Abstract
Purpose: Previously, we have shown that the SNP rs10932201 genotype of the cyclic AMP responsive element binding protein 1 gene (CREB1) contributes to individual differences in executive and memory function at the neural system and behavioral levels in healthy, young adults. However, longitudinal effects of CREB1 genotypes on cognition have not yet been addressed. Furthermore we were interested in replicating associations between CREB1 genotypes and human cognition in previous cross-sectional studies and explore whether APOE𝜀4 status might modify these relations.
Materials and Methods: We investigated whether common, independent tag SNPs within CREB1 (rs2253206, rs10932201, rs6785) influence individual differences in age-related longitudinal change and level of executive function and memory performance independent of baseline age, sex, APOE𝜀4 status, and education. Our analysis included data from cognitively unimpaired older adults participating in the Baltimore Longitudinal Study of Aging. Eleven measures from six cognitive tests (sample sizes range 617–786) were analyzed using linear mixed effects and generalized estimating equations models. Mean baseline age ranged from 50 to 69 years and mean time of follow-up (interval) ranged from 8 to 22 years.
Results: We found significant effects of all three CREB1 SNPs on performance level and/or longitudinal change in performance based on eight measures assessing semantic memory, episodic memory, or both executive function and semantic memory. SNP rs10932201 showed the most significant and largest effect (Cohen’s d = -0.70, p < 0.01) on age-related longitudinal decline of semantic memory. Additionally, we show interactions between all three CREB1 SNPs and APOE𝜀4 status on age-related longitudinal declines and levels of memory and executive function.
Conclusion: Our results suggest that CREB1 genotypes independently and by interactions with APOE𝜀4 status contribute to individual differences in cognitive aging.
Keywords: cognitive aging, memory, executive function, CREB1 genotypes, longitudinal study
Introduction
Human cognitive aging is highly variable across individuals (Lindenberger, 2014). Identifying genetic factors that contribute to interindividual variability in performance and age-related decline of specific cognitive functions can inform us about the underlying neuromolecular networks. Genetic variations in the CREB1 gene (HGNC: 2345) are of interest because CREB signaling pathways, by regulating gene expression, contribute to the adaptation of neuronal properties in brain regions important for memory and executive functions (Benito and Barco, 2010; Rapanelli et al., 2010; Pozzi et al., 2011; Sanchez-Huertas and Rico, 2011; Leckie et al., 2014). Memory and executive function are susceptible to age-related decline and early neurodegeneration (Ronnlund et al., 2005; Grober et al., 2008; Goh et al., 2012; Weintraub et al., 2012). Age-related changes in CREB signaling pathways have been reported in animal models, especially in relation to deficits in long-term and working memory, and brain regions such as the hippocampus and prelimbic prefrontal cortex (Brightwell et al., 2004; Kudo et al., 2005; Monti et al., 2005; Vandesquille et al., 2013). In young and aged adult mice, following inhibition the reactivation of CREB signaling pathways in the hippocampus has been consistently shown to increase CREB-dependent transcription and rescue deficient neuroplasticity and memory (Genoux et al., 2002; Smith et al., 2009; Yiu et al., 2011; Adlard et al., 2014). Similarly the aging-related decrease in long-term memory found in the control rats could be prevented by CREB gene transfer in the hippocampus (Mouravlev et al., 2006). Recent evidence also supports CREB activation levels as a potential biomarker for cognitive decline in Alzheimer’s disease (Bartolotti et al., 2016).
The APOE𝜀4 genotype has been related to lower memory and greater decline in cognitive performance or memory in cognitively unimpaired participants (Yaffe et al., 1997; Bretsky et al., 2003; Caselli et al., 2004, 2009, 2011; Packard et al., 2007; Beydoun et al., 2012; Schiepers et al., 2012; Samieri et al., 2014), which could be dependent on CREB signaling pathways (Chen et al., 2010; Qiao et al., 2014; Liu et al., 2015). For example, contrary to APOE3, APOE4 stimulates the transcriptional activity of CREB by activating the extracellular signal-regulated kinase (ERK) pathway in hippocampus neurons (Ohkubo et al., 2001). Accordingly, CREB may be important in elucidating pathways related to individual differences in cognitive decline.
Previously we reported cross-sectional effects of the CREB1 SNP rs10932201 genotype on executive and memory function at the neural system and behavioral levels in healthy, young adults (Wolf et al., 2015). Longitudinal effects of CREB1 genotypes on cognition have not yet been addressed. Here, we investigate whether common variants within the CREB1 gene, after adjustment for baseline age, sex, APOE𝜀4 status, and education, influence the performance level and longitudinal change in performance of executive and memory function during aging. Furthermore we address whether APOE𝜀4 status and CREB1 genotypes interactively influence cognitive aging.
Materials and Methods
Participants
Participants of the Baltimore Longitudinal Study of Aging (BLSA) are community-dwelling volunteers. Continuous recruitment into the BLSA has occurred since 1958. Exclusion criteria at enrollment were past or present psychiatric, neurological, cardiovascular, severe gastrointestinal, kidney or liver disease, diabetes, birth defects, established genetic diseases, inability to perform daily self-care without assistance, inability to walk independently for at least 400 m without assistance and without developing symptoms, inability to perform normal activities of daily living without shortness of breath, active cancer (any activity in the last 10 years), except for locally limited basal cell cancer, clinically significant hormonal dysfunction, muscle-skeletal conditions due to diseases or traumas, any long-term medical treatment, important sensory deficits, having severe English language difficulties, physical illness and weight over 136 kg. Specifically for our analysis we restricted the sample to Caucasian Americans with data available for the CREB1 SNPs of interest, APOE𝜀4 genotype status and education. Cognitive status of participants was assessed at every visit. Clinical consensus case conferences were performed if the Blessed Information Memory Concentration score was ≥4 (Fuld, 1978), the Clinical Dementia Rating score was ≥0.5 (Morris, 1993), or concerns were raised about a participant’s cognitive status, and upon death in case participants were in the autopsy study. Based on clinical and neuropsychological data, cognitive impairment and timing of onset were determined. Data after the onset of cognitive impairment for individuals who subsequently developed impairment were excluded from the current analyses.
The number of participants, time of follow-up and demographics varied depending on the cognitive measure (Supplementary Table S1). This study was carried out in accordance with the recommendations of NIH guidelines, the institutional review board National Institute of Environmental Health Sciences with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki at each study visit. The protocol was approved by the institutional review board National Institute of Environmental Health Sciences.
