Abstract
The RACK1 protein interacts with numerous proteins involved in signal transduction, the cytoskeleton, and mRNA splicing and translation. We used the 2-hybrid system to identify additional proteins interacting with RACK1 and isolated the RNA-binding protein SERBP1. SERPB1 shares amino acid sequence homology with HABP4 (also known as Ki-1/57), a component of the RNA spicing machinery that has been shown previously to interact with RACK1. Several different isoforms of SERBP1, generated by alternative mRNA splicing, interacted with RACK1 with indistinguishable interaction strength, as determined by a 2-hybrid beta-galactosidase assay. Analysis of deletion constructs of SERBP1 showed that the C-terminal third of the SERBP1 protein, which contains one of its two substrate sites for protein arginine N-methyltransferase 1 (PRMT1), is necessary and sufficient for it to interact with RACK1. Analysis of single amino acid substitutions in RACK1, identified in a reverse 2-hybrid screen, showed very substantial overlap with those implicated in the interaction of RACK1 with the cAMP-selective phosphodiesterase PDE4D5. These data are consistent with SERBP1 interacting selectively with RACK1, mediated by an extensive interaction surface on both proteins.
Keywords: RACK1, SERBP1, HABP4, 2-hybrid, PDE4D5, PDE4
1. Introduction
The WD-repeat β-propeller protein RACK1 interacts with an expanding list of protein “partners” and therefore has numerous and diverse functions in cells (see refs. (1–3) for reviews). First isolated as a partner of specific protein kinase C isoforms, for which it was named receptor for activated C-kinase (4–8), RACK1 has also been shown to interact with a considerable number of other signal transduction proteins, including the cAMP-specific phosphodiesterase PDE4D5 (9–17), the tyrosine kinase oncoproteins SRC (18–21), FLT1 (22), and FAK (23), protein serine phosphatase PP2A (24–26), the MAPKs JNK1 and ERK1/2 (27), VARP (28), phospholipase C (29), the NMDA receptor (30–33), cytokine and growth factor receptors (34–38), Gsβγ (39–42), selected 14.3.3 isoforms (43) and likely other signaling proteins (44–47). RACK1 also interacts with members of complexes that play important roles in protein degradation, such as the Cullin-2 based E3 ubiquitin ligases (48;48;49), as well as certain enzymes of unclear cellular function, such as inositol-requiring enzyme 1 (IRE1; refs, (50)). RACK1 also may interact with elements of the cytoskeleton, including β-integrins (51–56) and β-actin (21;21;57). In some of these contexts, RACK1 has been considered to be a scaffold protein, i.e., a protein that mediates the association of two other, separate proteins and thereby allows them to interact functionally (58).
Numerous investigators have shown that RACK1 has several important, possibly related, roles in mRNA splicing and in translation. RACK1 interacts directly with specific components of the 40S ribosomal subunit (specifically, the 18S rRNA and ribosomal proteins rpS16e, rpS17e, and rpS3e [new names: uS9, eS17 and uS3, respectively]; refs. (59;60)). It has been suggested that RACK1 may recruit signaling molecules, such as PKC and JNK, to the ribosome (61;62), although the precise function(s) of ribosomal RACK in complex eukaryotes remain an active area of investigation (3).
RACK1 also interacts with the HABP4 protein (synonym: Ki-1/57), which is involved in mRNA metabolism (63;64)). HABP4 interacts with the chromo-helicase-DNA-binding domain protein 3 (CHD-3), a nuclear protein involved in chromatin remodeling and transcription regulation (65). HABP4 is also a substrate of protein arginine N-methyltransferase 1, (PRMT1), which methylates two discreet regions on HABP4, both approximately 21 amino acids in length and consisting of repeats of the sequence RGG and/or RGR (66;67). HABP4 plays an important role in mRNA splicing, which is mediated by its interaction with the splicing proteins hnRNPQ and SFRS9 (68;69). It may also play a role in mRNA translation, as one group has shown that it interacts with ribosomal protein L38, although the physiologic significance of this finding is not clear (70). One group has also shown that RACK1 interacts with the polyA-binding protein LARP4 (71).
SERBP1 (synonyms: CGI-55, CHD3IP, HABP4L, PAI-RBP1), the focus of the present study, is an mRNA-binding protein (72). SERBP1 binds to a 134-nucleotide cyclic nucleotide-responsive sequence (CRS) located near the 3′ end of several mRNAs, most notably that encoding type-1 plasminogen activator inhibitor (PAI-1; refs. (72–74)). Studies in a number of cell lines that shown that elevation of intracellular cAMP levels markedly increases the rate of degradation of PAI-1 mRNA, which has been shown to be mediated by SERBP1 (73;75). Like HABP4, SERBP1 binds to CHD-3, which for SERBP1 involves two separate regions, located in its N- and C-termini (76;77). SERBP1 has also been shown to bind to the RNA splicing component U2AF (78). These data implicate SERBP1 in several nuclear functions, separate from its action on mRNA. Also like HABP4, SERBP1 is a substrate for PRMT1-mediated arginine methylation, which may regulate the nuclear-cytoplasmic distribution of the protein (79;80). SERBP1 and HABP4 target similar mRNA populations in cells, clearly suggesting some overlap in their physiological functions (81). SERBP1 also interacts with progesterone receptor membrane component 1 (PGRMC1), linking it to progesterone-signaling pathways (82).
Given the similarities between SERBP1 and HABP4 that have been reported to date, it would not be unexpected that they might share protein partners. As discussed above, both interact with CHD-3 (65;76;77). In the present study, we report that SERBP1, like HABP4, interacts with RACK1 in 2-hybrid assays. We also show that the interaction of SERBP1 with RACK1 requires an extensive region located in the C-terminal third of the SERBP1 protein. We also show that a large surface on RACK1, which overlaps substantially that needed for RACK1 to interact with PDE4D5, is necessary for it to interact with SERBP1. Our data provide additional impetus to search for additional functional roles of RACK1 in nuclear processes, such as chromatin remodeling, and possibly in mRNA splicing and stability.
2. Materials and Methods
2.1. Yeast 2-hybrid analyses
Yeast 2-hybrid techniques identical to those used previously by us were used to identify and analyze protein-protein interactions (10;14;17;83;84). In all experiments, one of the two interacting proteins was expressed as “bait”, in pLEXAN (a derivative of BTM116; refs. (10;85)), as a LexA DNA-binding domain fusion. The other interacting protein was expressed as “prey”, in pGADN (a derivative of pGAD.GH; ref. (85)), as a Saccharomyces cerevisiae GAL4 activation-domain protein fusion. For all experiments, the oncoproteins RAS and RAF1 (ref. (86)) were employed as standards. All proteins were targeted to the nucleus. Plasmids encoding the appropriate fusion pair were transfected into the S. cerevisiae strain L40 (ref. (86)). Yeast colonies expressing the fusions were obtained by selecting for the prototrophic markers encoded by the bait and prey plasmids. For filter β-galactosidase assay, colonies were streaked out as patches, transferred to nylon membranes (GE Healthcare), permeabilized with liquid nitrogen, and then incubated with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Xgal), as described previously (10;14;83;84). Positive patches were identified by their turning a blue color. Except where indicated otherwise, all experiments shown here have been repeated at least twice, in at least two different yeast transformants, with identical results.
