Abstract
Hypervirulent and multidrug resistant Klebsiella pneumoniae strains pose a significant threat to the public health. In the present study, 21 carbapenem-resistant K. pneumoniae isolates (CRKP) were determined by the string test as hypermucoviscous K. pneumoniae (HMKP), with the prevalence of 15.0% (21/140) among CRKP, and 1.1% (21/1838) among all K. pneumoniae isolates. Among them, 7 (33.3%), and 1 (4.76%) isolate belonged to capsular serotype K20 and K2 respectively, while 13 (61.9%, 13/21) weren't successfully typed by capsular serotyping. All the 21 isolates were carbapenemase-producers and were positive for blaKPC-2. In addition to blaKPC-2, all the 21 isolates except one harbor blaSHV-11, and 15 carry extended-spectrum β-lactamase gene blaCTX-M-65. The virulence-associated genes with more than 90% of positive rates among 21 isolates included ureA (100%, 21/21), wabG (100%, 21/21), fimH (95.2%, 20/21), entB (95.2%, 20/21), ycf (95.2%, 20/21), ybtS (95.2%, 20/21), and iutA (90.5%, 19/21). rmpA and aerobactin were found in 57.1% (12/21) isolates. Five sequence types (STs) were identified by multilocus sequence typing (MLST), including ST11 (11 K-non capsule typable and 5 K20 isolates), ST268 (1 K20 isolate and 1 K-non capsule typable isolate), ST65 (1 K2 isolate), ST692 (1 K-non capsule typable isolate), and ST595, a novel sequence type (1 K-non capsule typable isolate). Pulsed-field gel electrophoresis (PFGE) results showed two major PFGE clusters, of which cluster A accounts for 6 ST11 isolates (28.6%) and cluster B includes 8 ST11 isolates (38.1%, 8/21). Ten and six ST11 isolates were isolated from 2014 and 2015, respectively, while 8 were isolated from the same month of December in 2014. Ten isolates were collected from the intensive care unit (ICU), and all except one belonged to ST11. Additional 4 ST11 isolates were collected from patients in non-ICU wards, who had more than 10 days of ICU stay history in 2014 prior to transfer to their current wards where the isolates were recovered. Taken together, the present study showed a hospital outbreak and dissemination of ST11 HMKP with carbapenem resistance caused by KPC-2. Effective surveillance and strict infection control strategies should be implemented to prevent outbreak by HMKP with carbapenem resistance in hospitals.
Keywords: Klesiella pneumoniae, hypermucoviscous, carbapenem resistance, KPC-2, epidemiology
Introduction
Klebsiella pneumoniae is an important human pathogen causing numerous infections in hospitals and long-term care facilities as well as in the communities worldwide, including lung, urinary tract, surgical sites, soft tissues infections and bacteremia (Shon et al., 2013). A new variant of K. pneumoniae causing distinctive syndromes such as pyogenic liver abscesses (PLA), designated as hypervirulent (hypermucoviscous) K. pneumoniae (HMKP), was initially described from Taiwan in the mid-1980s and 1990s (Liu et al., 1986; Cheng et al., 1991; Wang et al., 1998). HMKP frequently caused severe and life-threatening infections, including endophthalmitis and meningitis in young and healthy individuals, and it is now becoming a global public health problem (Struve et al., 2015). The HMKP strains usually have a distinct hypermucoviscosity phenotype when grown on agar plates (Struve et al., 2015). Since its emergency in Southeast Asia, HMKP has caused sporadic infections in many countries from North America, Europe, South America, Middle East, Australia and Africa (Fang et al., 2005; Siu et al., 2012; Shon et al., 2013; Bialek-Davenet et al., 2014; Yang et al., 2014; Struve et al., 2015; Prokesch et al., 2016). A number of putative virulence genes, including mucoviscosity-associated gene A (magA), regulator of mucoid phenotype A (rmpA) and aerobactin genes (aerobactin), have been found to be associated with HMKP (Fang et al., 2004; Yu et al., 2006; Russo et al., 2014). HMKP is rarely resistant to commonly used antimicrobial agents except for an intrinsic resistance to ampicillin (Lin et al., 2010; Zhang et al., 2016). However, along with the dissemination of mobile genetic elements encoding carbapenemases, carbapenem-resistant HMKP isolates have been increasingly reported (Yang et al., 2014; Yao et al., 2015; Zhang, Y. et al., 2015). The emergence of carbapenem resistant, hypermucoviscous K. pneumoniae strains are of great concern as they may be capable of causing severe, untreatable infections in healthy individuals. Indeed, the convergence of enhanced virulence and acquired carbapenem resistance in K. pneumoniae strains pose an important threat to the public health. Although, many studies have reported HMKP infections, especially the PLA, the literatures about the prevalence, and molecular characteristics of carbapenem-resistant HMKP isolates are limited. The aim of the present study is to investigate the prevalence of HMKP among CRKP isolates, and to describe the molecular characteristics of carbapenem-resistant HMKP isolates in Wenzhou, eastern China.
