNA targets were selected based on within-subject comparisons and a more relaxed criteria of nominal p values of < 0.1 and |log2 FC| > 0.5. The microarray data has been deposited in the Gene Expression Omnibus (GEO), accession number GSE 83630 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83630.
Real-time PCR array
The expression levels of the 25 miRNA described above were tested in all 51 patients samples obtained on days 0, 3 and 7 using a custom quantitative real-time PCR array (Qiagen). The primers for the 25 miRNA targets were pre-coated on 384-well custom PCR array plates (Qiagen). Briefly, cDNA was prepared from 350 ng of total RNA including miRNA, using miScript II RT Kit as per manufacturer’s instructions (Qiagen). RNU6 miRNA was the control used to normalize the data (i.e. Δ Ct = −log 2 (relative abundance)). Heat maps were generated using an analytic program of scripts from the JMP statistical discovery software package (http://abs.cit.nih.gov/MSCLtoolbox/).
Interactions of expressed miRNA with mRNA associated with ARDS
We used a computational method to determine the potential binding sites of the differentially expressed miRNA from this study with mRNA described in previous studies of the transcriptomes of blood leukocytes from patients with ARDS (6–9). Top cited genes from these studies were analyzed (mRNA targets: n = 8 (7), n = 14 (9), n = 19 (6), n = 3 (8)). The authors chose six additional relevant targets. The program RNA22 version 2.0 (https://cm.jefferson.edu/rna22v2/) is a pattern-based methodology based on computational and experimental evidence for the identification of putative microRNA binding sites and the corresponding heteroduplexes (18). Nominal p-values are provided with the binding location to the leftmost position of the predicted target site, the folding energy (in –Kcal/mol) and a schematic of the heteroduplex (18). Gene ontology of the above mRNA was performed using MetaCore (Thomas Reuters, Alexandria, VA).
Statistical analysis
Categorical patient characteristics were compared using Pearson’s Chi-square test or Fisher’s exact test as appropriate. Continuous patient characteristics were compared using Welch’s t test or linear regression models. The normalized cycle threshold (Ct) numbers (i.e. Δ Ct = −log 2 (relative abundance)) were analyzed using linear mixed models with random subject effects (to account for correlation within each subject). The contrasts of interest were estimated to assess the differences between day 0, 3 and 7 in either the placebo/usual care or the corticosteroid/usual care group including the interaction of time and steroid administration. All analyses were conducted using R statistical package (version 2.15.1) unless otherwise noted.
Results
Patient clinical characteristics
The baseline characteristics of the 51 patients (37 treated with corticosteroids/usual care and 14 treated with placebo/usual care) were similar and are summarized in Table 1. These patients were similar to the remaining 41 study patients without available samples, based on age, sex, APACHE 3 score, PaO2/FiO2, total leukocyte count and mortality (Supplemental File Table 1). The conditions associated with ARDS included community-acquired pneumonia (n=23), aspiration pneumonia (n=8), post-operative respiratory failure (n=5), hospital-acquired pneumonia (n=4) and mixed etiologies (n = 11).
Table 1.
Baseline clinical characteristics of study patients treated with corticosteroids compared to those treated with usual care alone.
| Variables | Patients treated with corticosteroids n = 37 |
Patients treated with placebo n = 14 |
P value |
|---|---|---|---|
| Agea | 49.0 ± 17.3 | 54.1 ± 17.6 | 0.35 |
| Sexb (men/women) | 19/18 | 9/14 | 0.53 |
| APACHE 3a | 59.1 ± 16.0 | 63.9 ± 21.8 | 0.40 |
| PaO2/FiO2 day 0a | 116.5 ± 54.4 | 118.6 ± 30.9 | 0.89 |
| WBCa (× 103/mm3) | 15.7 ± 6.5 | 15.2 ± 9.3 | 0.85 |
| Hospital mortality (%)b | 7/37 (19) | 5/14 (36) | 0.27 |
Summary data displayed as mean ± sd,
– t-test,
– Fisher’s exact test
As previously described in the larger patient cohort (14, 19), the 51 patients described above had similar changes in systemic markers of inflammation; IL-6 and C-reactive protein (CRP) decreased significantly by days 3 and day 7 in patients treated with corticosteroids (p <0.001 for both on days 3 and 7). In contrast, IL-6 and CRP were unchanged at day 3 (p > 0.2) and then decreased by day 7 (p ≤ 0.004 for both) in the placebo/usual care patients (Figure 1). These findings suggest a significant anti-inflammatory effect of corticosteroid therapy as early as day 3 in the course of ARDS.