Acquisition of Cognitive Data
Data were collected for comprehensive medical, physiological, and neuropsychological evaluations approximately every 2 years and since 2000 every year for participants older than 80 years. We selected six cognitive tests that have previously been shown to be sensitive to individual differences in primarily memory (Golski et al., 1998; Balthazar et al., 2008; Goh et al., 2012) or both memory and executive function (Paula et al., 2013; Shao et al., 2014) during healthy cognitive aging. These tests included the Boston Naming Test (BNT), the Benton Visual Retention Test (BVRT), California Verbal Learning Test (CVLT), the Clock Drawing Test (CLOCKs), the Category and Letter Fluency Tests. The data used in our analysis was acquired for the BVRT between 1960 and 2014, for the CVLT between 1993 and 2014, and for the remaining three tests between 1984 and 2014. Until 2005 the CLOCKs and Fluency Tests were only obtained from participants with age ≥60 years instead of at every visit. The specific measures used from each cognitive test, the range for each measure and a summary of the cognitive processes contributing to each measure are shown in Supplementary Table S2.
Detailed Description of Cognitive Tests
The BVRT measures visual constructional skill and short-term figural memory. Participants study 10 line drawings including one to three geometric figures for 10 s each, and then immediately reproduce them from memory using pencil and paper. The designs become more difficult over the 10 trials. The BVRT has been administered to BLSA participants since 1960. The dependent variable was the total number of errors, which were scored independently by two trained technicians, employing a modified error scoring method, based on the method provided in the BVRT manual (Benton, 1974).
The BNT (Kaplan et al., 1983) is a measure of object recognition and semantic retrieval. Participants identify and name a series of line drawings of objects, beginning with common objects and ending with infrequent objects. Administration in the BLSA involves cueing incorrectly named objects, with either a stimulus cue for perceptual errors or a phonemic cue for semantic errors. This test has been administered to BLSA participants 70 years and older since 1986 and to BLSA participants 60 years and older since 1990. The dependent measure was the number of words out of 60 correctly named without cues.
The CVLT assesses verbal learning and memory (Delis et al., 1987). Five learning trials of 16 shopping items, with four items from each of four semantic categories are presented orally and the sum of the five trials provides a measure of immediate free recall. In addition, short- and long-delayed free recall, short- and long-delayed cued recall, and recognition memory are assessed. The CVLT has been administered to BLSA participants since 1993. Four dependent measures were used in the present analyses: total number of items recalled across the five immediate recall learning trials, short-delay free recall, long-delay free recall, and recognition memory with maximum scores of 80, 16, 16, and 16, respectively.
The Clock Drawing Test (CLOCK) is a test of constructional and visuospatial ability (Rouleau et al., 1992). Three clocks were drawn; two from memory (3:25, 11:10) and one was copied (11:10). Participants were asked to draw a clock, put in all of the numbers and set the hands to the specified time. The Clock Drawing task has been administered to BLSA participants 70 years and older since 1986 and to BLSA participants 60 years and older since 1990. The clock face was given up to 2 points, and the numbers and hands are given up to 4 points, for a total of 10 points.
Letter and category fluency (Fluencies) tests are measures of fluent language production and executive function. Participants were given 60 s to generate as many words as possible beginning with specific letters (F, A, S) (Benton, 1968) and from specific categories (fruits, animals, vegetables) (Newcombe, 1969). Fluency measures have been administered to BLSA participants 70 years and older since 1986, to BLSA participants 60 years and older since 1990 and to all participants since 2005. The total numbers of correct words generated in 60 s, across the three trials of letter and category fluency, were the dependent measures of interest.
Measuring Symptoms of Depression
We used the Center for Epidemiological Studies Depression (CESD) scale (Radloff, 1977) to screen for symptoms of depression. Based on our largest sample (N = 786), this score was unavailable for 50 participants (6.4%) partly due to the fact that the CESD scale was unavailable before 1977.
Genotyping
In BLSA, blood samples were collected for DNA extraction, and genome-wide genotyping was completed for 1231 subjects using Illumina 550K. Genotype data were available from 848 participants of European ancestry using a call rate of >98.5% without sex discrepancy based on homozygosity rates. 501,704 autosomal SNPs passed quality control (completeness > 99%, MAF > 1%, HWE > 10-4) were used for imputation (Price et al., 2006). Imputation of ∼2.5 million HapMap SNPs were imputed using CEU sample (Phase II, release 22, build 36) as a reference using MACH (Tanaka et al., 2009). Based on genotyping and imputing, genotype data for eight SNPs within the CREB1 gene (Supplementary Table S3a) were available for those participants with cognitive data.
APOE𝜀4 genotype status was determined by polymerase chain reaction amplification of leukocyte DNA followed by HhaI digestion and product characterization (Hixson and Vernier, 1990) and by the TaqMan assay systems based on several single nucleotide polymorphisms located in the ApoE gene (Koch et al., 2002).
Statistical Analysis of Genetic and Cognitive Data
We used the SPSS (IBM SPSS Statistics 23) and R (version 3.2.2) for Windows for our statistical analysis of genetic and behavioral data.
Bivariate Pearson’s correlation coefficients (two-tailed) were used to test pair-wise LD between SNPs. The cutoff for LD was set to an R2 value of 0.6. Chi-squared tests were used to check HWE [α-level 0.05; 2 degrees of freedom (df)] and differences in genotype groups between sex (α-level 0.05; 2 df). ANOVAs were used to analyze differences in age, education (α-level 0.05; two-tailed; 783 df) and score for symptoms of depression (733 df) between SNP-based genotype groups. Only SNPs rs2253206, rs10932201, and rs6785 identified as independent were further analyzed because they were not in high LD defined as an R2 of ≥0.6.
Calculation of Cognitive Performance Measures
Performance measures were based on the total sum of correct responses for the BNT, CLOCKs, CVLT free recall measures, Category and Letter Fluency and the total sum of errors for the BVRT. The CVLT-recognition correct was the sum of all correctly recognized targets (total hits). The CVLT-recognition discriminability is another performance accuracy measure, which was calculated as Z-score for the probability of hit [Z(p(hit))] minus Zscore for the probability of false alarm/incorrectly recognized as target [Z(p(FA))]. The CVLT-recognition response bias measures the degree of performance bias, which was calculated as Z-score for the probability of total hits plus Z-score for the probability of false alarms [-0.5∗Z(p(hit)) + Z(p(FA))]. Values closer to zero indicate less biased performance for the CVLT-recognition response bias measure, a value of zero indicates a completely unbiased or neutral response. Because of missing information on FA, the CVLT-recognition response bias and discriminability measures could not be calculated for one visit of a subject with multiple visits. Scores with zero total hits were excluded for nine visits from subjects with more than one visit and one subject with a single visit.