Materials and methods
Collection and identification of K. pneumoniae clinical isolates
From January 2013 to December 2015, a total of 1838 K. pneumoniae isolates were consecutively collected from various specimens of patients at the first Affiliated Hospital of Wenzhou Medical University located in Wenzhou, eastern China. These isolates were identified as K. pneumoniae by Gram-staining and a VITEK-2 automated microbiology analyzer (bioMérieux, Marcy l'Etoile, France). The control strains, including Staphylococcus aureus ATCC25923, Escherichia coli ATCC25922, and Pseudomonas aeruginosa ATCC27853, were used for control for the species identification. Patient information were extracted from the medical records. This study was approved by the ethical committee of the first Affiliated Hospital of Wenzhou Medical University. Informed consents were obtained from the patients involved in current study.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing for agents commonly used for clinical K. pneumoniae infection treatment, was examined using a VITEK-2 automated microbiology analyzer platform (bioMérieux, Marcy l'Etoile, France). The minimal inhibitory concentrations (MICs) of imipenem and colistin for imipenem-resistant HMKP were further verified by E-test method according to the guideline recommended by the Clinical and Laboratory Standards Institute (CLSI, 2016). E. coli ATCC25922 was used as a control strain for antimicrobial susceptibility testing.
Detection of ESBLs and KPC enzymes
A modified Hodge test was performed to detect carbapenemases, as described previously (CLSI, 2016). All the isolates were tested for ESBL production by the CLSI-recommended confirmatory double-disc combination test (CLSI, 2016). Escherichia coli ATCC 25922 as an indicator bacteria, with sterile saline to E. coli ATCC 25922 suspension was adjusted to 0.5 mihms turbidity, and then diluted 10 times with saline. The diluted E. coli ATCC 25922 bacterial solution was evenly coated on an MH agar plate and the plate was dried for 3–10 min. Imipenem tablets (10 μg/tablet) was placed in the center of the MH agar plate. Using the 1 μl inoculation ring to pick the test strain or quality control strains to the center of the plate of drug sensitive paper as a starting point, along the centrifugal direction of the line, length 20–25 mm. The plate was incubated at 35°C for 16–20 h.
Phenotypical identification of HMKP isolates
String test was used to detect hypermucoviscosity phenotype as described previously (Shon et al., 2013). The K. pneumoniae isolates with positive hypermucoviscosity phenotypes were designated as HMKP. Briefly, an inoculation loop or needle was used to stretch the bacterial colonies of K. pneumoniae isolate on Columbia blood agar plate (BIO-KONT, Wenzhou, China) from overnight culture. The formation of viscous string with >5 mm in length was considered to be positive for the string test.
Capsular serotyping and detection of resistance genes and virulence-associated genes
The primer sequences for capsular serotyping, and the detection of resistance and virulence genes were listed in Table 1. Capsular serotypes, including K1, K2, K5, K20, K54, and K57, were determined using the methods described previously (Fang et al., 2007; Turton et al., 2010). The presence of resistance genes were detected by PCR amplification followed by Sanger sequencing using the primers and conditions described elsewhere (Pagani et al., 2003; Jiang et al., 2005; Li, H. et al., 2014; Validi et al., 2016). Seventeen virulence-associated genes, including aerobactin, iroN, kfuB, rmpA, wcaG, alls, ybtS, ureA, uge, wabG, ycf, entB, iutA, vatD, magA, fimH, and mrkD, were determined by PCR using the primers described previously for carbapenem-resistant HMKP isolates (Yu et al., 2006, 2008; Turton et al., 2010; Candan and Aksoz, 2015). Bacterial strains harboring these virulence genes from our previous studies were used as positive controls in the PCRs.
Table 1.
The sequences of primers for capsular serotyping, resistance genes and virulence-associated genes.