Figure 1. A and B: Interleukin-6 and C-reactive protein levels during the first week of ARDS.
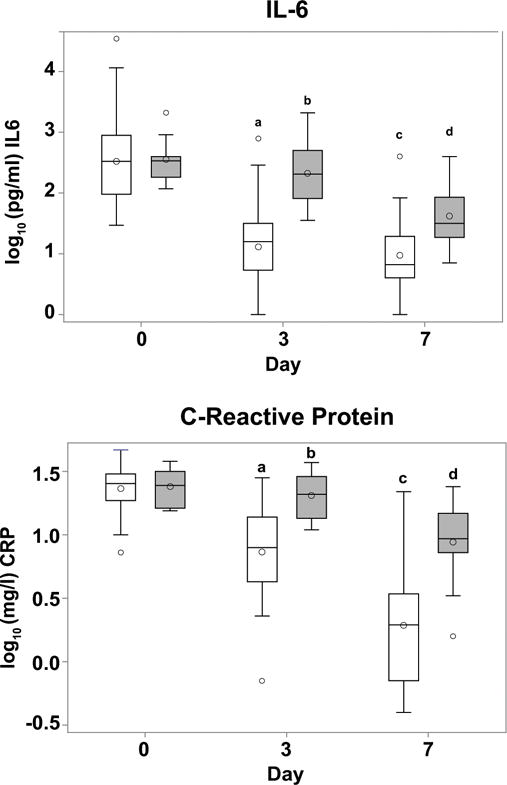
Box plots of log10 IL-6 (pg/ml) and C-reactive protein (mg/l) plasma concentrations on days 0, 3 and 7 in all 51 patients studied. Open boxes - corticosteroids/usual care, gray boxes - placebo/usual care. These data represent a subset of previously described data (19, 24). A: Plasma IL-6 levels during first week of ARDS. No significant change was found in IL-6 by day 3 in the placebo/usual care group compared to day 0 (b p=NS). In contrast, in the corticosteroid/usual care group, levels were significantly lower at ARDS day 3 compared to day 0 (a p<0.001). In both groups at day 7, levels were significantly lower than day 0 levels (c p<0.001 for corticosteroid/usual care group, d p=0.0001 for placebo/usual care group). B: Plasma C-reactive protein levels during first week of ARDS. No significant change was found in C-reactive protein by day 3 in the placebo/usual care group compared to day 0 (b p=NS). In contrast, in the corticosteroid/usual care group, levels were significantly lower at ARDS day 3 compared to day 0 (a p<0.001). In both groups at day 7, levels were significantly lower than day 0 levels (c p<0.001 for corticosteroid/usual care group, d p=0.004 for placebo/usual care group,).
RT-PCR analysis of 25 targeted miRNA species
Twenty-five miRNA were selected in an exploratory analysis from microarrays of leukocytes from 8 patients at baseline and day 3. These were further analyzed using RT-PCR. No significant differences in miRNA expression were found at day 0 among the 51 patients with ARDS (Figure 2A). With the exception of 3 miRNA (miR-335–5p, miR-151a-5p and miR-4454), the remaining 22 miRNA were significantly elevated in the 51 patients compared to the healthy controls (p ≤ 0.01 for all 22) (Figure 2A and B).
Figure 2. A and B. Baseline miRNA expression in leukocytes from patients with ARDS compared to healthy controls.