2.2. Yeast 2-hybrid screen
The yeast 2-hybrid screen used, as bait, the full open reading frame of human RACK1 (GenBank M24194) cloned into the NotI site of pLEXAN, to express it as a LexA DNA-binding domain fusion (i.e., pLexARACK1). pLexARACK1 was used to screen a cDNA library derived from the HeLa human cervical cancer cell line S3, cloned into the EcoRI and XhoI sites of the pGAD.GH vector; we have described this library previously (10). All interactions were evaluated in the S. cerevisiae strain L40 (ref. (86)). Positives were isolated by their ability to grow as colonies on plates lacking histidine, without the addition of 3-amino-triazole. Positive colonies were streaked as patches on plates selecting only for prototropic markers encoded by the bait and prey plasmids and then subjected to the filter β-galactosidase assay described above. The prey plasmid was then isolated from HIS+, β-gal+ colonies and sequenced, using the Sanger sequence method, on an ABI 3870 sequencer. DNA sequence data was analysed using the basic local alignment sequence tool (BLAST) at www.ncbi.nlm.nih.gov/blast/. All positive clones were verified for their ability to interact with pLexARACK1 by re-transforming them back into the L40 strain, with pLexRACK1, and repeating the filter β-galactosidase assay. All prey were also tested for non-specific interactions by testing them against pLEXAN (empty vector) and a panel of test bait, which included several transcription factors and several kinases; no non-specific interactions were noted.
3. Results
3.1. Identification of SERBP1 and ribosomal proteins as RACK1-interacting partners in a 2-hybrid screen
Previously, we isolated RACK1 in a 2-hybrid screen where PDE4D5 was used as bait (10). To identify additional proteins that interact with RACK1, we used RACK1 as bait to screen a HeLa-derived 2-hybrid library, using procedures described in Materials and Methods. Of the approximately 500 positive (i.e., HIS+ β-gal+) colonies obtained in the screen, the prey plasmids from 192 colonies were isolated and analysed in detail by restriction enzyme mapping and sequencing. The 192 positive prey encoded GAL4 fusions with SERBP1 (148 isolates), PDE4D5 (2 isolates), ribosomal protein S13 (1 isolate; rpS13e, new name: uS15), ribosomal protein S17 (4 isolates, rpS17e, new name: eS17, ref. (59)), and 22 isolates, generally seen only 1–3 times each, of uncertain significance. Fifteen isolates encoded non-physiological fusions (i.e., the physiological open reading frame [ORF] of the insert cDNA was not in frame with that of GAL4).
3.2. SERBP1 isoforms define regions required for its interaction with RACK1
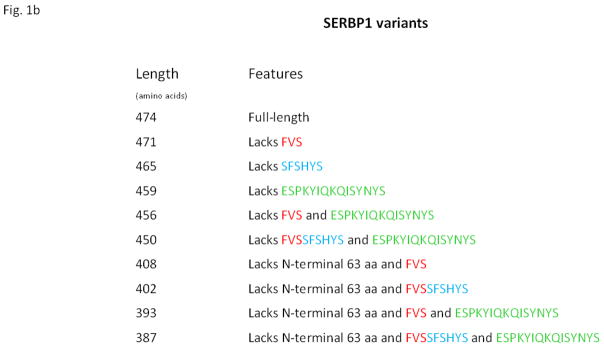
The human SERBP1 gene encodes a large number of protein isoforms, by transcripts that are generated by alternative mRNA splicing. The longest known isoform has 474 amino acids (Fig. 1a). In our screen, we isolated cDNAs that encoded each of the 402-, 393- and 387-amino acid isoforms (Fig. 1b). Since some of the isolates that encoded the 393- and 387-amino acid isoforms isolates contained inserts that were truncated at the 5′ ends of their ORFs, it is possible that their cDNAs were derived from mRNAs encoding the 450- and 456-amino acid isoforms (Fig. 1b). The regions of sequence FVSSFSHYS and ESPKYIQKQISYNYS, which are encoded by variably-spliced exons and are of unknown physiological significance, are not present in at least one of 402-, 393- and 387-amino acid isoforms (Fig. 1b). Since all of these isoforms were capable of interacting with RACK1, these regions of SERBP1 clearly are not essential for it to bind to RACK1. Conversely, at least some of the “core” regions (i.e., those found in all the known isoforms) are essential for the interaction.
Fig. 1. SERBP1 protein isoforms and key functional features.
1a. Amino acid sequence of the longest known (474-amino acid) human SERBP1 isoform. The single-letter amino acid code is used. Regions that are encoded by alternatively-spliced exons and therefore not present in some of the shorter isoforms are shown in gold (i.e., the 63-amino-acid N-terminal region), red, blue or green (see Fig. 1b for more details). The two regions that are targets for arginine methylation by PRMT1 (typically, repeats of sequence RGG or RGR) are shown in grey. The C-terminal region required for interaction with RACK1 is underlined.
1b. Human SERBP1 variants. BLAST searches of amino acid sequence data in GenBank using the 474-amino acid SERBP1 isoform as a query yielded 9 additional isoforms, all encoded by SERBP1 and generated by alternative mRNA splicing. Each of these isoforms differs from the 474-amino acid isoform in that it lacks specific blocks of sequence, as shown.
3.3. Deletion constructs define an extensive area on SERBP1 required for its interaction with RACK1
To define further the region(s) on SERBP1 required for it to interact with RACK1, a series of SERBP1 deletion constructs were prepared in pGADN (see Materials and Methods) and tested in 2-hybrid filter β-galactosidase assays for their ability to interact with pLexARACK1 (Fig. 2). These data show that amino acids 354 to 474 (using coordinates based on the full-length isoform, Fig. 1a) of SERBP1 are necessary and sufficient for it to interact with RACK1. This region of SERBP1 has 65 of 121, or 53.7%, amino acid identity with the corresponding region of HABP4 (Fig. 3). It contains one of the arginine methylation sites (Figs. 3 and 1a) but also extensive areas of undetermined function. It has no obvious amino acid sequence homology to the regions of PDE4D5 required for its interaction with RACK1 (10;11;14–16).
Fig. 2. Determination of the region of SERBP1 required for it to interact with RACK1.