| Genes | Primers | Sequence (5′ → 3′) | T (°C) | Fragment (bp) | Refrences |
|---|---|---|---|---|---|
| K1 | F | GGTGCTCTTTACATCATTGC | 54 | 1,283 | Fang et al., 2007 |
| R | GCAATGGCCATTTGCGTTAG | Turton et al., 2010 | |||
| K2 | F | GACCCGATATTCATACTTGACAGAG | 64 | 641 | Fang et al., 2007 |
| R | CCTGAAGTAAAATCGTAAATAGATGGC | Turton et al., 2010 | |||
| K20 | F | CGGTGCTACAGTGCATCATT | 56 | 741 | Fang et al., 2007 |
| R | GTTATACGATGCTCAGTCGC | Turton et al., 2010 | |||
| blaKPC | F | ATGTCACTGTATCGCCGTCT | 55 | 893 | Li, H. et al., 2014 |
| R | TTTTCAGAGCCTTACTGCCC | Validi et al., 2016 | |||
| blaSHV | F | AGCCGCTTGAGCAAATTAAAC | 56 | 713 | Jiang et al., 2005 |
| R | ATCCCGCAGATAAATCACCAC | ||||
| blaCTX-M-65 | F | ATGGTGACAAAGAGAGTGCA | 54 | 870 | Jiang et al., 2005 |
| R | CCCTTCGGCGATGATTCTC | Pagani et al., 2003 | |||
| ureA | F | GACAAGCTGTTGCTGTTTACC | 58 | 270 | Candan and Aksoz, 2015 |
| R | CGGGTTGTGAACGGTGAC | ||||
| wabG | F | ACCATCGGCCATTTGATAGA | 58 | 683 | Candan and Aksoz, 2015 |
| R | CGGACTGGCAGATCCATATC | ||||
| fimH | F | TGCTGCTGGGCTGGTCGATG | 62 | 909 | Candan and Aksoz, 2015 |
| R | GGGAGGGTGACGGTGACATC | ||||
| entB | F | ATTTCCTCAACTTCTGGGGC | 56 | 371 | Candan and Aksoz, 2015 |
| R | AGCATCGGTGGCGGTGGTCA | ||||
| ycf | F | ATCAGCAGTCGGGTCAGC | 58 | 160 | Candan and Aksoz, 2015 |
| R | CTTCTCCAGCATTCAGCG | ||||
| ybtS | F | CACCGCAAACGCAATCTG | 56 | 782 | Candan and Aksoz, 2015 |
| R | GCCATAGACGCTGTTGTTGA | ||||
| iutA | F | GGCTGGACATCATGGGAACTGG | 66 | 300 | Candan and Aksoz, 2015 |
| R | CGTCGGGAACGGGTAGAATCG | ||||
| rmpA | F | ACTGGGCTACCTCTGCTTCA | 58 | 516 | Candan and Aksoz, 2015 |
| R | CTTGCATGAGCCATCTTTCA | Turton et al., 2010 | |||
| aerobactin | F | GCATAGGCGGATACGAACAT | 58 | 556 | Yu et al., 2008 |
| R | CACAGGGCAATTGCTTACCT | ||||
| IroN | F | GGCTACTGATACTTGACTATTC | 58 | 992 | Candan and Aksoz, 2015 |
| R | CAGGATACAATAGCCCATAG | ||||
| KfuB | F | GAAGTGACGCTGTTTCTGGC | 58 | 960 | Yu et al., 2008 |
| R | TTTCGTGTGGCCAGTGACTC | ||||
| wcaG | F | GGTTGGKTCAGCAATCGTA | 54 | 169 | Candan and Aksoz, 2015 |
| R | ACTATTCCGCCAACTTTTGC | Turton et al., 2010 | |||
| alls | F | CCGAAACATTACGCACCTTT | 58 | 508 | Candan and Aksoz, 2015 |
| R | ATCACGAAGAGCCAGGTCAC | ||||
| uge | F | TCTTCACGCCTTCCTTCACT | 60 | 534 | Candan and Aksoz, 2015 |
| R | GATCATCCGGTCTCCCTGTA | ||||
| vatD | F | GAAGGAAACAAATCAGTA | 48 | 463 | Candan and Aksoz, 2015 |
| R | GTTTTATTTCGTTAGCAG | ||||
| magA | F | GGTGCTCTTTACATCATTGC | 48 | 1,149 | Candan and Aksoz, 2015 |
| R | GCAATGGCCATTTGCGTTAG | Yu et al., 2006 |
Multilocus sequence typing (MLST)
MLST was performed on all carbapenem-resistant HMKP isolates by amplifying the seven standard housekeeping loci, including gapA, infB, mdh, pgi, phoE, rpoB, and tonB, as described previously (Diancourt et al., 2005). Sequence types (STs) were assigned using the online database on the Pasteur Institute MLST website (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Pulsed-field gel electrophoresis (PFGE)
PFGE was performed on all carbapenem-resistant HMKP isolates. In brief, genomic DNA was prepared by embedding K. pneumoniae cells in agarose plugs, followed by XbaI digestion for 12 h at 37°C. Electrophoresis was performed at 14°C for 20 h using the Bio-Rad CHEF III system (120° angle, 6V/cm, switch times of 6 and 36 s). Salmonella serotype Braenderup strain H9812 was used as the molecular marker, and DNA bands were stained with ethidium bromide (0.5 μg/mL). DNA band profiles were interpreted by the criteria of Tenover et al. (1995). Analysis of the PFGE patterns was performed with Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice Similarity coefficient. The isolates sharing more than 80% similarity were defined as the same PFGE cluster.
Results
Prevalence of carbapenem-resistant HMKP isolates
Among 1838 K. pneumoniae isolates, 140 (7.6%) were resistant to both imipenem and ertapenem, and were referred as carbapenem-resistant K. pneumoniae (CRKP). Among 140 CRKP isolates, 21 (15.0%) were positive for the string test, and were identified as HMKP. 18, 2, and 1 HMKP had imipenem MICs of >32 mg/L, 8 mg/L, and 4 mg/L, respectively. The prevalence of carbapenem-resistant HMKP among all K. pneumoniae isolates was 1.1% (21/1838). Interestingly, carbapenem-resistant HMKP was not found in any of the 23 CRKP isolates in 2013. By contrast, 27.9% (12/43) and (12.2%, 9/74) CRKP isolates in 2014 and 2015 were HMKP, respectively. Twenty-one carbapenem-resistant HMKP isolates were recovered from sputum (14 isolates), pus (2 isolates), urine (2 isolates), blood (2 isolates), and drainage (1 isolate), respectively.