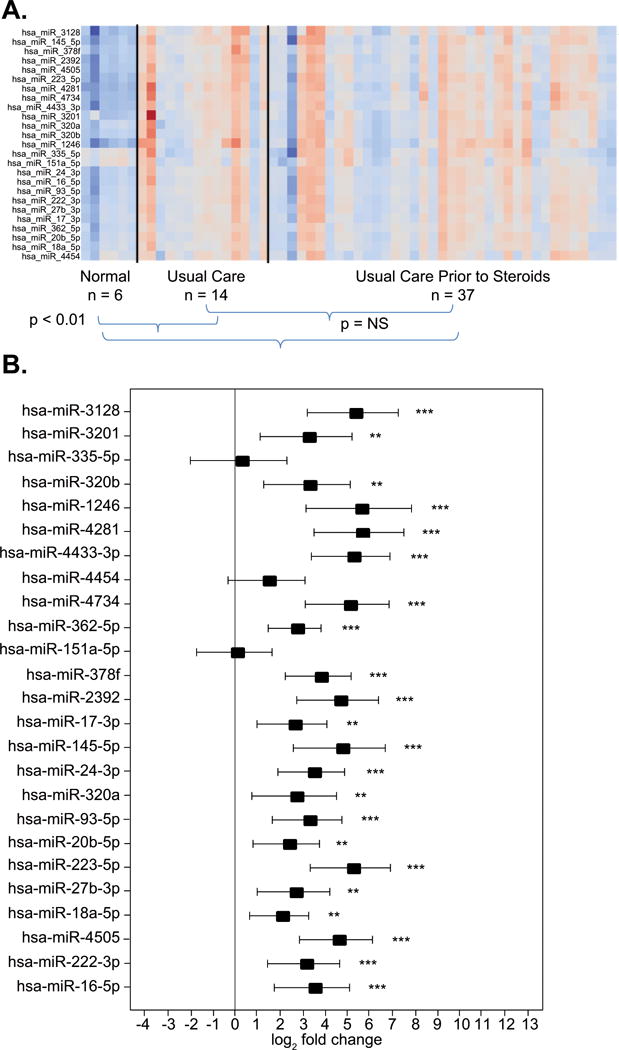
A: Each column of the heat map represents an individual and each row represents a specific miRNA. Red depicts upregulation and blue downregulation of miRNA. The heat map shows the miRNA expression profiles among all 51 patients with ARDS at day 0 prior to either placebo or corticosteroid randomization and the expression profiles in healthy controls (n = 6). No differences were found in baseline miRNA expression among the 51 patients with ARDS prior to randomization to placebo/usual care (n=14) or steroid/usual care (n=37) (p=NS). At the onset of ARDS, miRNA expression differed in 22 of 25 miRNA compared to healthy subjects (p<0.01). The three miRNA without significant difference in expression in healthy volunteers versus ARDS were miR-335-5p, miR-151a-5p and miR-4454. B: A summary graph of the changes in miRNA expression comparing patients with ARDS and healthy controls. The x-axis shows log2 fold change and the y-axis shows each of the 25 individual miRNA comparing the baseline expression to that of healthy controls. The point estimates of fold change with 95% confidence intervals are shown. Significant changes in miRNA expressions are represented graphically as: * p<0.05, ** p<0.01 and *** p<0.001.
We next examined the temporal sequence of changes in blood leukocyte miRNA expression on day 0 compared to day 3 and day 7. No significant changes were found comparing day 3 to baseline (day 0) (Figure 3A), whereas significant increased expression was found on day 7 compared to either day 3 or day 0 (Figure 3 B and C respectively). In the 51 patients, 21 of the 25 miRNA targets were significantly increased compared to their respective baselines on day 7 (Figure 3C).
Figure 3. Changes in miRNA expression based on RT-PCR analysis of leukocytes in 51 patients with ARDS.
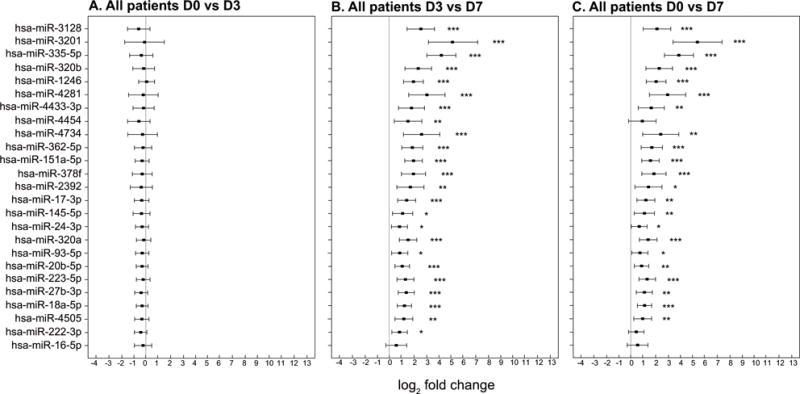
The x-axis shows log2 fold change and the y-axis shows each of the 25 individual miRNA. The point estimates of fold change with 95% confidence intervals are shown. Significant changes in miRNA expressions are represented graphically as: * p<0.05, ** p<0.01 and *** p<0.001 a: No significant changes in miRNA expression were found between day 0 and day 3 for any of the 25 miRNA evaluated. b: A significant increase in miRNA expression between day 3 and day 7 was observed for 21 of the 25 miRNA evaluated. c: A significant increase in miRNA expression between day 0 and day 7 day was observed for 21 of the 25 miRNA evaluated.