These 11 cognitive measures were used as dependent variables in our models. Baseline age, education, and time of follow-up (interval) were continuous predictors, and sex, APOE𝜀4 status, and SNP were nominal predictors. There were 15 APOE𝜀4/4 carriers in our largest sample, which unfortunately is insufficient to carry out a subgroup specific analysis. Therefore APOE𝜀4 status was defined as APOE𝜀4 positive for participants with at least one APOE𝜀4 allele, while other participants were defined as APOE𝜀4 negative. For the CREB1 SNPs, the genotype group homozygous for the global major allele was used as the reference genotype group (rs2253206 → AA, rs10932201 → GG, and rs6785 → GG). We modeled the genotype effect using either all three genotype groups or two groups by combining heterozygous and homozygous genotype groups for the global minor allele versus the genotype group homozygous for the global major allele (reference group).
We used generalized estimating equations (GEE) models with Poisson distribution for CLOCK based measures after their transformation (maximal possible score minus observed score) to model the counts. Accordingly, originally high performance scores were turned into low performance scores. These transformed scores were back-transformed for the graphical presentations. Linear mixed effects models (LME) with [unstructured covariance for the random effects] were used for all other cognitive measures. We included as fixed effects baseline age, sex, APOE𝜀4 status and education, time of follow-up (interval), SNP and interactions of interest: baseline age × interval, sex × interval, APOE𝜀4 status × interval, and SNP × interval, into both the LME and GEE models. Intercept and interval for each subject were also included as random effects with the unstructured covariance in the LME models, and exchangeable covariance was used for repeated measures in the GEE models. We mean centered baseline age, sex, APOE𝜀4 status and education for simultaneous interpretation of main effects and interactions in the models. The main effect of interval is the slope for the mean of baseline age, sex, APOE𝜀4 status, and education for the SNP genotype reference group. Using these models allowed us to analyze cross-sectional effects (baseline age, sex, APOE𝜀4 status, education and each SNP) as well as interactions of interest with the longitudinal effect of interval simultaneously.
We used Cohen’s d effect size definition implemented in LME models, where the group difference is estimated based on the beta coefficient from LME regression models and standard deviations of the data are estimated from the covariance structure of the random effects. For the GEE models, Cohen’s d effect size was calculated by using the estimates of variance-covariance matrix from the random effects in generalized linear mixed models.
We also tested for putative interactions between the CREB1 SNPs and APOE𝜀4 status by adding to all our models the interaction between SNP and APOE𝜀4 status as well as the interaction between SNP, APOE𝜀4 status and interval. For those models showing significant interactions, we used post hoc test to identify group differences that underlie the overall effect.
Results
CREB1 Genotypes
Allele frequencies for eight SNPs within the CREB1 gene are shown in Supplementary Table S3a. Based on MAF < 0.05 in our largest sample (N = 786) SNP rs16839883 (MAF = 0.01) and rs2709393 (MAF = 0.02) were excluded from further analysis. Results of pairwise linkage disequilibrium analysis (Supplementary Table S3b) revealed strong linkage between rs2253206 and rs2254137 (R2 = 0.6) as well as rs2551928, rs1045780, and rs6785. Therefore we selected rs2253206, rs10932201, and rs6785 (range of R2 = 0.1–0.2) for further analysis as independent tag SNPs. None of the CREB1 SNPs were in linkage disequilibrium with APOE𝜀4 genotype status (all R2 = 0). All three independent SNP genotypes were distributed according to HWE (Supplementary Table S3c) and were not significantly different by sex, baseline age, education or symptoms of depression score (Supplementary Table S3d).
Sensitivity of Cognitive Measures to Aging and Baseline Age
We observed significant longitudinal declines (effect of interval) in performance for all our measures of memory and executive function as well as lower cross-sectional performance levels and greater longitudinal declines in performance with increased baseline age for most of these measures (Supplementary Table S4).
Cross-Sectional and Longitudinal Effects of CREB1 Genotypes on Individual Differences in Memory and Executive Function
Performance of executive function and semantic memory was significantly affected by SNP rs2253206, showing cross-sectional effects for both CLOCK-based measures and a longitudinal effect for the CLOCK-11:10-based measure (Figures 1A,B and Supplementary Tables S5, S6). The AA genotype was associated with beneficial effects on both performance level and change in executive function and semantic memory, while the GG genotype was associated with lower performance level and the GA genotype with greater decline in performance. Significantly less longitudinal decline in performance of executive function and semantic memory measured by Category Fluency was observed in the genotype group AA compared to the group with GG/GA genotype for SNP rs2553206 (Figure 1C and Supplementary Table S7). None of the SNPs showed a significant effect on letter fluency (Supplementary Table S8). In addition to the significant effects of SNP rs2253206, at trend level SNP rs6785 also affected performance level and SNP rs10932201 influenced the decline in executive function and semantic memory (Supplementary Table S5).
FIGURE 1.
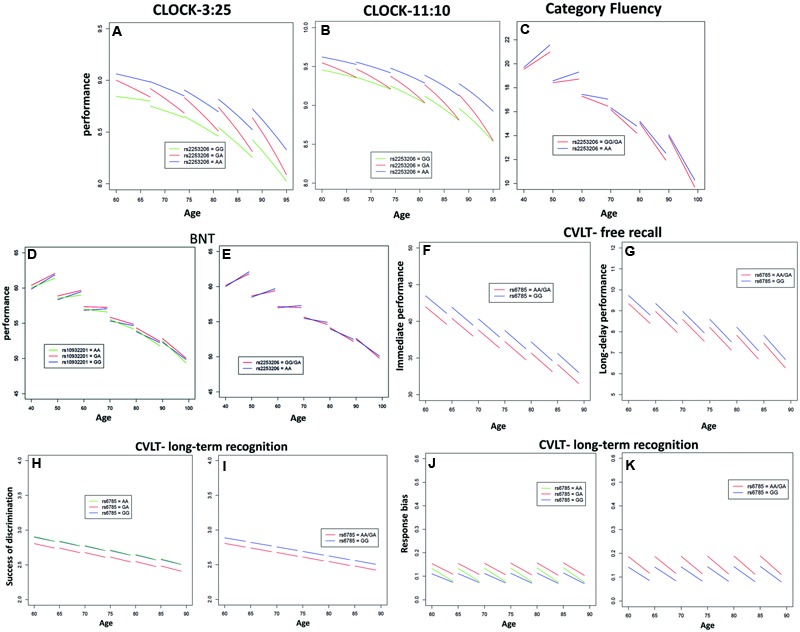
Significant effects of CREB1 genotypes on the longitudinal population-average trajectories for performance measures of executive function and/or memory (A) CLOCK-3:25, (B) CLOCK-11:10, (C) Category Fluency, (D,E) BNT, (F) California Verbal Learning Test (CVLT)-immediate free recall, (G) CVLT-long-delay free recall, (H,I) CVLT-recognition discriminability, and (J,K) CVLT-recognition response bias based on the predicted values after adjusting for baseline age, sex, education, and APOE𝜀4 status. All the effects were estimated using linear mixed effects (LME) models except for CLOCK measures where generalized estimating equations (GEE) models were used.