Antimicrobial susceptibility and resistance mechanism
The antimicrobial resistance rates of the 21 carbapenem-resistant HMKP isolates were shown in Table 2. They were all resistant to ampicillin, ampicillin/sulbactam, aztreonam, cefazolin, piperacillin/tazobactam and ceftriaxone. Nineteen (90.5%) of 21 isolates were resistant to ceftazidime and cefotetan, and 80.9% (17/21) were resistant to ciprofloxacin, levofloxacin, gentamicin, tobramycin and cefepime. Sixteen (76.2%, 16/21) and fifteen (71.4%, 15/21) isolates were resistant to amikacin and fosfomycin, respectively. Only one isolate was resistant to sulfamethoxazole/trimethoprim and they were all susceptible to colistin.
Table 2.
The antimicrobial resistance profiling of 21 carbapenem-resistant HMKP isolates.
| Antimicrobials | HMKP (n = 21) | |
|---|---|---|
| No. | % | |
| Amikacin | 16 | 76.2 |
| Ampicillin | 21 | 100.0 |
| Amicillin/sulbatam | 21 | 100.0 |
| Aztreonam | 21 | 100.0 |
| Cefazolin | 21 | 100.0 |
| Cefotetan | 19 | 90.5 |
| Ceftriaxone | 21 | 100.0 |
| Cefepime | 17 | 80.9 |
| Ceftazidime | 19 | 90.5 |
| Ceftriaxone | 21 | 100.0 |
| Ciprofloxacin | 17 | 80.9 |
| Colistin | 0 | 0 |
| Imipenem | 21 | 100.0 |
| Levofloxacin | 17 | 80.9 |
| Fosfomycin | 15 | 71.4 |
| TZPa | 21 | 100.0 |
| Tobramycin | 17 | 80.9 |
| Gentamicin | 17 | 80.9 |
| SXTb | 1 | 4.8 |
TZP, Piperacillin/tazobactam.
SXT, trimethoprim/sulfamethoxazole.
All 21 HMKP isolates with carbapenem resistance were found to be carbapenemase-producers determined by modified Hodge test. In accordance with the phenotypic results, all isolates were positive for blaKPC-2.In addition to blaKPC-2, all except one isolates harbored blaSHV-11, and 15 (71.4%) harbored extended-spectrum β-lactamase gene blaCTX-M-65.
Clinical characteristics of 21 patients infected with carbapenem-resistant HMKP isolates
Clinical characteristics of 21 patients infected by carbapenem-resistant HMKP were summarized in Table 3. Among them, 9 were female and 12 were male, with age ranging from 22 to 82 years old. They were from 8 wards including intensive care unit (ICU) (10 cases), neurology ICU (2 cases), gastrointestinal surgery ward (2 cases), traumatology ward (2 case) and other four 4 wards (one case each). However, 4 patients from non-ICU wards had 10–40 days ICU stay history before transfer to their current wards. Fourteen isolates were collected from sputum samples of cases who had pneumonia. Hyperglycaemia was found among 9 cases (42.9%, 9/21). Thirteen cases (61.9%, 13/21) had been treated by carbapenems (imipenem, meropenem or panipenem), and 11 cases (52.4%, 11/21) had surgery. Mechanical ventilation was performed on 15 patients (71.4%, 15/21), and various drainage systems were installed in 14 patients (66.7%, 14/21).
Table 3.
Clinical characteristics of 21 patients infected with carbapenem-resistant HMKP isolatesa.
| PFGE type | Resistance genes | Virulence genes | Isolation site | Underlying conditions | Treatment | Invasive procedures | Outcome | |
|---|---|---|---|---|---|---|---|---|
| B23 | A | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Sputum | Cancer, encephalorrhagia pneumonia | Panipenem | Mechanical ventilation, drainage system, surgery | Survived |
| B27 | blaKPC-2, blaSHV-11 | ureA, wabG, fimH, entB, ycf, ybtS, rmpA, A | Sputum | Encephalorrhagia, pneumonia, hyperglycaemia | TZP | Mechanical ventilation, drainage system, surgery | Giving up treatment | |
| B29 | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS | Sputum | encephalorrhagia pneumonia | TZP, meropenem | Mechanical ventilation, drainage system, surgery | Giving up treatment | |
| B41 | C | blaKPC-2, blaSHV-11 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Sputum | Pneumonia, hypertension, hyperglycaemia | meropenem | Mechanical ventilation | Survived |
| B44 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Urine | acute peritonitis, colon cancer, Urinal tract infection | TZP | Mechanical ventilation, drainage system, surgery | Giving up treatment |
| B49 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Sputum | Head injury pneumonia | SXT, meropenem, levofloxacin | Mechanical ventilation, drainage system | Survived |