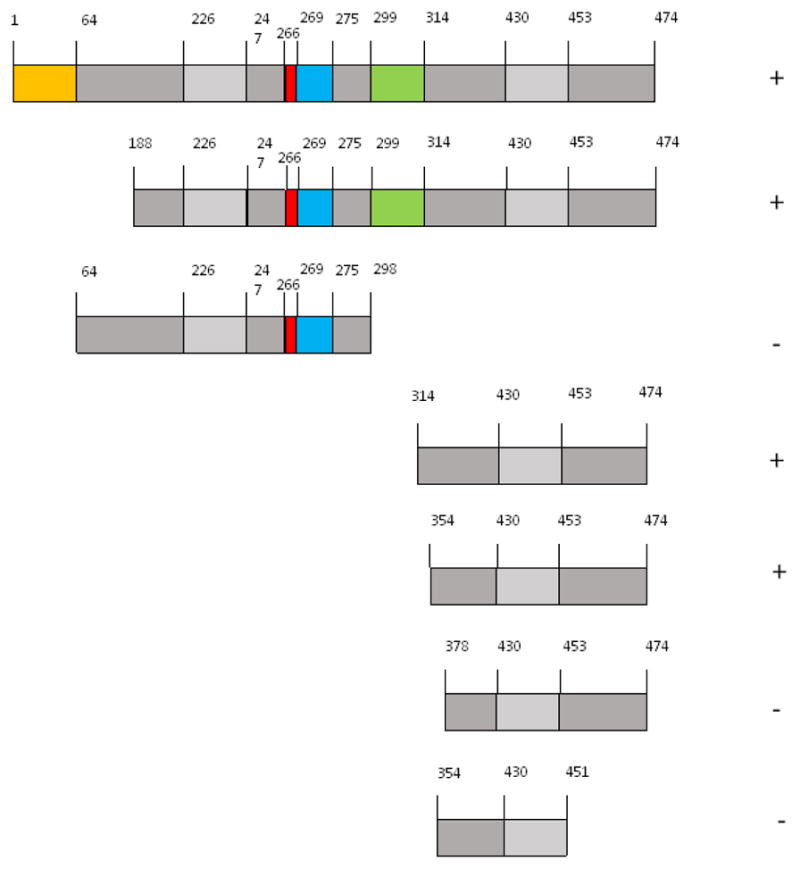
Yeast 2-hybrid experiments were performed using RACK1 as bait and various regions of the SERBP1 cDNA as prey. For bait, RACK1 was expressed in S. cerevisiae L40 as a LexA fusion (i.e., as pLexARACK1). For prey, the SERBP1 clones were expressed as GAL4 fusions (i.e., in pGADN containing full-length SERBP1, or various deletion subclones thereof). The strength of the Interactions were determined by a filter β-galactosidase assay. Interactions that were indistinguishable from that of RACK1 and full-length SERBP1 were scored as positive (symbol to the right of each construct); those that were indistinguishable from the corresponding empty vectors were scored as negative. All data were obtained in a least 3 independent experiments, using separate transformations. Discrete regions of amino acid sequence encoded by each construct are shown, using the same color scheme as in Fig. 1a.
Fig. 3. Alignment of the amino acid sequence of the long SERBP1 isoform with that of the longest known HABP4 isoform.
BLAST searches of human amino acid sequence data in GenBank using the 413-amino acid HABP1 isoform as a query yielded at least 2 additional isoforms, all encoded by HABP4 and generated by alternative mRNA splicing. The longest HABP4 isoform (413 amino acids) was then aligned with the longest SERBP1 isoform (474 amino acids) using BLASTP at www.ncbi.nlm.nih.gov/blast/. The alignment is shown using the single letter amino acid code. Dashes indicate gaps inserted by the alignment program, amino acid identities are indicated by the presence of that amino acid in the line between the 2 sequences, and related amino acids are indicated by the (+) sign. The region of SERBP1 essential for its interaction with RACK1 (amino acids 354 to 474, Fig. 2) is underlined.
3.4. Specific amino acids in RACK1 are required for its interactions with both SERPB1 and PDE4D5
RACK1 is a WD-repeat, β-propeller protein that has extensive amino acid sequence homology to the trimeric G-protein Gsβ subunit (refs. (87–90); see ref. (12) for an alignment). The crystal structure of RACK1 from Arabidopsis thaliana demonstrates its β-propeller structure (91) and in particular shows the presence of highly conserved surface residues that could play critical roles in protein-protein interactions (91). Human RACK1 has high amino acid sequence homology to A. thaliana RACK1 (92) and its solution structure demonstrates high structural homology to A. thaliana RACK1, as well as overall similarity to Gsβ and other β-propeller domains (64;93). Therefore, it would be of interest to determine whether amino acid side chains in RACK1 that are critical for it to interact with one partner also play a role in interacting with other partner(s). We used 2-hybrid approaches to test this hypothesis for RACK1.
Previously, we have used a “reverse” 2-hybrid approach to identify amino acids of RACK1 essential for it to interact with PDE4D5 (12). Briefly, we used PCR with a low-fidelity polymerase to perform random mutagenesis of a cDNA encoding WD-repeats 5 through 7 of RACK1 and then used a 2-hybrid assay to isolate mutants that attenuated the interaction of the WD-repeats with PDE4D5. With this approach, we identified 11 amino acids in RACK1 that, when individually mutated, blocked its interaction with PDE4D5. Molecular modeling showed that all 11 mutations mapped to a single surface on the RACK1 protein, presumably a “face” necessary for it to interact with PDE4D5 (12).
We therefore tested the effects of 8 of these 11 mutants on the ability of RACK1 to interact with SERBP1, using a 2-hybrid assay (Fig. 4). The results show that a number of the mutations (e.g., H191R, I236T, F241S, C249R, W259R, and W310R) significantly attenuated the interaction of RACK1 with both PDE4D5 and SERBP1. The N237D mutant affected the interactions with either protein to a roughly similar extent (although to a lesser extent than other mutations), while the C288G mutant attenuated the interaction with SERBP1 much more than it did with PDE4D5. These results are consistent with the concept that a discrete set of amino acids on one surface of RACK1 is necessary for it to interact with multiple partners, with additional amino acids contributing to the specificity of interactions with individual partners, such as PDE4D5 or SERBP1. Our approach cannot rule out the possibility that that some of the mutations could destabilize RACK1 generally, rather than contribute directly to the binding surface.
Fig. 4. Analysis of mutations in RACK1 that attenuate its interaction with SERBP1 and PDE4D5.
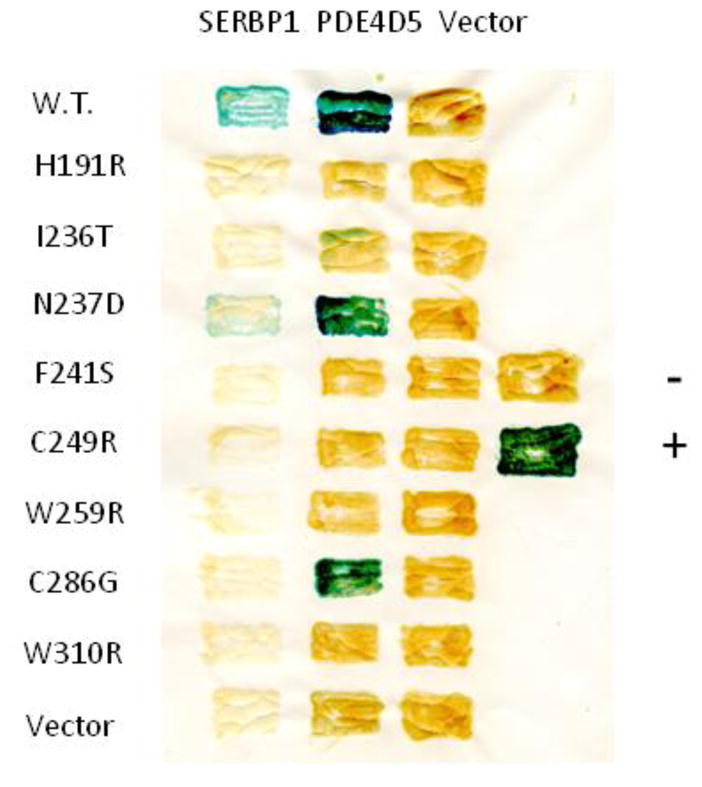
Various mutations in RACK1 that attenuated its interaction with PDE4D5 were created by random mutagenesis and identified in a reverse 2-hybrid screen, as we have described previously (12). These mutations were then tested for their ability to attenuate the interaction of RACK1 with SERBP1, as described in Materials and Methods. Yeast S. cerevisiae L40 cells in any given row contained the same bait and those in any given column contained the same prey. Positive interactions, assessed with a filter β-galactosidase assay, produce blue patches, while negative interactions produce amber patches. Controls are vectors alone. Standards are the oncoproteins RASV12 and RAF1. This figure shows data typical of experiments performed at least 3 times.