Genotype groups AA or GA compared to GG for SNP rs10932201 (Figure 1D and Supplementary Table S9) and GG/GA compared to AA for SNP rs2253206 (Figure 1E and Supplementary Table S9) showed significantly greater longitudinal decline in performance of semantic memory and language measured by BNT.
The performance level of verbal episodic memory during immediate as well as delayed free recall measured by CVLT was significantly lower in genotype group AA/GA compared to GG for SNP rs6785 (Figures 1F,G and Supplementary Tables S11, S12). We found also a significant main effect of this SNP on the performance of episodic long-term recognition memory measured by CVLT-recognition discriminability, which reflects the success of discriminating target from non-target items (Figures 1H,I and Supplementary Table S14). Again, group AA/GA compared to GG performed at a lower level for this cognitive measure. The performance level of episodic long-term recognition memory measured by CVLT-recognition response bias was significantly lower indicated by greater response bias (further away from zero) in genotype group AA/GA compared to GG for SNP rs6785 (Figures 1J,K and Supplementary Table S15). At trend level cross-sectional performance of episodic short-term memory was also influenced by SNP rs6785 (Supplementary Table S13). Trends for cross-sectional and longitudinal effects of SNP rs10932201 were also observed on visual immediate episodic memory (Supplementary Table S10). SNP rs2253206 showed an effect on the performance level of verbal immediate episodic memory at trend level (Supplementary Table S11). Please see Table 1 for an overview of CREB1 SNPs significantly affecting cognitive measures.
Table 1.
Cross-sectional and longitudinal effects of CREB1 genotypes on cognitive measures controlling for baseline age, sex, APOE𝜀4 status, and education.
| Cognitive functions | Cognitive measure | SNP and grouping (group size) | Estimate SNP main effect (Cohen’s d) | p-value SNP main effect | Estimate interaction SNPx interval (Cohen’s d) | p-value interaction SNPx interval |
|---|---|---|---|---|---|---|
| Executive function and semantic memory | CLOCK-3:25 trans-formed | rs2253206 overall | 0.018∗ | 0.053 | ||
| GG (193) vs. AA (129) | 0.209 (0.50) | 0.015∗ | -0.006 (-0.88) | 0.569 | ||
| GG vs. GA (312) | -0.145 (-0.35) | 0.022∗ | 0.016 (0.36) | 0.016∗ | ||
| GA vs. AA | 0.064 (0.15) | 0.457 | 0.010 (1.47) | 0.334 | ||
| CLOCK-11:10 trans-formed | rs2253206 overall | 0.031∗ | 0.029∗ | |||
| GG vs. AA | 0.364 (0.36) | 0.013∗ | -0.008 (-0.23) | 0.529 | ||
| GG vs. GA | -0.182 (-0.18) | 0.089 | 0.025 (0.72) | 0.010∗ | ||
| GA vs AA | 0.182 (0.18) | 0.222 | 0.017 (0.49) | 0.147 | ||
| Category Fluency | rs2253206 | -0.146 (-0.05) | 0.583 | -0.050 (-0.35) | 0.025∗ | |
| GG/GA (N = 575) vs AA (150) | ||||||
| Semantic memory and language | BNT | rs10932201 overall | 0.226 | 0.004∗∗ | ||
| AA (138) vs. GG (180) | 0.191 (0.05) | 0.672 | -0.069 (-0.70) | 0.013∗ | ||
| AA vs. GA (303) | -0.353 (-0.10) | 0.322 | 0.003 (0.03) | 0.894 | ||
| GA vs. GG | 0.544 (0.15) | 0.096 | -0.072 (-0.70) | 0.002∗∗ | ||
| rs10932201 | 0.434 (0.12) | 0.158 | -0.071 (-0.70) | 0.001∗∗ | ||
| AA/GA vs. GG | ||||||
| rs2253206 | 0.172 (0.05) | 0.346 | -0.054 (-0.50) | 0.039∗ | ||
| GG/GA (494) vs. AA (127) | ||||||
| Episodic memory | CVLT-immediate free recall | rs6785 | -1.512 (-0.16) | 0.021∗ | 0.003 (-0.01) | 0.960 |
| AA/GA (265) vs. GG (468) | ||||||
| CVLT-long-delay free recall | rs6785 | -0.384 (-0.15) | 0.045∗ | -0.004 (-0.03) | 0.809 | |
| AA/GA vs. GG | ||||||
| CVLT-recognition discriminability | rs6785 overall | 0.038∗ | 0.872 | |||
| AA (32) vs. GG (468) | 0.020 (0.05) | 0.825 | -0.004 (-0.21) | 0.601 | ||
| AA vs. GA (233) | 0.116 (0.26) | 0.206 | -0.004 (-0.21) | 0.624 | ||
| GA vs. GG | -0.961 (-0.22) | 0.013∗ | -0.000 (-0.01) | 0.967 | ||
| rs6785 | -0.082 (-0.18) | 0.026∗ | -0.006 (-0.32) | 0.862 | ||
| AA/GA vs. GG | ||||||
| CVLT-recognition response bias | rs6785 overall | 0.021∗ | 0.182 | |||
| AA vs. GG | 0.013 (0.08) | 0.743 | -0.003 (-0.30) | 0.480 | ||
| AA vs. GA | -0.034 (-0.22) | 0.393 | 0.000 (0.02) | 0.967 | ||
| GA vs. GG | 0.047 (0.30) | 0.005∗∗ | -0.003 (-0.30) | 0.074 | ||
| rs6785 | 0.042 (0.27) | 0.008∗∗ | -0.003 (-0.30) | 0.064 | ||
| AA/GA vs. GG |
The reference genotype group is underlined. Significant effects are shown in bold with ∗ indicating significance at p = 0.05 and ∗∗ at p = 0.01.
APOE𝜀4 Status-Dependent Longitudinal CREB1 Genotype Effects on Individual Differences in Executive Function and Semantic Memory
We observed a significant interaction (p < 0.01) between rs2253206 genotype and APOE𝜀4 status for the decline in executive function and semantic memory performance, measured by CLOCK-3:25 (Table 2 and Figure 2A). Following-up this overall interaction effect with post hoc tests (Supplementary Table S16), we found significantly greater longitudinal decline in participants with the genotype AA and APOE𝜀4 positive compared to negative status. There were no differences in the longitudinal declines for the GG/GA genotype group by APOE𝜀4 status. For the same measure of executive function and semantic memory, we also observed that longitudinal decline in performance was significantly (p < 0.05) affected by the interaction between SNP rs6785 and APOE𝜀4 status (Table 2 and Figure 2B). According to the post hoc tests (Supplementary Table S16), the effect of APOE𝜀4 status on longitudinal decline in performance was not significant in participants with either GG or AA/GA genotype.