| B50 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Sputum | Cirrhosis, pneumonia | imipenem | Mechanical ventilation, drainage system, | Giving up treatment |
| B51 | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Sputum | Hemorrhagic shock, pneumonia | SCF, minocycline moxifloxacin | Mechanical ventilation, drainage system | Giving up treatment | |
| B52 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ybtS, iutA, rmpA, A | Urine | Urinal tract infection hypertension | TZP | drainage system, surgery | Survived |
| B53 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Sputum | Encephalorrhagia, pneumonia, hyperglycaemia | Tigecycline SXT, TZP | Mechanical ventilation, drainage system, surgery | Giving up treatment |
| B56 | A | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Sputum | Pneumonia, colon cancer, heart disease, diabetes mellitus | Meropenem, SXT, teicoplanin | Mechanical ventilation, drainage system | Giving up treatment |
| B72 | blaKPC-2, blaSHV-11 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A, IroNB | Sputum | Heart disease, pneumonia, diabetes mellitus | imipenem | Mechanical ventilation: | Giving up treatment | |
| B73 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Sputum | Head injury, pneumonia, hyperglycaemia | Cefuroxime | Mechanical ventilation, drainage system, surgery | Giving up treatment |
| B75 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, ycf, ybtS, iutA, rmpA, A | Sputum | Pneumonia, congenital heart disease | Tigecycline | Mechanical ventilation, surgery | Survived |
| B77 | B | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Sputum | Pneumonia | Panipenem, Ofloxacin | Survived | |
| B92 | blaKPC-2 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A, Kfu, wcaG | Sputum | Multiple injuries, pneumonia | Imipenem, levofloxacin | Survived | ||
| B98 | A | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Blood | Septic shock, Multiple organ failure | − | Mechanical ventilation | Death |
| B101 | A | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Pus | acute pancreatitis hyperglycaemia | Meropenem, levofloxacin, linezolid | Mechanical ventilation, drainage system | Giving up treatment |
| B102 | A | blaKPC-2, blaSHV-11, blaCTX-M-65 | ureA, wabG, fimH, entB, ycf, ybtS, iutA | Pus | Chronic kidney disease peritonitis | Linezolid, SXT meropenem | Drainage system | Giving up treatment |
| B103 | A | blaKPC-2, blaSHV-11 | ureA, wabG, fimH, entB, ycf, ybtS, iutA, rmpA, A | Drainage | Perforation of sigmoid colon | imipenem | Drainage system, surgery | Survived |
| B104 | C | blaKPC-2, blaSHV-11 | ureA, wabG, entB, ycf, iutA, rmpA, A | Blood | Diabetes mellitus, hypertension | Imipenem, levofloxacin | Survived |
TZP, piperacillin/tazobactam; SXT, sulfamethoxazole /trimethoprim; A, aerobactin.
Capsular serotyping for carbapenem-resistant HMKP isolates
Among the 21 carbapenem-resistant HMKP isolates, 7 (33.3%) belonged to capsular serotype K20, and another one belonged to K2 (4.5%, 1/22). The remaining 13 isolates (61.9%) were not successfully typed, and were therefore defined as K-nontypable.
Prevalence of virulence-associated genes among carbapenem-resistant HMKP isolates
The virulence-associated genes with more than 90% of detection rates among the 21 isolates included ureA (100%, 21/21), wabG (100%, 21/21), fimH (95.2%, 20/21), entB (95.2%, 20/21), ycf (95.2%, 20/21), ybtS (95.2%, 20/21), and iutA (90.5%, 19/21).
Molecular characteristics of carbapenem-resistant HMKP isolates
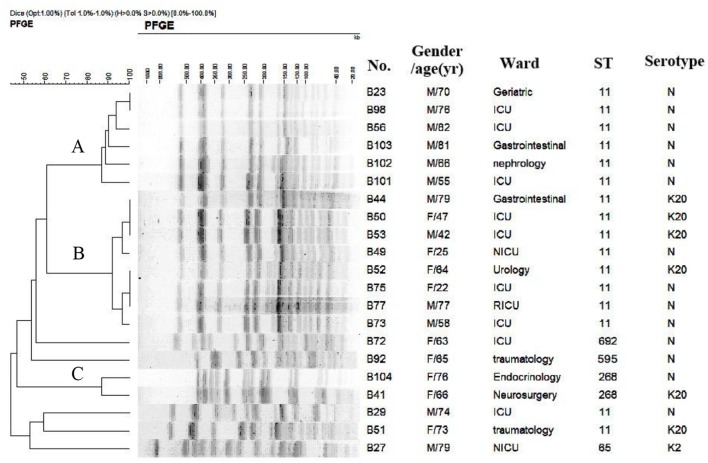
Among the 21 carbapenem-resistant HMKP isolates, 5 STs were identified, including ST11 (16 isolates), ST268 (2 isolates), ST65 (1 isolate), ST692 (1 isolate), and ST595, a novel type of STs (1 isolate). PFGE results showed that 16 ST11 isolates were divided into two different PFGE clusters, with cluster A accounting for 6 ST11 isolates (28.6%) and cluster B including 8 ST11 isolates (38.1%, 8/21) (Figure 1). Two ST268 isolates belonged to the same cluster. Each of the remaining 3 isolates formed a singleton.
Figure 1.
PFGE results for 21 carbapenem-resistant HMKP isolates.