In a subgroup analysis, the effects of corticosteroid therapy on miRNA expression were evaluated. In the placebo/usual care group, 19 miRNA were significantly increased from day 3 to 7 and 14 miRNA increased from day 0 to 7 (Figure 4A and B respectively). In the corticosteroid/usual care group, 24 miRNA were increased comparing day 3 to 7 and 21 miRNA were increased comparing day 0 to day 7. (Figure 5 A and B respectively). Heat maps of these data are found in Supplemental Figure 1 and 2. The interaction of time and steroid administration was not significant (p>0.05) suggesting that corticosteroid therapy had no effect on the expression of these miRNA.
Figure 4. Changes in miRNA in patients who received placebo/usual care.
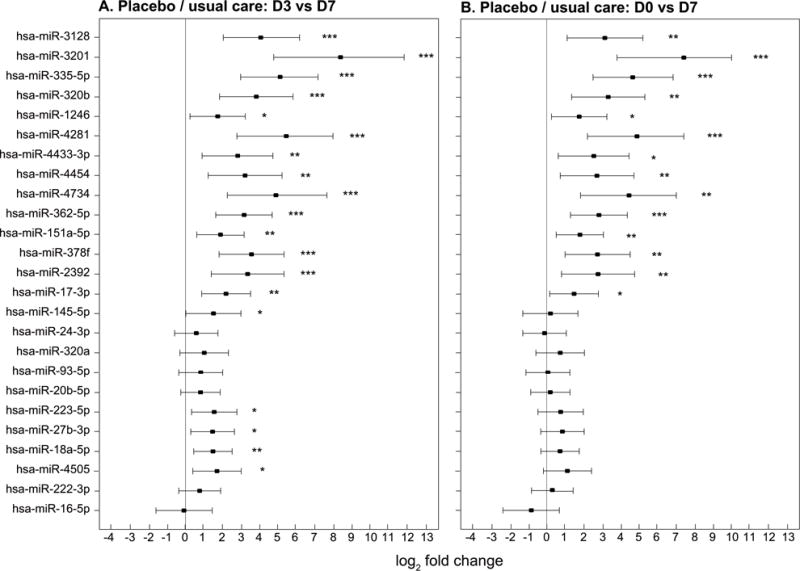
The x-axis shows miRNA fold change in RT-PCR values represented by log2 fold change and the y-axis shows each of the 25 individual miRNA. The point estimates of fold change with 95% confidence intervals are shown. Significant temporal changes in miRNA expressions are represented graphically as: * p<0.05, ** p<0.01 and *** p<0.001. a: Changes in miRNA expression from ARDS day 3 to day 7 for 14 patients who received placebo/usual care. There was significant increase in miRNA expression (p<0.05) between day 3 and day 7 for 19 of the 25 miRNA evaluated. b: Changes in miRNA expression from ARDS day 0 to day 7 for 14 patients who received placebo/usual care. There was significant increase in miRNA expression between day 0 and day 7 for 14 of the 25 miRNA evaluated.
Figure 5. Changes in miRNA in patients who were treated with corticosteroids/usual care.