Table 2.
Interactions between interval, CREB1 genotypes and APOE𝜀4 status significantly influence decline in executive function and semantic memory controlling for baseline age, sex, and education.
| SNP and grouping | Cognitive measure | Estimate/s, p-value, Cohen’s d SNP main effect | Estimate/s, p-value, Cohen’s d SNPx interval interaction | Estimate, p-value, Cohen’s d APOE𝜀4 main effect | Estimate, p-value, Cohen’s d APOE𝜀4x interval interaction | Estimate/s, p-value, Cohen’s d SNPx APOE𝜀4 interaction | Estimate/s, p-value, Cohen’s d interval SNPx APOE𝜀4 interaction |
|---|---|---|---|---|---|---|---|
| rs2253206 GG/GA vs AA | CLOCK-3:25 transformed | 0.131 | 0.003 | -0.223 | 0.054 | 0.263 | -0.064 |
| 0.109 | 0.716 | 0.241 | 0.003∗∗ | 0.194 | 0.001∗∗ | ||
| 0.20 | 0.22 | -0.34 | 1.19 | 0.41 | -1.45 | ||
| rs6785 AA/GA vs GG | CLOCK-3:25 transformed | 0.111 | -0.007 | -0.029 | 0.012 | 0.052 | -0.028 |
| 0.079 | 0.275 | 0.969 | 0.787 | 0.717 | 0.045∗ | ||
| 0.22 | -0.20 | -0.04 | 0.29 | 0.09 | -0.76 |
Significant effects are shown in bold with ∗ indicating significance at p = 0.05 and ∗∗ at p = 0.01. The reference genotype group is underlined.
FIGURE 2.
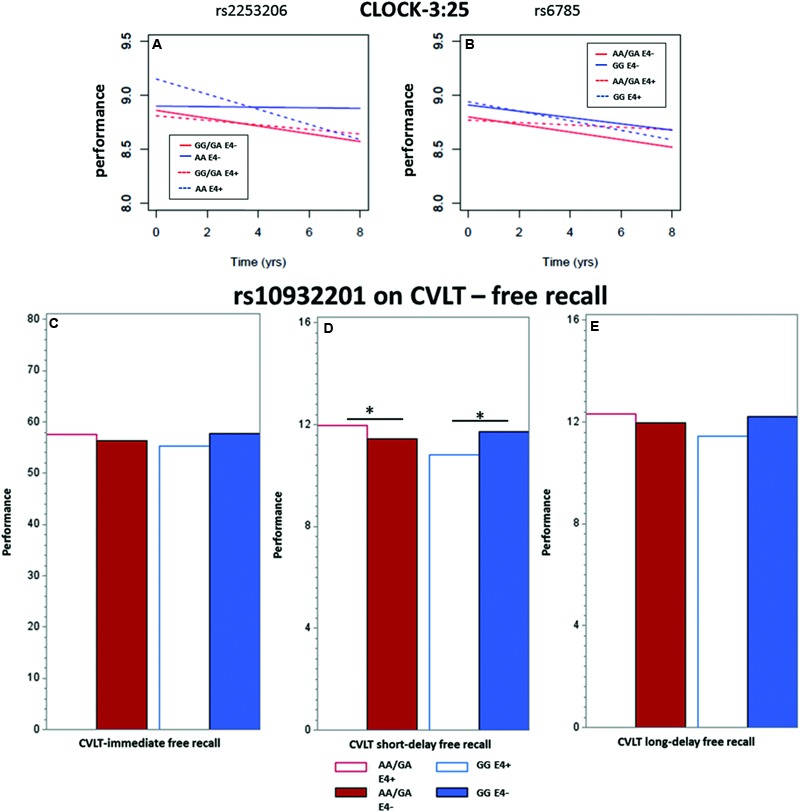
Showing the significant interactions between CREB1 SNP genotypes and APOE𝜀4 status on the longitudinal population-average trajectories for executive function and memory performance (A,B) CLOCK-3:25 based on the predicted values after adjusting for baseline age, sex and education, and on population-average cross-sectional memory performance (C) immediate recall, (D) short-term and (E) long-term episodic memory based on the predicted values after adjusting for baseline age, sex, and education. All the effects were estimated using LME models except for CLOCK measures where GEE models were used. ∗ indicates significance at p = 0.05.
APOE𝜀4 Status-Dependent Cross-Sectional CREB1 Genotype Effects of SNP rs10932201 on Individual Differences in Episodic Memory
The interaction between the SNP rs10932201 genotype and APOE𝜀4 status significantly affected the performance level of immediate recall, long-delay (both p < 0.05) and short-delay (p < 0.01) episodic memory (Table 3 and Figures 2C–E). Post hoc testing (Supplementary Table S16) revealed that the level of performance was significantly lower in genotype group GG comparing APOE𝜀4 positive with negative status for short-term episodic memory (p < 0.05) and there was a tendency for similar effects for immediate and long-delay recall. In the other genotype group AA/GA, APOE𝜀4 positive compared to negative status was associated with significantly (p < 0.05) higher performance level for short-term episodic memory. No significant effects of APOE𝜀4 status were seen on performance levels for either immediate or long-term episodic memory in this genotype group.
Table 3.
Interactions between CREB1 genotype and APOE𝜀4 status significantly influence episodic memory controlling for baseline age, sex, and education.
| SNP and grouping | Cognitive measure | Estimate/s, p-value, Cohen’s d SNP main effect | Estimate/s, p-value, Cohen’s d SNPx interval interaction | Estimate, p-value, Cohen’s d APOE𝜀4 main effect | Estimate, p-value, Cohen’s d APOE𝜀4x interval interaction | Estimate/s, p-value, Cohen’s d SNPx APOE𝜀4 interaction | Estimate/s, p-value, Cohen’s d interval SNPx APOE𝜀4 interaction |
|---|---|---|---|---|---|---|---|
| rs10932201 AA/GA vs GG | CVLT-immediate free recall | -0.145 | -0.057 | -2.486 | 0.010 | 3.564 | -0.214 |
| 0.83 | 0.438 | 0.388 | 0.251 | 0.029∗ | 0.201 | ||
| -0.02 | -0.11 | -0.27 | 0.02 | 0.38 | -0.35 | ||
| CVLT-short-delay free recall | 0.194 | -0.017 | -0.899 | 0.004 | 1.428 | -0.070 | |
| 0.365 | 0.392 | 0.453 | 0.168 | 0.004∗∗ | 0.113 | ||
| 0.07 | -0.14 | -0.34 | 0.03 | 0.54 | -0.49 | ||
| CVLT-long-delay free recall | 0.133 | -0.023 | -0.753 | -0.014 | 1.094 | -0.025 | |
| 0.522 | 0.198 | 0.389 | 0.204 | 0.022∗ | 0.555 | ||
| 0.05 | -0.14 | -0.29 | 0.07 | 0.43 | -0.21 |
The reference genotype group is underlined. Significant effects are shown in bold with ∗ indicating significance at p = 0.05 and ∗∗ at p = 0.01.