Discussion
HMKP has been frequently described in several Chinese hospitals, however, carbapenem-resistant HMKP was rarely found in many previous reports (Li, W. et al., 2014; Liu et al., 2014; Zhang et al., 2016). Here we described the prevalence of HMKP among CRKP in a large tertiary hospital in China, and genetically and phenotypically characterized the carbapenem-resistant HMKP isolates. In this study, the prevalence of HMKP among CRKP isolates is higher than that of 7.4% from two hospitals in Zhejiang Province in China in 2013 (Zhang, R. et al., 2015), but it is similar to the prevalence of 17.9% (5/28) from an another previous report in China (Zhang, Y. et al., 2015). Our study and other reports suggested that acquiring high virulence and carbapenem resistance in K. pneumoniae is increasingly becoming a concern in the hospitals in China.
Diabetes mellitus has been considered as a significant risk factor for HMKP infection (Cheng et al., 1991; Wang et al., 1998; Shon et al., 2013; Zhang et al., 2016). In this study, we also identified that 42.9% (9/21) of the patients with HMKP infections had hyperglycaemia. The outcome of patients with infections by carbapenem-resistant HMKP is uncertain. Zhang et al. reported all 5 patients with HMKP infections died of septic shock (Zhang, R. et al., 2015), while another Chinese report showed 4 of 5 patients with similar infections survived (Zhang, Y. et al., 2015). In the present study, 9 patients survived and 1 patient die of septic shock and multiple organ failure, while the remaining 11 patients refused further treatments and requested to be discharged due to the progress of uncontrolled infections or their underlying diseases.
Zhang et al. reported that among 5 carbapenem-resistant HMKP isolates, only one belonged to K2 and the remaining 4 were nontypable (Zhang, R. et al., 2015; Zhang, Y. et al., 2015). Similarly, in this study we only found that one carbapenem-resistant HMKP isolate belonged to K2 (4.5%, 1/22). By contrast, Yao et al. reported that 6 out of 7 carbapenem-resistant HMKP isolates belonged to K2 (Yao et al., 2015). Another Chinese study found that 5 carbapenem-resistant HMKP strains causing fatal infections belonged to K1. K20 hasn't been found to be associated with carbapenem-resistant HMKP isolates before, however, we identified that 33.3% (7/21) of carbapenem-resistant HMKP isolates belonged to K20 in the present study, suggesting that K20 may be an important capsular serotype associated with carbapenem-resistant HMKP.
Although, carbapenem-resistant HMKP isolates were multi-resistant to clinically common-used antimicrobial agents in the present study, sulfamethoxazole/trimethoprim and colistin still had efficient antimicrobial activity in vitro against these isolates, indicating that sulfamethoxazole/trimethoprim and colistin could be valuable treatment choices against carbapenem-resistant HMKP infections. blaKPC-2, which is the main molecular mechanism of carbapenem resistance in K. pneumoniae in China, has been previously found to be responsible for carbapenem resistance among HMKP isolates (Zhang, R. et al., 2015). Similarly, all carbapenem-resistant HMKP isolates in the present study were positive for blaKPC-2.
Previous studies showed that there was a correlation between the presence of rmpA gene and virulence in terms of abscess formation of HMKP isolates (Yu et al., 2006; Yang et al., 2014; Yan et al., 2016; Zhang et al., 2016). A Chinese report described that rmpA was found in all K. pneumoniae isolates causing PLA (Qu et al., 2015). In the present study, 57.1% (12/21) of the carbapenem-resistant HMKP isolates harbored rmpA. Meanwhile, previous studies reported that magA was associated with the hypermucoviscosity phenotype of K. pneumoniae, which was characteristic of the K1 capsular operon (Fang et al., 2004, 2005; Chuang et al., 2006). Yu et al. reported that there was a strong association between kfuB and alls and K1 isolates, with all K1 isolates positive for kfuB and alls, and all K2 isolates negative for these two genes (Yu et al., 2008). In this study, all carbapenem-resistant HMKP isolates were negative for magA, alls, and kfuB, which is consistent with the result that no K1 capsular serotype was identified among the HMKP isolates. aerobactin is an important virulence determinant for HMKP, and it has been used as a marker for the identification of HMKP (Zhang et al., 2016). 57.1% (12/21) of the carbapenem-resistant HMKP isolates were found to carry aerobactin in the present study, while iorN and wcaG were only found in one isolate each.
ST11 is the predominant clone of KPC-producing K. pneumoniae isolates in China (Qi et al., 2011). However, this epidemic clone was rarely found in HMKP isolates (Liu et al., 2014; Zhang et al., 2016). Zhang et al. reported that ST11 was found in 3 carbapenem-resistant HMKP isolates with unidentified K types (Zhang, Y. et al., 2015). In the present study, ST11 was found to be the most common clone (76.2%, 16/21). Similar to the previous report (Zhang, Y. et al., 2015), 11 out of 16 ST11 isolates were nontypable by the capsular serotyping. However, 5 ST11 isolates belonged to K20, which was firstly reported in current study. Epidemic ST11 strains have been associated with multidrug-resistance and now become hypermucoviscous, which may make the treatment and control for K. pneumoniae infections more difficult and poses a major clinical problem for the future. ST23 was the most commonly described ST among HMKP isolates, and was strongly correlated with capsular serotype K1 and liver abscess in several previous studies (Turton et al., 2007; Chung et al., 2008; Shon et al., 2013). A report from China showed that among 5 KPC-2-producing HMKP with K1 serotype, 2 belong to ST23, and 3 belonged to ST1797 (Zhang, R. et al., 2015). In this study, ST23 was not identified, which is consistent with the finding of absence of K1 serotype. Previous studies showed that ST268 was only found among K20 isolates (Liu et al., 2014; Lin et al., 2015; Yan et al., 2015, 2016; Zhang et al., 2016). Similarly, 1 ST268 isolate was found to carry K20 serotype in the present study. In this study, one K2 strain belonged to ST65, which is in accordance with previous study that ST65 was the most common ST associated with K2 serotype in K. pneumoniae (Liao et al., 2014). Although, ST692, a two-locus variant of ST65, has been deposited in the K. pneumoniae MLST database, it is not reported in published literatures. Our study is the first report of ST692 K. pneumoniae with carbapenem resistance associated with clinical infection.