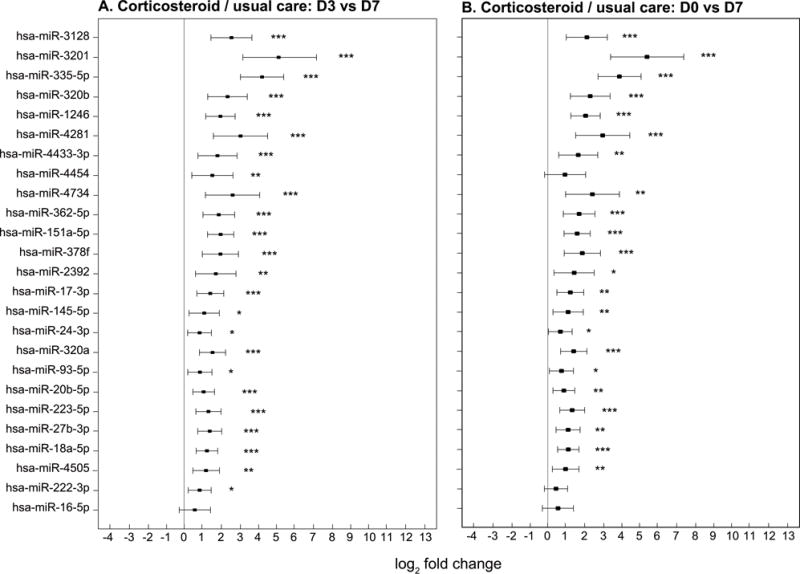
The x-axis shows miRNA fold change in RT-PCR values represented by log2 fold change and the y-axis shows each of the 25 individual miRNA. The point estimates of fold change with 95% confidence intervals are shown. Significant temporal changes in miRNA expressions are represented graphically as: * p<0.05, ** p<0.01 and *** p<0.001 a: Changes in miRNA expression from ARDS day 3 to day 7 for 37 patients who received corticosteroid/usual care. There was significant increase in miRNA expression between day 0 and day 7 for 24 of the 25 miRNA evaluated. b: Changes in miRNA expression from ARDS day 0 to day 7 for 37 patients who received corticosteroid/usual care. There was significant increase in miRNA expression between day 0 and day 7 for 22 of the 25 miRNA evaluated.
miRNA heteroduplex formation with mRNA
We analyzed our miRNA expression data with previous reports of mRNA altered in either alveolar macrophages or blood leukocytes of patients with ARDS. Among the top 50 mRNA species described from studies of blood mRNA signatures in ARDS, the miRNA that were differentially expressed in our study have the potential to form heterodimer complexes with 74% (37 of 50) of these mRNA (Supplemental Figure 3). In the original studies (6–9), 21 mRNA were upregulated and 16 were repressed. The gene ontology of these selected mRNA includes themes of acute inflammation, defense responses, activation of stress kinases, responses to hypoxia, regulation of cell mobility, tissue remodeling, and apoptosis. Some individual mRNA (e.g. CEACAM6, CCDC50) had 8–10 miRNA described in our study forming potential complexes with 19 – 21 binding sites on their respective targeted mRNA (Supplemental Figure 3).
Discussion
MiRNA play a fundamental role in the control of gene expression in health and disease. While several studies have shown alterations in blood leukocyte mRNA during the systemic inflammatory response during ARDS, no data are available describing changes in blood leukocyte miRNA. We identified 21 miRNA that are expressed at increased levels at the onset of ARDS, remain elevated at day 3 and increase further by day 7. Attributing a direct cause and biologic effect to the induction of miRNA is challenging because of their pleiotropic effects on mRNA translation. The temporal pattern of miRNA expression suggests that the underlying inflammatory processes that led to ARDS remained active at day 3 and the enhanced miRNA expression by day 7 may have a role in the resolution of inflammation. The greatest increase in miRNA occurred by day 7, a time of diminishing systemic inflammation as manifested by decreasing IL-6 and CRP levels. Notably, corticosteroids resulted in decreases in IL-6 and CRP on day 3, a response not seen in the placebo/usual care group. Yet, steroid therapy had no effect on the elevated miRNA species observed on days 3 or 7. These data suggest the presence of steroid-responsive and steroid-independent inflammatory axes during the course of ARDS.