Discussion
In this study, we examined whether three independent CREB1 SNP genotypes contribute to individual differences in the performance level and longitudinal change of memory and executive function in older adults free of cognitive impairment. Consistently across our various measures the minor allele AA genotype for SNP rs2253206 and major allele GG genotype for SNP rs10932201 and rs6785 were associated with beneficial performance effects. The associations observed were significant adjusting for the already established risk factors such as age, sex, APOE𝜀4 status, and education. Our findings support a contribution of genetic variability in CREB1 to individual differences in cognitive measures tapping primarily immediate recall and long-term episodic memory, semantic memory as well as both executive function and semantic memory.
In a recent genetic neuroimaging study, we found an association between the CREB1 SNP rs10932201 genotype and individual differences in performance level and neural correlates of reward- and associative memory-based decision-making (Wolf et al., 2015). More specifically, the AA/GA compared to the GG genotype was associated with lower performance in memory and executive function as well as a decrease in task-related activation in, e.g., the hippocampus and cingulate gyrus. Here, we show increased longitudinal decline in performance of memory and executive function comparing AA/GA versus GG genotype groups. Accordingly, the common genotype (GG) for rs10932201 in Caucasians was consistently associated with beneficial effects on memory and executive function across our measures and studies. Importantly, allele frequencies were similar for this genotype in our American and European Caucasian samples (A = 0.46/0.48).
The minor allele A for SNP rs6785 has been associated with the risk for depression/bipolar disorder in Caucasians as well as smaller hippocampus volume and increased CREB1 mRNA level in the prefrontal cortex of healthy Caucasian and African Americans (Li et al., 2014). Consistent with these previous observations, in our sample of healthy older Caucasian Americans, the A allele (AA/GA genotypes) for SNP rs6785 was associated with reduced immediate and long-term episodic memory performance.
We also found associations between the GG/GA versus AA genotype for SNP rs2253206 and lower performance level as well as increased longitudinal decline in performance of memory and executive function based on several cognitive measures (BNT, Category Fluency, CLOCKs, and CVLT-long-delay free recall). Previous research has linked this CREB1 promoter SNP rs2253206 with differences in BDNF mRNA levels in the human hippocampus as well as with other SNPs in the genes encoding BDNF and G protein-activated K+ channel 2 (Juhasz et al., 2011; Lazary et al., 2011). Moreover increased rumination, thought rumination and current depression severity have been connected with the G major allele of rs2253206 (Juhasz et al., 2011; Lazary et al., 2011).
Our results also agree with a previous cross-sectional study in older, cognitively healthy adults, reporting associations of other CREB1 SNPs, SNPs in the CREB1 binding protein gene (CREBBP), and another gene affecting the CREB1 pathway with episodic memory performance (Barral et al., 2014). Interestingly, these associations were, similarly to our findings, influenced by APOE𝜀4 status.
Consistently across those four measures affected by CREB1–APOE–interactions, the direction of cross-sectional and longitudinal effects of CREB1 genotypes on performance was unchanged in combination with APOE𝜀4 negative but reversed with positive status. Reversal of the direction of genotype effect means the genotype formerly associated with better performance or less decline in performance changes to be associated with lower performance or more decline in performance. Respectively the genotype formerly associated with lower performance or more decline in performance changes to being associated with the opposite effects on performance level and decline. Such reversal of CREB1 genotype effects on some measures of executive function, semantic and episodic memory in the presence of APOE𝜀4 positive status supports the idea of APOE𝜀4 positive status contributing to the deregulation of CREB1 signaling.
Our results converge with previous findings in rodent models indicating the disturbance of CREB function in the presence of APOE𝜀4 genotype or protein. Specifically, it was shown that level of CREB activation as well as signaling up-stream and down-stream of CREB was decreased in cortex and hippocampus of transgenic mice with APOE𝜀4 positive compared to negative genotype status (Liu et al., 2015), and in rats injected with apoe4 compared to apoe2 protein (Qiao et al., 2014). Moreover these APOE𝜀4 genotype and apoe4 isoform-specific alterations of CREB signaling pathways were shown to enhance age-related decline in memory and impair in vivo hippocampal long-term potentiation.
The strengths of our analysis include comprehensive cognitive assessment over a mean follow-up period of 11 years, a mean sample size of 707 and a mean number of 4095 observations across 11 cognitive measures. In addition, our analysis incorporated the adjustment for already established risk factors including APOE𝜀4 status, sex, age, and education. As mentioned SNPs rs6785 and rs2253206 have been associated with depression and depression-related phenotypes (Juhasz et al., 2011; Lazary et al., 2011; Li et al., 2014), the prevalence of depression increases with aging (Arve et al., 1999; Stek et al., 2004) and there is a relation between depression and cognitive deficits (Lichtenberg et al., 1995; Trivedi and Greer, 2014). However, adding a measure of symptoms of depression (CESD) as a covariate did not change our findings. Our sample was homogenous regarding ethnicity, education and cognitive status. We found that the performance level and/or longitudinal decline in performance for the majority of the cognitive measures examined were affected by CREB1 genotypes in adults during aging. Although significant effects were mostly of small size, the effect size for significant effects of these common polymorphisms on cognition varied ranging from Cohen’s d = -0.15 to -0.70. Small effects of SNPs are expected to be more frequent because of the multitude of genetic and environmental factors as well as their interactions that together contribute to individual differences in cognition (Wolf and Linden, 2012; Davies et al., 2015), cognitive aging and the risk of cognitive impairment.
Our current analyses were limited to a selection of only a few important factors that contribute to differences in human cognition during aging. Another concern is that we did not adjust for the number of SNPs and measures for executive function and memory tested. We argue that Bonferroni adjustment based on the number of analyses in our study is too conservative for the following reasons. (1) Our study was motivated by prior evidence for CREB1 genotype effects on brain function and cognition based on different sample populations. (2) The direction of genotype effects is the same across samples and measures. (3) The direction of genotype effects is the same across baseline performance and longitudinal decline. (4) The measures were not independent because there is an overlap between the measure-based samples and measures were selected to target two cognitive phenotypes (memory and executive function), with some measures being more correlated than others, e.g., measures for both memory and executive function versus measures for memory only. (5) Even if we use the Bonferroni corrected significance level of 0.01 according to three independent SNPs and two independent phenotypes (memory and executive function) we find multiple significant effects of CREB1 SNPs and APOE4-CREB1 interactions on cognition. There are several sources that may limit the generalizability of our findings. First we restricted the analysis to only Caucasians and unimpaired visits, which left us for our largest sample with only 6104 observations because 206 visits were excluded due to the presence of dementia, 96 visits due to MCI, 5 visits due to impairment but not MCI, and 21 visits due to unknown type of impairment. Furthermore the BLSA is a sample with good health, high education and socioeconomic status levels on average.