4. Discussion
In this study, we show that RACK1 interacts with SERBP1, a protein implicated in diverse functions, including transcription, genome integrity, and the regulation of mRNA stability. Our observation should stimulate additional investigation into RACK1’s biophysical and biochemical role in these processes. Given that RACK1 has numerous protein partners in cells, it also potentially links SERBP1 to some of RACK1’s other partners and to their physiological roles. It also provides insights into how RACK1 interacts with its partners and, in particular, the precise structural features on RACK1 that provide specificity to its interactions and determine the number of partners with which it can interact at any given time.
SERBP1 has homology to several related proteins, most notably HABP4 (synonym: Ki-1/57), a protein that also interacts with RNA and has several potential nuclear functions. However, SERBP1 differs from HABP4 in that it interacts avidly with highly-specific regions of sequence in the 3′ termini of several mRNAs, regulating their stability in response to specific stimuli (72–74). Both SERBP1 and HABP4 have 2 sites for arginine methylation by PRMT (Figs. 1 and 3) and interact with CHD-3, a nuclear protein involved in chromatin remodeling and transcription regulation (65;76;77). These common functions may provide an explanation for their significant amino acid sequence homology (Fig. 3); however, the precise region(s) of the 2 proteins responsible for many of their functions are not known, especially in the absence of structural clues. HABP4 is felt to be an intrinsically-disordered protein (94); to the best of our knowledge, there is no structural data on SERBP1.
We have shown that an extensive region in SERBP1 (amino acids 354 to 474, using co-ordinates based on the full-length isoform, Figs. 1a and 2) is necessary for it to interact with RACK1; since this region has substantial homology to the corresponding region of HABP4 (Fig. 3) it is a reasonable hypothesis that this region of HABP4 is also essential for it to interact with RACK1. More generally, it appears that many of RACK1’s partners employ a large region(s) of their respective proteins in their interaction with RACK1; for example, we and our collaborators have identified 2 extensive regions in PDE4D5 that are required for it to interact with RACK1 (10–16;95). The large size of the interacting regions of PDE4D5 and RACK1 is consistent with the high avidity of their interaction; in this regard, we and our collaborators have shown previously that the interaction between RACK1 and PDE4D5 has an affinity in the low nanomolar range (10). Although there is no obvious amino acid sequence homology between the RACK1-interaction regions of PDE4D5 and that of SERBP1 (and, by extension, that of HABP4), it is possible that similarities in the overall protein conformation of these regions might be apparent upon appropriate structural study.
Given the large region(s) on RACK1’s partners that seem to be essential for many of them to interact with RACK1, a reasonable hypothesis would be that a corresponding large region on RACK1 is also essential for the interaction. We have shown previously that a large surface (WD-repeats 5 through 7) on RACK1 is essential for it to interact with PDE4D5 and have identified a number of amino acids on RACK1 that, when individually mutated, markedly attenuate their interaction (12). In the present study, we have shown that many of these RACK1 mutants also attenuate its interaction with SERBP1 (Fig. 4). These observations suggest, but do not prove, that a large region of RACK1 interacts with SERBP1 and that this region overlaps substantially with the region of RACK1 that is essential for it to interact with PDE4D5. Potential alternative explanations for our data could be that some of the mutations alter the conformation of RACK1 generally (rather than affecting the interaction face itself), or destabilize the protein in cells. Finally, we cannot exclude the possibility that some of our observations reflect artifacts of the yeast 2-hybrid system; however, our S. cerevisiae-based system has, in a very large number of cases, been shown to be a reasonably physiologic system for these studies.
One important issue in interpreting data on the interaction of RACK1 and its partners is the stoichiometry of the complex. In our 2-hybrid studies, RACK1 and its partners each interact as monomers. Some studies have suggested that RACK1 is dimeric in cells (96–99), while other data (100) are more consistent with it existing in solution primarily as a monomer. It is clearly monomeric in the ribosome (101). If RACK1 exists primarily as a monomer in cells, then our mutagenesis data are consistent with the surface on WD repeats 5 through 7 directly interacting with PDE4D5 and SERBP1; however, if RACK1 exists as a dimer, then the mutations could disrupt RACK1 dimerization and not affect the actual interaction site(s) with its partners. Additional structural or interaction data will be essential to distinguish these possibilities. Finally, we cannot exclude the possibility that many of RACK1’s partners could also dimerize or multimerize; in this regard, we, our collaborators, and a number of other groups have clearly shown that PDE4D5 can dimerize (83;84;102–105), although the regions of PDE4D5 that are essential for dimerization are quite separate from those necessary for it to interact with RACK1 (see ref. (17) for a more complete discussion).
Our 2-hybrid screen data also provide some additional insight into the interaction of RACK1 with the small (40S) ribosomal subunit. A large number of biochemical studies have shown previously that RACK1 interacts directly with the 40S subunit. A structural study employing X-ray diffraction has shown that RACK1 interacts directly with rpS16e, rpS17e, and rpS3e [new names: uS9, eS17 and uS3, respectively, ref. (59)] and the 18S rRNA of the human 40S subunit (101). In our 2-hybrid screen, we isolated fusions encoding the rpS13e and rpS17e proteins (new names uS15 and eS17, respectively; ref. (59)). Our data confirm the importance of rpS17e in the interaction of RACK1 with the 40S subunit; however, the significance of our rpS13e isolate is unclear, as this protein is located some distance away from RACK1 (101). The functional significance of the interaction of RACK1 with the ribosome, especially in complex eukaryotes, remains uncertain (3).
Collectively, the studies reported here provide additional impetus for the study of the role(s) of RACK1 in nuclear processes, such as gene expression, RNA processing and the maintenance of genome integrity, and also in ribosomal composition and the regulation of translation. They also provide additional insights into the interaction face(s) that mediate the interaction of RACK1 with its partners. Further structural and functional experiments should provide insights into the physiological significance of our observations. One possible experiment would be to over-express (or, alternatively knock down) RACK1 in an assay of SERBP1 action on PAI1 mRNA stability.
5. Conclusion
We have identified SERBP1, ribosomal protein S17 (rpS17e) and potentially ribosomal protein S13 (rpS13e) as partners for RACK1 in a 2-hybrid screen.
The interaction of RACK1 and SERBP1 is mediated by large region(s) on both proteins, located in the C-terminal third of SERBP1 and a large “face” on one side of WD-repeats 5 through 7 of RACK1, respectively.