The miRNA detected in the cohort have biologic relevance as they have theoretical high-affinity binding to mRNA previously described as being expressed in leukocytes during ARDS (6–9). MiRNA can modulate stress signals as either a critical intermediate within a pathway or by titrating a key component of a signal pathway (20). Further, the miRNA can function in a negative or positive feedback loop to modify a signal or simultaneously target both positive and negative regulators of a pathway, buffering the pathway from extremes of activation or repression (20). These mechanisms may have relevance to our observations of multiple potential binding sites of the miRNA found at 7 days with previously described leukocyte mRNA from patients with ARDS (Figure 6). The regulatory impact of multiple mRNA binding sites is suggested from single cell experiments where a threshold level of miRNA regulation on mRNA targets is achieved, repressing protein production (21). If the target mRNA increases in quantity, the pool of miRNA available for repression becomes limiting, allowing for graded mRNA expression. This model suggests a spectrum of responses from full repression to a titrated response depending on the relative abundance of each RNA species (21).
MiRNAs clusters are transcribed as a single unit (polycistronic) generating multiple miRNA from a single primary transcript . Their coordinate expression suggests a synergistic role in regulating biologic processes. We found 3 of the 6 members of the miR-17~92 cluster (miR-17-3p, 18a-5p, and 20b-5p) up-regulated in the cohort at 7 days. This cluster is widely expressed in tissues and modulates cell proliferation and apoptosis (22). Their relationship to these two processes in ARDS is unknown.
A secondary analysis of our study assessed the effects of steroids in the expression of cell- associated miRNA. We did not observe a suppressive effect of methylprednisolone on the miRNA species analyzed in blood leukocytes. The interaction of time and steroid administration was not significant, suggesting that the 14 miRNA on day 7 in the placebo/usual care group were induced independent of steroid administration and that limited power may have contributed to the absence of change in the remaining eight miRNA (Figure 4B and D). These observations contradict many reports of glucocorticoid effects on miRNA expression in vitro. Glucocorticoids may affect miRNA by direct effects on miRNA promoters as well as the proteins that regulate the biogenesis and processing of miRNA after transcription (23–25). Dexamethasone, a glucocorticoid receptor (GR) agonist, reduced miRNAs in leukemic cells and directly inhibited the miRNA-17 host gene by an increase in GR binding to an upstream promoter of miR-17HG (25). Further, dexamethasone repressed miR-17 expression in leukemic cells and apoptotic osteoblasts (25, 26). In contrast, miRNA members of the miRNA 17~92 cluster were upregulated in our study.
MiRNAs induced by endotoxin stimulation of a murine macrophage cell line, were suppressed by dexamethasone (27). Other reports have shown that glucocorticoids induce miRNA in some leukemic primary cells and cell lines (28). Levels of GR may also be influenced by miRNA. In multiple myeloma cells, miR-130b inhibits GR and protein expression (29). Hydrocortisone-induction of miR-124 expression in lymphocytes is associated with the downregulation of the GR alpha (30). Lastly, glucocorticoids may suppress miRNA processing enzymes. Dexamethasone-induced apoptosis of primary murine cells and human leukemia cells suppressed the majority of miRNAs and decreased the mRNA and proteins of miRNA-processing enzymes (i.e., Drosha, DGCR8, Dicer) (31).
In contrast to the suppressive effects of glucocorticoids on miRNA expression in cell lines and primary cells in culture, organ miRNA responses to inflammatory stimuli are variable. In mice genetically resistant to tumor necrosis factor, miR-511 is induced by glucocorticoids and results in decreased tumor necrosis factor receptor type 1 expression and protects against TNF toxicity (32). Mice exposed to aerosolized endotoxin express multiple miRNA in the lung after six hours. However, pretreatment with dexamethasone had no effect on the endotoxin-induced miRNA expression (33). These data are similar to the results of our study suggesting that miRNA and corticosteroids may have similar but relatively independent mechanisms that modulate inflammation. Further, these results suggest that extrapolation of glucocorticoid effects from in vitro data is context dependent and affected by the cell target and its physiologic state.