One potentially important interaction partner for CREB appears to be BACE1, which besides cleaving APP may also regulate synaptic plasticity and myelination (Kandalepas and Vassar, 2014). Upregulation of BACE1 protein level has been reported to downregulate the cAMP-PKA-CREB signaling pathway independent of BACE1 activity for Aβ generation (Chen et al., 2012). CREB might influence BACE1 level because the human BACE1 promoter contains a potential CREB binding site (Sambamurti et al., 2004). Genetic variants in CREB binding protein (CBP) have been associated with cognitive decline in human aging (Trompet et al., 2011; Barral et al., 2014). Other genes, besides being implicated in AD, are suspected to be linked with age-related cognitive decline in healthy aging, e.g., APP and TOMM40 (Caselli et al., 2012; Jonsson et al., 2012; Davies et al., 2015; Payton et al., 2016) also may be regulated by CREB (Ge et al., 2004; Zhang et al., 2005; Tanis et al., 2008; Rodriguez-Tornos et al., 2013).
Importantly, CREB1 also mediates interactions between genetic and environmental factors by regulating the transcription of a large number of neuroplasticity-related genes dependent on neuronal activation during, e.g., social and cognitive activities (Wolf and Linden, 2012; Benito and Barco, 2015). Such experience-dependent regulation of CREB-signaling is involved in learning and memory formation. For example the learning experience-dependent activation of CREB by reelin has been shown to affect synaptic plasticity in the hippocampus (Brai et al., 2015), the regulation of synaptic plasticity genes and hippocampus-dependent associative learning (Telese et al., 2015).
We provide the first evidence for the influence of CREB on age-related longitudinal decline in memory and executive function in humans. The results from our previous (Wolf et al., 2015) and the current study suggest that CREB1 gene variants contribute to individual differences in the level of memory and executive function performance in both young and older cognitively healthy adults. In addition to APOE𝜀4 status independent effects of CREB1 genotypes, we also provide evidence for interactive effects between APOE𝜀4 status and CREB1 genotypes on human cognition. The coexistence of APOE𝜀4 status dependent and independent effects of CREB1 genotypes is supported by studies in animals showing APOE dependent and independent CREB signaling pathways.
In the future we will consider interactions between variants of CREB1 and CREB1 target genes or individual differences in environment-related factors such as cognitive and social activities.
Author Contributions
CW designed the project, organized, analyzed and interpreted the genetic and behavioral data, and wrote the manuscript. YA, MB, and CG substantially contributed to the organization and statistical analysis of the data. TT carried out the genotyping. MK substantially contributed to the data collection. SR supervised the project. All authors critically revised and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Elizabeth Burke, Angela Dabney, and Wendy Elkins for participant management and data acquisition, Yu Peng for help with figures and effect size calculations as well as all our participants.
Footnotes
Funding. This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fnagi.2017.00142/full#supplementary-material
References
- Adlard P. A., Sedjahtera A., Gunawan L., Bray L., Hare D., Lear J., et al. (2014). A novel approach to rapidly prevent age-related cognitive decline. Aging Cell 13 351–359. 10.1111/acel.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arve S., Tilvis R. S., Lehtonen A., Valvanne J., Sairanen S. (1999). Coexistence of lowered mood and cognitive impairment of elderly people in five birth cohorts. Aging (Milano) 11 90–95. [PubMed] [Google Scholar]
- Balthazar M. L., Cendes F., Damasceno B. P. (2008). Semantic error patterns on the boston naming test in normal aging, amnestic mild cognitive impairment, and mild Alzheimer’s disease: is there semantic disruption? Neuropsychology 22 703–709. 10.1037/a0012919 [DOI] [PubMed] [Google Scholar]
- Barral S., Reitz C., Small S. A., Mayeux R. (2014). Genetic variants in a ’cAMP element binding protein’ (CREB)-dependent histone acetylation pathway influence memory performance in cognitively healthy elderly individuals. Neurobiol. Aging 35 2881.e7–2881.e10. 10.1016/j.neurobiolaging.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolotti N., Bennett D. A., Lazarov O. (2016). Reduced pCREB in Alzheimer’s disease prefrontal cortex is reflected in peripheral blood mononuclear cells. Mol. Psychiatry 21 1158–1166. 10.1038/mp.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E., Barco A. (2010). CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 33 230–240. 10.1016/j.tins.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Benito E., Barco A. (2015). The neuronal activity-driven transcriptome. Mol. Neurobiol. 51 1071–1088. 10.1007/s12035-014-8772-z [DOI] [PubMed] [Google Scholar]
- Benton A. L. (1968). Differential behavioral effects in frontal lobe disease. Neuropsychologia 6 53–60. 10.1016/0028-3932(68)90038-9 [DOI] [Google Scholar]
- Benton A. L. (1974). Revised Visual Retention Test: Clinical and Experimental Applications 4th Edn. New York: Psychological Corporation. [Google Scholar]
- Beydoun M. A., Boueiz A., Abougergi M. S., Kitner-Triolo M. H., Beydoun H. A., Resnick S. M., et al. (2012). Sex differences in the association of the apolipoprotein E epsilon 4 allele with incidence of dementia, cognitive impairment, and decline. Neurobiol. Aging 33 720–731.e4. 10.1016/j.neurobiolaging.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brai E., Marathe S., Astori S., Fredj N. B., Perry E., Lamy C., et al. (2015). Notch1 regulates hippocampal plasticity through interaction with the reelin pathway, glutamatergic transmission and CREB signaling. Front. Cell Neurosci. 9:447 10.3389/fncel.2015.00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretsky P., Guralnik J. M., Launer L., Albert M., Seeman T. E., MacArthur Studies of Successful Aging. (2003). The role of APOE-epsilon4 in longitudinal cognitive decline: MacArthur studies of successful aging. Neurology 60 1077–1081. 10.1212/01.WNL.0000055875.26908.24 [DOI] [PubMed] [Google Scholar]