These data provide additional impetus for investigation of the role(s) of RACK1 in nuclear processes, such as gene expression, RNA processing and the maintenance of genome integrity, and also in ribosomal composition and the regulation of translation.
Highlights.
SERBP1 and ribosomal protein S17 (rpS17e) are partners for RACK1 in 2-hybrid analyses.
The interaction of RACK1 and SERBP1 is mediated by large region(s) on both proteins.
The interaction region on SERBP1 is located in the C-terminal third of the protein.
The interaction region on RACK1 includes its WD-repeats 5 through 7.
The data provide evidence for roles of RACK1 in nuclear processes and RNA processing.
Acknowledgments
Competing interests: The author certifies that he has no conflicts of interest.
Funding: The Bolger Prostate Cancer Research Fund (no grant number), the National Institute of General Medical Sciences of the National Institutes of Health (1-R01-GM58553, G. B. Bolger, principal investigator), and the National Cancer Institute of the NIH to the University of Alabama at Birmingham Comprehensive Cancer Center under award number P30 CA013148 (for DNA sequencing).
The content is solely the responsibility of the author and does not necessarily represent the official views of the NIH, which played no role on the in study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; or in the decision to submit the manuscript for publication.
Authors’ contributions: GBB provided overall supervision and planned and performed all experiments.
Availability of data and materials: The authors agree to share all materials and will deposit data into public-available databases according to Journal policy.
Abbreviations
- RACK1
Receptor for activated C-Kinase
- PRMT1
Protein arginine methyl transferase 1
- CHD-3
Chromo-helicase-DNA-binding domain protein 3
- PGRMC1
Progesterone receptor membrane component 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–73. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 2.Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallo S, Manfrini N. Working hard at the nexus between cell signaling and the ribosomal machinery: An insight into the roles of RACK1 in translational regulation. Translation (Austin) 2015;3:e1120382. doi: 10.1080/21690731.2015.1120382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mochly Rosen D, Khaner H, Lopez J, Smith BL. Intracellular receptors for activated protein kinase C. Identification of a binding site for the enzyme. J Biol Chem. 1991;266:14866–8. [PubMed] [Google Scholar]
- 5.Mochly Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci US A. 1991;88:3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–43. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ron D, Luo J, Mochly-Rosen D. C2 region-derived peptides inhibit translocation and function of beta protein kinase C in vivo. J Biol Chem. 1995;270:24180–7. doi: 10.1074/jbc.270.41.24180. [DOI] [PubMed] [Google Scholar]
- 8.Ron D, Mochly-Rosen D. An autoregulatory region in protein kinase C: the pseudoanchoring site. Proc Natl Acad Sci USA. 1995;92:492–6. doi: 10.1073/pnas.92.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger GB, Erdogan S, Jones RE, Loughney K, Scotland G, Hoffmann R, Wilkinson I, Farrell C, Houslay MD. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem J. 1997;328(Pt 2):539–48. doi: 10.1042/bj3280539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarwood SJ, Steele MR, Scotland G, Houslay MD, Bolger GB. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. Journal of Biological Chemistry. 1999;274:14909–17. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 11.Bolger GB, McCahill A, Yarwood SJ, Steele MS, Warwicker J, Houslay MD. Delineation of RAID1, the RACK1 interaction domain located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. BMC Biochem. 2002;3:24. doi: 10.1186/1471-2091-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steele MR, McCahill A, Thompson DS, MacKenzie C, Isaacs NW, Houslay MD, Bolger GB. Identification of a surface on the beta-propeller protein RACK1 that interacts with the cAMP-specific phosphodiesterase PDE4D5. Cell Signal. 2001;13:507–13. doi: 10.1016/s0898-6568(01)00167-x. [DOI] [PubMed] [Google Scholar]
- 13.Bird RJ, Baillie GS, Yarwood SJ. Interaction with receptor for activated C-kinase 1 (RACK1) sensitizes the phosphodiesterase PDE4D5 towards hydrolysis of cAMP and activation by protein kinase C. Biochem J. 2010;432:207–16. doi: 10.1042/BJ20101010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger GB, Baillie GS, Li X, Lynch MJ, Herzyk P, Mohamed A, Mitchell LH, McCahill A, Hundsrucker C, Klussmann E, Adams DR, Houslay MD. Scanning peptide array analyses identify overlapping binding sites for the signalling scaffold proteins, beta-arrestin and RACK1, in cAMP-specific phosphodiesterase PDE4D5. Biochemical Journal. 2006;398:23–36. doi: 10.1042/BJ20060423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baillie GS, Adams DR, Bhari N, Houslay TM, Vadrevu S, Meng D, Li X, Dunlop A, Milligan G, Bolger GB, Klussmann E, Houslay MD. Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochemical Journal. 2007;404:71–80. doi: 10.1042/BJ20070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith KJ, Baillie GS, Hyde EI, Li X, Houslay TM, McCahill A, Dunlop AJ, Bolger GB, Klussmann E, Adams DR, Houslay MD. 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1. Cell Signal. 2007;19:2612–24. doi: 10.1016/j.cellsig.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Bolger GB. RACK1 and beta-arrestin2 attenuate dimerization of PDE4 cAMP phosphodiesterase PDE4D5. Cell Signal. 2016;28:706–12. doi: 10.1016/j.cellsig.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang BY, Conroy KB, Machleder EM, Cartwright CA. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245–56. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang BY, Chiang M, Cartwright CA. The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J Biol Chem. 2001;276:20346–56. doi: 10.1074/jbc.M101375200. [DOI] [PubMed] [Google Scholar]
- 20.Doan AT, Huttenlocher A. RACK1 regulates Src activity and modulates paxillin dynamics during cell migration. Exp Cell Res. 2007;313:2667–79. doi: 10.1016/j.yexcr.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceci M, Welshhans K, Ciotti MT, Brandi R, Parisi C, Paoletti F, Pistillo L, Bassell GJ, Cattaneo A. RACK1 is a ribosome scaffold protein for beta-actin mRNA/ZBP1 complex. PLoS One. 2012;7:e35034. doi: 10.1371/journal.pone.0035034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F, Yamauchi M, Muramatsu M, Osawa T, Tsuchida R, Shibuya M. RACK1 regulates VEGF/Flt1-mediated cell migration via activation of a PI3K/Akt pathway. J Biol Chem. 2011;286:9097–106. doi: 10.1074/jbc.M110.165605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiely PA, Baillie GS, Barrett R, Buckley DA, Adams DR, Houslay MD, O’Connor R. Phosphorylation of RACK1 on tyrosine 52 by c-Abl is required for insulin-like growth factor I-mediated regulation of focal adhesion kinase. J Biol Chem. 2009;284:20263–74. doi: 10.1074/jbc.M109.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mourton T, Hellberg CB, Burden-Gulley SM, Hinman J, Rhee A, Brady-Kalnay SM. The PTPmu protein-tyrosine phosphatase binds and recruits the scaffolding protein RACK1 to cell-cell contacts. J Biol Chem. 2001;276:14896–901. doi: 10.1074/jbc.M010823200. [DOI] [PubMed] [Google Scholar]