Among 16 ST 11 isolates, the first and second isolates were emerged in January and June, 2014 in our hospital, while 8 (50%) were identified in December, 2014. In 2015, 6 ST11 isolates were identified, and the last isolate was found in October. All 10 except one (ST692) isolates from ICU belonged to ST11. Although, other 4 ST11 isolates were collected from patients in non-ICU wards, they had more than 10 days of ICU stay history in 2014 prior to transfer to their current wards where the isolates were recovered. We suspected that the 4 patients may be initially infected by ST11 carbapenem-resistant HMKP during their stay in ICU, followed by transfer to other wards. The remaining three ST11 isolates were found in neurology ICU (NICU), respiratory ICU (RICU) and nephrology ward. Our data indicated that there was an outbreak of ST11 HMKP with carbapenem resistance in the ICU of our hospital between 2014 and 2015.
In conclusion, the present study reported an outbreak of ST11 HMKP with carbapenem resistance caused by KPC-2 in our hospital. Effective surveillance and strict infection control strategies should be implemented to prevent the dissemination of HMKP with carbapenem resistance, especially outbreak caused by this important pathogen.
Author contributions
LZ, SW, YG, YJ, JD, ZH, JL, and XQ collected bacteria and performed the experiments. FY, LW, and JP made substantial contributions to conception and design. RZ, LC, and BK revised the manuscript critically for important intellectual content. LH and LC participated experimental design and data analysis. FY drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported in part by National Institutes of Health (NIH) Grant R01AI090155 (to BK) and R21AI117338 (to LC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents to FY.
References
- Bialek-Davenet S., Criscuolo A., Ailloud F., Passet V., Jones L., Delannoy-Vieillard A. S., et al. (2014). Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 20, 1812–1820. 10.3201/eid2011.140206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candan E. D., Aksoz N. (2015). Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim. Polonica 62, 867–874. 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- Cheng D. L., Liu Y. C., Yen M. Y., Liu C. Y., Wang R. S. (1991). Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch. Intern. Med. 151, 1557–1559. 10.1001/archinte.1991.00400080059010 [DOI] [PubMed] [Google Scholar]
- Chuang Y. P., Fang C. T., Lai S. Y., Chang S. C., Wang J. T. (2006). Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193, 645–654. 10.1086/499968 [DOI] [PubMed] [Google Scholar]
- Chung D. R., Lee H. R., Lee S. S., Kim S. W., Chang H. H., Jung S. I., et al. (2008). Evidence for clonal dissemination of the serotype K1 Klebsiella pneumoniae strain causing invasive liver abscesses in Korea. J. Clin. Microbiol. 46, 4061–4063. 10.1128/JCM.01577-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing, 26th Informational Supplement (M100-S26). Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Diancourt L., Passet V., Verhoef J., Grimont P. A., Brisse S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C. T., Chuang Y. P., Shun C. T., Chang S. C., Wang J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. 10.1084/jem.20030857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C. T., Lai S. Y., Yi W. C., Hsueh P. R., Liu K. L., Chang S. C. (2007). Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45, 284–293. 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- Fang F. C., Sandler N., Libby S. J. (2005). Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 43, 991–992. 10.1128/JCM.43.2.991-992.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Ni Y., Jiang Y., Yuan F., Han L., Li M., et al. (2005). Outbreak of infection caused by Enterobacter cloacae producing the novel VEB-3 beta-lactamase in China. J. Clin. Microb. 43, 826–831. 10.1128/JCM.43.2.826-831.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhang J., Liu Y., Zheng R., Chen H., Wang X., et al. (2014). Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn. Microbiol. Infect. Dis. 78, 63–65. 10.1016/j.diagmicrobio.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Li W., Sun G., Yu Y., Li N., Chen M., Jin R., et al. (2014). Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58, 225–232. 10.1093/cid/cit675 [DOI] [PubMed] [Google Scholar]
- Liao C. H., Huang Y. T., Chang C. Y., Hsu H. S., Hsueh P. R. (2014). Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur. J. Clin. Microbiol. Infect. Dis. 33, 365–369. 10.1007/s10096-013-1964-z [DOI] [PubMed] [Google Scholar]
- Lin Y. T., Jeng Y. Y., Chen T. L., Fung C. P. (2010). Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: clinical and microbiological characteristics in Taiwan, 2001-2008. BMC infect. Dis. 10:307. 10.1186/1471-2334-10-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. T., Wang Y. P., Wang F. D., Fung C. P. (2015). Community-onset Klebsiella pneumoniae pneumonia in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front. Microbiol. 9:122. 10.3389/fmicb.2015.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. C., Cheng D. L., Lin C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Int. Med. 146, 1913–1916. 10.1001/archinte.1986.00360220057011 [DOI] [PubMed] [Google Scholar]