Our study is limited by several considerations. This was a subgroup of patients from a larger study cohort, chosen because of the availability of peripheral blood samples for analysis. The clinical characteristics of this subgroup were not different from the clinical study cohort (Supplemental table 1), suggesting that the responses found in our study were representative of the group at large . The study is retrospective and focused on a limited number of miRNA for sequential analysis. A more comprehensive microarray analysis of a greater number of patients would have likely identified more differentially expressed miRNA and some of those miRNA may be influenced by corticosteroid therapy. The cells used in our study were frozen archived samples. In contrast to mRNA, miRNA stability in frozen tissues, cell lines and serum has been shown to be relatively robust (34, 35). However, it is possible that less abundant miRNA may be prone to detection loss with prolonged storage (35). Degradation of samples was minimized by storing all samples in −80 °C freezer and limiting freeze/thaw cycles to the initial sample collection and RNA isolation. The potential heterocomplex formation of miRNA and mRNA is based on prior reports of mRNA expression rather than simultaneous measures of both in the same patient cohort. We did not compare day 0 miRNA expression values with critically ill non-ARDS patients. However, compared to their respective day 0 values, the temporal increase in miRNA expression in these critically ill patients suggests a role in modulating gene expression during the acute phase of ARDS. Lastly, identifying the association of miRNA expression with a specific cell type (e.g., neutrophil, lymphocyte, monocyte) was not feasible with the available samples.
Conclusions
We have shown that elevated levels of leukocyte miRNA expression occur early in the course of ARDS and increase in intensity after one week during the waning of the acute inflammatory response. The increased expression of miRNA appears independent of corticosteroid therapy suggesting they have a role in steroid-independent mechanisms that contribute to the resolution of inflammation.
Supplementary Material
Acknowledgments
Funding
This research was supported by the Intramural Research Program of the Critical Care Medicine Department of the National Institutes of Health in conjunction with the Veterans Affairs Medical Center, Memphis, TN, the Baptist Memorial Health Care Foundation and the Assisi Foundation of Memphis. The contents of this manuscript do not necessarily represent the views of the Department of Veterans Affairs or the US Department of Health and Human Services.
Footnotes
Competing interests
The authors have no competing interests to declare.
References
- 1.Han S, Mallampalli RK. The acute respiratory distress syndrome: from mechanism to translation. J Immunol. 2015;194(3):855–60. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meduri GU, Headley S, Kohler G, Stentz F, Tolley E, Umberger R, Leeper K. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS. Plasma IL-1 beta and IL-6 levels are consistent and efficient predictors of outcome over time. Chest. 1995;107(4):1062–73. doi: 10.1378/chest.107.4.1062. [DOI] [PubMed] [Google Scholar]
- 3.McClintock D, Zhuo H, Wickersham N, Matthay MA, Ware LB. Biomarkers of inflammation, coagulation and fibrinolysis predict mortality in acute lung injury. Crit Care. 2008;12(2):R41. doi: 10.1186/cc6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meduri GU, Bridges L, Shih MC, Marik PE, Siemieniuk RA, Kocak M. Prolonged glucocorticoid treatment is associated with improved ARDS outcomes: analysis of individual patients’ data from four randomized trials and trial-level meta-analysis of the updated literature. Intensive Care Med. 2016;42:829–840. doi: 10.1007/s00134-015-4095-4. [DOI] [PubMed] [Google Scholar]
- 5.Seam N, Suffredini AF. Steroids are part of rescue therapy in ARDS patients with refractory hypoxemia: we are not sure. Intensive Care Med. 2016;42:924–927. doi: 10.1007/s00134-015-4160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovach MA, Stringer KA, Bunting R, Wu X, San Mateo L, Newstead MW, Paine R, Standiford TJ. Microarray analysis identifies IL-1 receptor type 2 as a novel candidate biomarker in patients with acute respiratory distress syndrome. Respir Res. 2015;16:29. doi: 10.1186/s12931-015-0190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howrylak JA, Dolinay T, Lucht L, Wang Z, Christiani DC, Sethi JM, Xing EP, Donahoe MP, Choi AM. Discovery of the gene signature for acute lung injury in patients with sepsis. Physiol Genomics. 2009;37(2):133–9. doi: 10.1152/physiolgenomics.90275.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinay T, Kim YS, Howrylak J, Hunninghake GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L, Nakahira K, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med. 2012;185(11):1225–34. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kangelaris KN, Prakash A, Liu KD, Aouizerat B, Woodruff PG, Erle DJ, Rogers A, Seeley EJ, Chu J, Liu T, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol. 2015;308(11):L1102–13. doi: 10.1152/ajplung.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425(19):3582–600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster PS, Plank M, Collison A, Tay HL, Kaiko GE, Li J, Johnston SL, Hansbro PM, Kumar RK, Yang M, et al. The emerging role of microRNAs in regulating immune and inflammatory responses in the lung. Immunol Rev. 2013;253(1):198–215. doi: 10.1111/imr.12058. [DOI] [PubMed] [Google Scholar]
- 13.Rajasekaran S, Pattarayan D, Rajaguru P, Sudhakar Gandh PS, Thimmulappa RK. MicroRNA Regulation of Acute Lung Injury and Acute Respiratory Distress Syndrome. J Cell Physiol. 2016;231(10):2097–106. doi: 10.1002/jcp.25316. [DOI] [PubMed] [Google Scholar]
- 14.Meduri GU, Golden E, Freire AX, Taylor E, Zaman M, Carson SJ, Gibson M, Umberger R. Methylprednisolone infusion in early severe ARDS: results of a randomized controlled trial. Chest. 2007;131(4):954–63. doi: 10.1378/chest.06-2100. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 16.Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Front Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 18.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Seam N, Meduri GU, Wang H, Nylen ES, Sun J, Schultz MJ, Tropea M, Suffredini AF. Effects of methylprednisolone infusion on markers of inflammation, coagulation, and angiogenesis in early acute respiratory distress syndrome. Crit Care Med. 2012;40(2):495–501. doi: 10.1097/CCM.0b013e318232da5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43(9):854–9. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Concepcion CP, Bonetti C, Ventura A. The microRNA-17–92 family of microRNA clusters in development and disease. Cancer J. 2012;18(3):262–7. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1(1):31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cochrane DR, Cittelly DM, Richer JK. Steroid receptors and microRNAs: relationships revealed. Steroids. 2011;76(1–2):1–10. doi: 10.1016/j.steroids.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Harada M, Pokrovskaja-Tamm K, Soderhall S, Heyman M, Grander D, Corcoran M. Involvement of miR17 pathway in glucocorticoid-induced cell death in pediatric acute lymphoblastic leukemia. Leuk Lymphoma. 2012;53(10):2041–50. doi: 10.3109/10428194.2012.678004. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Xu J, Qi J, Zhang L, Wang J, Liang J, Qian N, Zhou H, Wei L, Deng L. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J Cell Sci. 2013;126(Pt 4):978–88. doi: 10.1242/jcs.117515. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Xiong S, Jiang P, Liu R, Liu X, Qian J, Zheng X, Chu Y. Glucocorticoids inhibit lipopolysaccharide-mediated inflammatory response by downregulating microRNA-155: a novel anti-inflammation mechanism. Free Radic Biol Med. 2012;52(8):1307–17. doi: 10.1016/j.freeradbiomed.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Rainer J, Ploner C, Jesacher S, Ploner A, Eduardoff M, Mansha M, Wasim M, Panzer-Grumayer R, Trajanoski Z, Niederegger H, et al. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. 2009;23(4):746–52. doi: 10.1038/leu.2008.370. [DOI] [PubMed] [Google Scholar]
- 29.Tessel MA, Benham AL, Krett NL, Rosen ST, Gunaratne PH. Role for microRNAs in regulating glucocorticoid response and resistance in multiple myeloma. Horm Cancer. 2011;2(3):182–9. doi: 10.1007/s12672-011-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ledderose C, Mohnle P, Limbeck E, Schutz S, Weis F, Rink J, Briegel J, Kreth S. Corticosteroid resistance in sepsis is influenced by microRNA-124–induced downregulation of glucocorticoid receptor-alpha. Crit Care Med. 2012;40(10):2745–53. doi: 10.1097/CCM.0b013e31825b8ebc. [DOI] [PubMed] [Google Scholar]
- 31.Smith LK, Shah RR, Cidlowski JA. Glucocorticoids modulate microRNA expression and processing during lymphocyte apoptosis. J Biol Chem. 2010;285(47):36698–708. doi: 10.1074/jbc.M110.162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puimege L, Van Hauwermeiren F, Steeland S, Van Ryckeghem S, Vandewalle J, Lodens S, Dejager L, Vandevyver S, Staelens J, Timmermans S, et al. Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol Med. 2015;7(8):1004–17. doi: 10.15252/emmm.201405010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, Jung K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56(6):998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 35.Becker N, Lockwood CM. Pre-analytical variables in miRNA analysis. Clin Biochem. 2013;46(10–11):861–8. doi: 10.1016/j.clinbiochem.2013.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