- 25.Kiely M, Adams DR, Hayes SL, O’Connor R, Baillie GS, Kiely PA. RACK1 stabilises the activity of PP2A to regulate the transformed phenotype in mammary epithelial cells. Cell Signal. 2016 doi: 10.1016/j.cellsig.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Kiely PA, Baillie GS, Lynch MJ, Houslay MD, O’Connor R. Tyrosine 302 in RACK1 is essential for insulin-like growth factor-I-mediated competitive binding of PP2A and beta1 integrin and for tumor cell proliferation and migration. J Biol Chem. 2008;283:22952–61. doi: 10.1074/jbc.M800802200. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Bergami P, Huang C, Goydos JS, Yip D, Bar-Eli M, Herlyn M, Smalley KS, Mahale A, Eroshkin A, Aaronson S, Ronai Z. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–60. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marubashi S, Ohbayashi N, Fukuda M. A Varp-Binding Protein, RACK1, Regulates Dendrite Outgrowth through Stabilization of Varp Protein in Mouse Melanocytes. J Invest Dermatol. 2016;136:1672–80. doi: 10.1016/j.jid.2016.03.034. [DOI] [PubMed] [Google Scholar]
- 29.Disatnik MH, Hernandez-Sotomayor SM, Jones G, Carpenter G, Mochly-Rosen D. Phospholipase C-gamma 1 binding to intracellular receptors for activated protein kinase C. Proc Natl Acad Sci USA. 1994;91:559–63. doi: 10.1073/pnas.91.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaka R, Thornton C, Vagts AJ, Phamluong K, Bonci A, Ron D. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc Natl Acad Sci USA. 2002;99:5710–5. doi: 10.1073/pnas.062046299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaka R, He DY, Phamluong K, Ron D. Pituitary adenylate cyclase-activating polypeptide (PACAP(1–38)) enhances N-methyl-D-aspartate receptor function and brain-derived neurotrophic factor expression via RACK1. J Biol Chem. 2003;278:9630–8. doi: 10.1074/jbc.M209141200. [DOI] [PubMed] [Google Scholar]
- 32.Yaka R, Phamluong K, Ron D. Scaffolding of Fyn kinase to the NMDA receptor determines brain region sensitivity to ethanol. J Neurosci. 2003;23:3623–32. doi: 10.1523/JNEUROSCI.23-09-03623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macdonald DS, Weerapura M, Beazely MA, Martin L, Czerwinski W, Roder JC, Orser BA, MacDonald JF. Modulation of NMDA receptors by pituitary adenylate cyclase activating peptide in CA1 neurons requires G alpha q, protein kinase C, and activation of Src. J Neurosci. 2005;25:11374–84. doi: 10.1523/JNEUROSCI.3871-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geijsen N, Spaargaren M, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ. Association of RACK1 and PKCbeta with the common beta-chain of the IL-5/IL-3/GM-CSF receptor. Oncogene. 1999;18:5126–30. doi: 10.1038/sj.onc.1202896. [DOI] [PubMed] [Google Scholar]
- 35.Croze E, Usacheva A, Asarnow D, Minshall RD, Perez HD, Colamonici O. Receptor for activated C-kinase (RACK-1), a WD motif-containing protein, specifically associates with the human type I IFN receptor. J Immunol. 2000;165:5127–32. doi: 10.4049/jimmunol.165.9.5127. [DOI] [PubMed] [Google Scholar]
- 36.Usacheva A, Smith R, Minshall R, Baida G, Seng S, Croze E, Colamonici O. The WD motif-containing protein receptor for activated protein kinase C (RACK1) is required for recruitment and activation of signal transducer and activator of transcription 1 through the type I interferon receptor. J Biol Chem. 2001;276:22948–53. doi: 10.1074/jbc.M100087200. [DOI] [PubMed] [Google Scholar]
- 37.Hermanto U, Zong CS, Li W, Wang LH. RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting protein, modulates IGF-I-dependent integrin signaling and promotes cell spreading and contact with extracellular matrix. Mol Cell Biol. 2002;22:2345–65. doi: 10.1128/MCB.22.7.2345-2365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiely PA, Sant A, O’Connor R. RACK1 is an IGF-1 Receptor interacting protein that can regulate IGF-1- mediated Akt activation and protection from cell death. J Biol Chem. 2002:8. doi: 10.1074/jbc.M201758200. [DOI] [PubMed] [Google Scholar]
- 39.Dell EJ, Connor J, Chen S, Stebbins EG, Skiba NP, Mochly-Rosen D, Hamm HE. The betagamma subunit of heterotrimeric G proteins interacts with RACK1 and two other WD repeat proteins. J Biol Chem. 2002;277:49888–95. doi: 10.1074/jbc.M202755200. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Spiegelberg BD, Lin F, Dell EJ, Hamm HE. Interaction of Gbetagamma with RACK1 and other WD40 repeat proteins. J Mol Cell Cardiol. 2004;37:399–406. doi: 10.1016/j.yjmcc.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 41.Chen S, Dell EJ, Lin F, Sai J, Hamm HE. RACK1 regulates specific functions of Gbetagamma. J Biol Chem. 2004;279:17861–8. doi: 10.1074/jbc.M313727200. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Lin F, Hamm HE. RACK1 binds to a signal transfer region of G betagamma and inhibits phospholipase C beta2 activation. J Biol Chem. 2005;280:33445–52. doi: 10.1074/jbc.M505422200. [DOI] [PubMed] [Google Scholar]
- 43.Neasta J, Kiely PA, He DY, Adams DR, O’Connor R, Ron D. Direct interaction between scaffolding proteins RACK1 and 14–3–3 zeta regulates brain-derived neurotrophic factor (BDNF) transcription. J Biol Chem. 2012;287:322–36. doi: 10.1074/jbc.M111.272195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigas AC, Ozanne DM, Neal DE, Robson CN. The scaffolding protein RACK1 interacts with androgen receptor and promotes cross-talk through a protein kinase C signaling pathway. J Biol Chem. 2003;278:46087–93. doi: 10.1074/jbc.M306219200. [DOI] [PubMed] [Google Scholar]
- 45.Kraus S, Gioeli D, Vomastek T, Gordon V, Weber MJ. Receptor for activated C kinase 1 (RACK1) and Src regulate the tyrosine phosphorylation and function of the androgen receptor. Cancer Res. 2006;66:11047–54. doi: 10.1158/0008-5472.CAN-06-0596. [DOI] [PubMed] [Google Scholar]
- 46.Cao G, Thebault S, van der Wijst J, van der Kemp A, Lasonder E, Bindels RJ, Hoenderop JG. RACK1 inhibits TRPM6 activity via phosphorylation of the fused alpha-kinase domain. Curr Biol. 2008;18:168–76. doi: 10.1016/j.cub.2007.12.058. [DOI] [PubMed] [Google Scholar]