- Liu Y. M., Li B. B., Zhang Y. Y., Zhang W., Shen H., Li H., et al. (2014). Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob. Agents Chemother. 58, 5379–5385. 10.1128/AAC.02523-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L., Dell'Amico E., Migliavacca R., D'Andrea M. M., Giacobone E., Amicosante G., et al. (2003). Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy J. Clin. Microb. 41, 4264–4269. 10.1128/JCM.41.9.4264-4269.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokesch B. C., TeKippe M., Kim J., Raj P., TeKippe E. M., Greenberg D. E. (2016). Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 16, e190–e195. 10.1016/S1473-3099(16)30021-4 [DOI] [PubMed] [Google Scholar]
- Qi Y., Wei Z., Ji S., Du X., Shen P., Yu Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- Qu T. T., Zhou J. C., Jiang Y., Shi K. R., Li B., Shen P., et al. (2015). Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15:161. 10.1186/s12879-015-0899-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Olson R., Macdonald U., Metzger D., Maltese L. M., Drake E. J., et al. (2014). Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82, 2356–2367. 10.1128/IAI.01667-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon A. S., Bajwa R. P., Russo T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu L. K., Yeh K. M., Lin J. C., Fung C. P., Chang F. Y. (2012). Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887. 10.1016/S1473-3099(12)70205-0 [DOI] [PubMed] [Google Scholar]
- Struve C., Roe C. C., Stegger M., Stahlhut S. G., Hansen D. S., Engelthaler D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 6:e00630. 10.1128/mBio.00630-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microb. 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J. F., Englender H., Gabriel S. N., Turton S. E., Kaufmann M. E., Pitt T. L. (2007). Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J. Med. Microbiol. 56(Pt 5), 593–597. 10.1099/jmm.0.46964-0 [DOI] [PubMed] [Google Scholar]
- Turton J. F., Perry C., Elgohari S., Hampton C. V. (2010). PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59(Pt 5), 541–547. 10.1099/jmm.0.015198-0 [DOI] [PubMed] [Google Scholar]
- Validi M., Soltan Dallal M. M., Douraghi M., Fallah Mehrabadi J., Rahimi Foroushani A. (2016). Identification of Klebsiella pneumoniae Carbapenemase-producing Klebsiella oxytoca in Clinical Isolates in Tehran Hospitals, Iran by Chromogenic Medium and Molecular Methods. Osong Public Health Res. Perspect. 7, 301–306. 10.1016/j.phrp.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H., Liu Y. C., Lee S. S., Yen M. Y., Chen Y. S., Wang J. H., et al. (1998). Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438. [DOI] [PubMed] [Google Scholar]
- Yan J. J., Zheng P. X., Wang M. C., Tsai S. H., Wang L. R., Wu J. J. (2015). Allocation of Klebsiella pneumoniae Bloodstream Isolates into four distinct groups by ompK36 typing in a Taiwanese University Hospital. J. Clin. Microb. 53, 3256–3263. 10.1128/JCM.01152-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Zhou M., Zou M., Liu W. E. (2016). Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 387–396. 10.1007/s10096-015-2551-2 [DOI] [PubMed] [Google Scholar]
- Yang Z., Liu W., Cui Q., Niu W., Li H., Zhao X., et al. (2014). Prevalence and detection of Stenotrophomonas maltophilia carrying metallo-beta-lactamase blaL1 in Beijing, China. Front. Microbiol. 5:692. 10.3389/fmicb.2014.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Xiao X., Wang F., Zhou L., Zhang X., Zhang J. (2015). Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. 10.1016/j.ijid.2015.06.023 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Ko W. C., Cheng K. C., Lee C. C., Lai C. C., Chuang Y. C. (2008). Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62, 1–6. 10.1016/j.diagmicrobio.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Ko W. C., Cheng K. C., Lee H. C., Ke D. S., Lee C. C., et al. (2006). Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 42, 1351–1358. 10.1086/503420 [DOI] [PubMed] [Google Scholar]
- Zhang R., Lin D., Chan E. W., Gu D., Chen G. X., Chen S. (2015). Emergence of Carbapenem-Resistant Serotype K1 Hypervirulent Klebsiella pneumoniae strains in China. Antimicrob. Agents Chemother. 60, 709–711. 10.1128/AAC.02173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng J., Liu W., Zhao F., Hu Z., Zhao C., et al. (2015). Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Infect. 71, 553–560. 10.1016/j.jinf.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao C., Wang Q., Wang X., Chen H., Li H., et al. (2016). High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–620. 10.1128/AAC.01127-16 [DOI] [PMC free article] [PubMed] [Google Scholar]