- 47.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327:463–6. doi: 10.1126/science.1180067. [DOI] [PubMed] [Google Scholar]
- 48.Cai W, Yang H. The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions. Cell Div. 2016;11:7. doi: 10.1186/s13008-016-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu YV, Semenza GL. RACK1 vs. HSP90: competition for HIF-1 alpha degradation vs stabilization. Cell Cycle. 2007;6:656–9. doi: 10.4161/cc.6.6.3981. [DOI] [PubMed] [Google Scholar]
- 50.Liu D, Liu X, Zhou T, Yao W, Zhao J, Zheng Z, Jiang W, Wang F, Aikhionbare FO, Hill DL, Emmett N, Guo Z, Wang D, Yao X, Chen Y. IRE1-RACK1 axis orchestrates ER stress preconditioning-elicited cytoprotection from ischemia/reperfusion injury in liver. J Mol Cell Biol. 2016;8:144–56. doi: 10.1093/jmcb/mjv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liliental J, Chang DD. Rack1, a receptor for activated protein kinase C, interacts with integrin beta subunit. J Biol Chem. 1998;273:2379–83. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- 52.Buensuceso CS, Woodside D, Huff JL, Plopper GE, O’Toole TE. The WD protein Rack1 mediates protein kinase C and integrin-dependent cell migration. J Cell Sci. 2001;114:1691–8. doi: 10.1242/jcs.114.9.1691. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez MM, Ron D, Touhara K, Chen CH, Mochly-Rosen D. RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry. 1999;38:13787–94. doi: 10.1021/bi991055k. [DOI] [PubMed] [Google Scholar]
- 54.Besson A, Wilson TL, Yong VW. The anchoring protein RACK1 links protein kinase Cepsilon to integrin beta chains. Requirements for adhesion and motility. J Biol Chem. 2002;277:22073–84. doi: 10.1074/jbc.M111644200. [DOI] [PubMed] [Google Scholar]
- 55.Feng C, Li YF, Yau YH, Lee HS, Tang XY, Xue ZH, Zhou YC, Lim WM, Cornvik TC, Ruedl C, Shochat SG, Tan SM. Kindlin-3 mediates integrin alphaLbeta2 outside-in signaling, and it interacts with scaffold protein receptor for activated-C kinase 1 (RACK1) J Biol Chem. 2012;287:10714–26. doi: 10.1074/jbc.M111.299594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qu J, Ero R, Feng C, Ong LT, Tan HF, Lee HS, Ismail MH, Bu WT, Nama S, Sampath P, Gao YG, Tan SM. Kindlin-3 interacts with the ribosome and regulates c-Myc expression required for proliferation of chronic myeloid leukemia cells. Sci Rep. 2015;5:18491. doi: 10.1038/srep18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neasta J, Fiorenza A, He DY, Phamluong K, Kiely PA, Ron D. Activation of the cAMP Pathway Induces RACK1-Dependent Binding of beta-Actin to BDNF Promoter. PLoS One. 2016;11:e0160948. doi: 10.1371/journal.pone.0160948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–80. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 59.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A, Yusupov M. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–9. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aseffa A, Gumy A, Launois P, MacDonald HR, Louis JA, Tacchini-Cottier F. The early IL-4 response to Leishmania major and the resulting Th2 cell maturation steering progressive disease in BALB/c mice are subject to the control of regulatory CD4+CD25+ T cells. J Immunol. 2002;169:3232–41. doi: 10.4049/jimmunol.169.6.3232. [DOI] [PubMed] [Google Scholar]
- 61.Sharma G, Pallesen J, Das S, Grassucci R, Langlois R, Hampton CM, Kelly DF, des GA, Frank J. Affinity grid-based cryo-EM of PKC binding to RACK1 on the ribosome. J Struct Biol. 2013;181:190–4. doi: 10.1016/j.jsb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gandin V, Gutierrez GJ, Brill LM, Varsano T, Feng Y, Aza-Blanc P, Au Q, McLaughlan S, Ferreira TA, Alain T, Sonenberg N, Topisirovic I, Ronai ZA. Degradation of newly synthesized polypeptides by ribosome-associated RACK1/c-Jun N-terminal kinase/eukaryotic elongation factor 1A2 complex. Mol Cell Biol. 2013;33:2510–26. doi: 10.1128/MCB.01362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nery FC, Passos DO, Garcia VS, Kobarg J. Ki-1/57 interacts with RACK1 and is a substrate for the phosphorylation by phorbol 12-myristate 13-acetate-activated protein kinase C. J Biol Chem. 2004;279:11444–55. doi: 10.1074/jbc.M306672200. [DOI] [PubMed] [Google Scholar]
- 64.Nery FC, Bressan GC, Alborghetti MR, Passos DO, Kuniyoshi TM, Ramos CH, Oyama S, Jr, Kobarg J. A spectroscopic analysis of the interaction between the human regulatory proteins RACK1 and Ki-1/57. Biol Chem. 2006;387:577–82. doi: 10.1515/BC.2006.074. [DOI] [PubMed] [Google Scholar]
- 65.Lemos TA, Passos DO, Nery FC, Kobarg J. Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 2003;533:14–20. doi: 10.1016/s0014-5793(02)03737-7. [DOI] [PubMed] [Google Scholar]
- 66.Passos DO, Bressan GC, Nery FC, Kobarg J. Ki-1/57 interacts with PRMT1 and is a substrate for arginine methylation. FEBS J. 2006;273:3946–61. doi: 10.1111/j.1742-4658.2006.05399.x. [DOI] [PubMed] [Google Scholar]
- 67.Kobarg J, Schnittger S, Fonatsch C, Lemke H, Bowen MA, Buck F, Hansen HP. Characterization, mapping and partial cDNA sequence of the 57-kD intracellular Ki-1 antigen. Exp Clin Immunogenet. 1997;14:273–80. [PubMed] [Google Scholar]
- 68.Bressan GC, Quaresma AJ, Moraes EC, Manfiolli AO, Passos DO, Gomes MD, Kobarg J. Functional association of human Ki-1/57 with pre-mRNA splicing events. FEBS J. 2009;276:3770–83. doi: 10.1111/j.1742-4658.2009.07092.x. [DOI] [PubMed] [Google Scholar]
- 69.Bressan GC, Kobarg J. From protein interaction profile to functional assignment: the human protein Ki-1/57 is associated with pre-mRNA splicing events. RNA Biol. 2010;7:268–71. doi: 10.4161/rna.7.3.11489. [DOI] [PubMed] [Google Scholar]
- 70.Goncalves KA, Bressan GC, Saito A, Morello LG, Zanchin NI, Kobarg J. Evidence for the association of the human regulatory protein Ki-1/57 with the translational machinery. FEBS Lett. 2011;585:2556–60. doi: 10.1016/j.febslet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 71.Yang R, Gaidamakov SA, Xie J, Lee J, Martino L, Kozlov G, Crawford AK, Russo AN, Conte MR, Gehring K, Maraia RJ. La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol Cell Biol. 2011;31:542–56. doi: 10.1128/MCB.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plasminogen activator inhibitor mRNA. J Biol Chem. 2001;276:3341–7. doi: 10.1074/jbc.M006538200. [DOI] [PubMed] [Google Scholar]
- 73.Tillmann-Bogush M, Heaton JH, Gelehrter TD. Cyclic nucleotide regulation of PAI-1 mRNA stability. Identification of cytosolic proteins that interact with an a-rich sequence. J Biol Chem. 1999;274:1172–9. doi: 10.1074/jbc.274.2.1172. [DOI] [PubMed] [Google Scholar]
- 74.Ahn JW, Kim S, Na W, Baek SJ, Kim JH, Min K, Yeom J, Kwak H, Jeong S, Lee C, Kim SY, Choi CY. SERBP1 affects homologous recombination-mediated DNA repair by regulation of CtIP translation during S phase. Nucleic Acids Res. 2015;43:6321–33. doi: 10.1093/nar/gkv592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iwaki S, Yamamura S, Asai M, Sobel BE, Fujii S. Posttranscriptional regulation of expression of plasminogen activator inhibitor type-1 by sphingosine 1-phosphate in HepG2 liver cells. Biochim Biophys Acta. 2012;1819:1132–41. doi: 10.1016/j.bbagrm.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 76.Lemos TA, Passos DO, Nery FC, Kobarg J. Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 2003;533:14–20. doi: 10.1016/s0014-5793(02)03737-7. [DOI] [PubMed] [Google Scholar]
- 77.Lemos TA, Kobarg J. CGI-55 interacts with nuclear proteins and co-localizes to p80-coilin positive-coiled bodies in the nucleus. Cell Biochem Biophys. 2006;44:463–74. doi: 10.1385/CBB:44:3:463. [DOI] [PubMed] [Google Scholar]
- 78.Prigge JR, Iverson SV, Siders AM, Schmidt EE. Interactome for auxiliary splicing factor U2AF(65) suggests diverse roles. Biochim Biophys Acta. 2009;1789:487–92. doi: 10.1016/j.bbagrm.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee YJ, Hsieh WY, Chen LY, Li C. Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution. J Cell Biochem. 2012;113:2721–8. doi: 10.1002/jcb.24151. [DOI] [PubMed] [Google Scholar]
- 80.Lee YJ, Wei HM, Chen LY, Li C. Localization of SERBP1 in stress granules and nucleoli. FEBS J. 2014;281:352–64. doi: 10.1111/febs.12606. [DOI] [PubMed] [Google Scholar]
- 81.Costa FC, Saito A, Goncalves KA, Vidigal PM, Meirelles GV, Bressan GC, Kobarg J. Ki-1/57 and CGI-55 ectopic expression impact cellular pathways involved in proliferation and stress response regulation. Biochim Biophys Acta. 2014;1843:2944–56. doi: 10.1016/j.bbamcr.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 82.Peluso JJ, Yuan A, Liu X, Lodde V. Plasminogen activator inhibitor 1 RNA-binding protein interacts with progesterone receptor membrane component 1 to regulate progesterone’s ability to maintain the viability of spontaneously immortalized granulosa cells and rat granulosa cells. Biol Reprod. 2013;88:20. doi: 10.1095/biolreprod.112.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beard MB, Olsen AE, Jones RE, Erdogan S, Houslay MD, Bolger GB. UCR1 and UCR2 Domains Unique to the cAMP-specific Phosphodiesterase Family Form a Discrete Module via Electrostatic Interactions. Journal of Biological Chemistry. 2000;275:10349–58. doi: 10.1074/jbc.275.14.10349. [DOI] [PubMed] [Google Scholar]
- 84.Bolger GB, Dunlop AJ, Meng D, Day JP, Klussmann E, Baillie GS, Adams DR, Houslay MD. Dimerization of cAMP phosphodiesterase-4 (PDE4) in living cells requires interfaces located in both the UCR1 and catalytic unit domains. Cell Signal. 2015;27:756–69. doi: 10.1016/j.cellsig.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bartel PL, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–63. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 86.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–14. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 87.Guillemot F, Billault A, Auffray C. Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc Natl Acad Sci USA. 1989;86:4594–8. doi: 10.1073/pnas.86.12.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wall MA, Coleman DE, Lee E, Iniguez Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–58. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 89.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–9. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 90.Clapham DE, Neer EJ. G protein beta gamma subunits. Annu Rev Pharmacol Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- 91.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 2008;17:1771–80. doi: 10.1110/ps.035121.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006;57:2697–708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- 93.Goncalves KA, Borges JC, Silva JC, Papa PF, Bressan GC, Torriani IL, Kobarg J. Solution structure of the human signaling protein RACK1. BMC Struct Biol. 2010;10:15. doi: 10.1186/1472-6807-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bressan GC, Silva JC, Borges JC, Dos Passos DO, Ramos CH, Torriani IL, Kobarg J. Human regulatory protein Ki-1/57 has characteristics of an intrinsically unstructured protein. J Proteome Res. 2008;7:4465–74. doi: 10.1021/pr8005342. [DOI] [PubMed] [Google Scholar]
- 95.Sondek J, Siderovski DP. Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem Pharmacol. 2001;61:1329–37. doi: 10.1016/s0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- 96.Thornton C, Tang KC, Phamluong K, Luong K, Vagts A, Nikanjam D, Yaka R, Ron D. Spatial and temporal regulation of RACK1 function and N-methyl-D-aspartate receptor activity through WD40 motif-mediated dimerization. J Biol Chem. 2004;279:31357–64. doi: 10.1074/jbc.M402316200. [DOI] [PubMed] [Google Scholar]
- 97.Liu YV, Hubbi ME, Pan F, McDonald KR, Mansharamani M, Cole RN, Liu JO, Semenza GL. Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J Biol Chem. 2007;282:37064–73. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yatime L, Hein KL, Nilsson J, Nissen P. Structure of the RACK1 dimer from Saccharomyces cerevisiae. J Mol Biol. 2011;411:486–98. doi: 10.1016/j.jmb.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 99.Chu LY, Chen YH, Chuang NN. Dimerize RACK1 upon transformation with oncogenic ras. Biochem Biophys Res Commun. 2005;330:474–82. doi: 10.1016/j.bbrc.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 100.Goncalves KA, Borges JC, Silva JC, Papa PF, Bressan GC, Torriani IL, Kobarg J. Solution structure of the human signaling protein RACK1. BMC Struct Biol. 2010;10:15. doi: 10.1186/1472-6807-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–6. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 102.Cedervall P, Aulabaugh A, Geoghegan KF, McLellan TJ, Pandit J. Engineered stabilization and structural analysis of the autoinhibited conformation of PDE4. Proc Natl Acad Sci USA. 2015;112:E1414–E1422. doi: 10.1073/pnas.1419906112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Richter W, Conti M. Dimerization of the type 4 cAMP-specific phosphodiesterases is mediated by the upstream conserved regions (UCRs) J Biol Chem. 2002;277:40212–21. doi: 10.1074/jbc.M203585200. [DOI] [PubMed] [Google Scholar]
- 104.Richter W, Conti M. The oligomerization state determines regulatory properties and inhibitor sensitivity of type 4 cAMP-specific phosphodiesterases. J Biol Chem. 2004;279:30338–48. doi: 10.1074/jbc.M312687200. [DOI] [PubMed] [Google Scholar]
- 105.Xie M, Blackman B, Scheitrum C, Mika D, Blanchard E, Lei T, Conti M, Richter W. The upstream conserved regions (UCRs) mediate homo- and hetero-oligomerization of type 4 cyclic nucleotide phosphodiesterases (PDE4s) Biochemical Journal. 2014;459:539–50. doi: 10.1042/BJ20131681. [DOI] [PMC free article] [PubMed] [Google Scholar]