- Brightwell J. J., Gallagher M., Colombo P. J. (2004). Hippocampal CREB1 but not CREB2 is decreased in aged rats with spatial memory impairments. Neurobiol. Learn. Mem. 81 19–26. 10.1016/j.nlm.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Caselli R. J., Dueck A. C., Huentelman M. J., Lutz M. W., Saunders A. M., Reiman E. M., et al. (2012). Longitudinal modeling of cognitive aging and the TOMM40 effect. Alzheimers Dement 8 490–495. 10.1016/j.jalz.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli R. J., Dueck A. C., Locke D. E., Hoffman-Snyder C. R., Woodruff B. K., Rapcsak S. Z., et al. (2011). Longitudinal modeling of frontal cognition in APOE epsilon4 homozygotes, heterozygotes, and noncarriers. Neurology 76 1383–1388. 10.1212/WNL.0b013e3182167147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli R. J., Dueck A. C., Osborne D., Sabbagh M. N., Connor D. J., Ahern G. L., et al. (2009). Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. N. Engl. J. Med. 361 255–263. 10.1056/NEJMoa0809437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli R. J., Reiman E. M., Osborne D., Hentz J. G., Baxter L. C., Hernandez J. L., et al. (2004). Longitudinal changes in cognition and behavior in asymptomatic carriers of the APOE e4 allele. Neurology 62 1990–1995. 10.1212/01.WNL.0000129533.26544.BF [DOI] [PubMed] [Google Scholar]
- Chen Y., Durakoglugil M. S., Xian X., Herz J. (2010). ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc. Natl. Acad. Sci. U.S.A. 107 12011–12016. 10.1073/pnas.0914984107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Huang X., Zhang Y. W., Rockenstein E., Bu G., Golde T. E., et al. (2012). Alzheimer’s beta-secretase (BACE1) regulates the cAMP/PKA/CREB pathway independently of beta-amyloid. J. Neurosci. 32 11390–11395. 10.1523/JNEUROSCI.0757-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G., Armstrong N., Bis J. C., Bressler J., Chouraki V., Giddaluru S., et al. (2015). Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53949). Mol. Psychiatry 20 183–192. 10.1038/mp.2014.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis D. C., Kramer J. H., Kaplan E., Ober B. A. (1987). California Verbal Learning Test. San Antonio, TX: The Psychological Corporation. [Google Scholar]
- Fuld P. A. (1978). “Psychological testing in the differential diagnosis of the dementias,” in Alzheimer’s Disease: Senile Dementia and Related Disorders eds Katzman R., Terry R. D., Bock K. L. (New York: Raven Press; ). [Google Scholar]
- Ge Y. W., Ghosh C., Song W., Maloney B., Lahiri D. K. (2004). Mechanism of promoter activity of the beta-amyloid precursor protein gene in different cell lines: identification of a specific 30 bp fragment in the proximal promoter region. J. Neurochem. 90 1432–1444. 10.1111/j.1471-4159.2004.02608.x [DOI] [PubMed] [Google Scholar]
- Genoux D., Haditsch U., Knobloch M., Michalon A., Storm D., Mansuy I. M. (2002). Protein phosphatase 1 is a molecular constraint on learning and memory. Nature 418 970–975. 10.1038/nature00928 [DOI] [PubMed] [Google Scholar]
- Goh J. O., An Y., Resnick S. M. (2012). Differential trajectories of age-related changes in components of executive and memory processes. Psychol. Aging 27 707–719. 10.1037/a0026715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golski S., Zonderman A. B., Malamut B. L., Resnick S. M. (1998). Verbal and figural recognition memory: task development and age associations. Exp. Aging Res. 24 359–385. 10.1080/036107398244193 [DOI] [PubMed] [Google Scholar]
- Grober E., Hall C. B., Lipton R. B., Zonderman A. B., Resnick S. M., Kawas C. (2008). Memory impairment, executive dysfunction, and intellectual decline in preclinical Alzheimer’s disease. J. Int. Neuropsychol. Soc. 14 266–278. 10.1017/S1355617708080302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hixson J. E., Vernier D. T. (1990). Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 31 545–548. [PubMed] [Google Scholar]
- Jonsson T., Atwal J. K., Steinberg S., Snaedal J., Jonsson P. V., Bjornsson S., et al. (2012). A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488 96–99. 10.1038/nature11283 [DOI] [PubMed] [Google Scholar]
- Juhasz G., Dunham J. S., McKie S., Thomas E., Downey D., Chase D., et al. (2011). The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. Biol. Psychiatry 69 762–771. 10.1016/j.biopsych.2010.11.019 [DOI] [PubMed] [Google Scholar]
- Kandalepas P. C., Vassar R. (2014). The normal and pathologic roles of the Alzheimer’s beta-secretase, BACE1. Curr. Alzheimer Res. 11 441–449. 10.2174/1567205011666140604122059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E., Goodglass H., Weintraub S. (1983). The Boston Naming Test. Philadelphia, PA: Lea & Febiger. [Google Scholar]
- Koch W., Ehrenhaft A., Griesser K., Pfeufer A., Muller J., Schomig A., et al. (2002). TaqMan systems for genotyping of disease-related polymorphisms present in the gene encoding apolipoprotein E. Clin. Chem. Lab. Med. 40 1123–1131. 10.1515/CCLM.2002.197 [DOI] [PubMed] [Google Scholar]
- Kudo K., Wati H., Qiao C., Arita J., Kanba S. (2005). Age-related disturbance of memory and CREB phosphorylation in CA1 area of hippocampus of rats. Brain Res. 1054 30–37. 10.1016/j.brainres.2005.06.045 [DOI] [PubMed] [Google Scholar]
- Lazary J., Juhasz G., Anderson I. M., Jacob C. P., Nguyen T. T., Lesch K. P., et al. (2011). Epistatic interaction of CREB1 and KCNJ6 on rumination and negative emotionality. Eur. Neuropsychopharmacol. 21 63–70. 10.1016/j.euroneuro.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Leckie R. L., Oberlin L. E., Voss M. W., Prakash R. S., Szabo-Reed A., Chaddock-Heyman L., et al. (2014). BDNF mediates improvements in executive function following a 1-year exercise intervention. Front. Hum. Neurosci. 8:985 10.3389/fnhum.2014.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Luo X. J., Rietschel M., Lewis C. M., Mattheisen M., Muller-Myhsok B., et al. (2014). Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Mol. Psychiatry 19 452–461. 10.1038/mp.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg P. A., Ross T., Millis S. R., Manning C. A. (1995). The relationship between depression and cognition in older adults: a cross-validation study. J. Gerontol. B Psychol. Sci. Soc. Sci. 50 25–32. 10.1093/geronb/50B.1.P25 [DOI] [PubMed] [Google Scholar]
- Lindenberger U. (2014). Human cognitive aging: corriger la fortune? Science 346 572–578. 10.1126/science.1254403 [DOI] [PubMed] [Google Scholar]
- Liu D. S., Pan X. D., Zhang J., Shen H., Collins N. C., Cole A. M., et al. (2015). APOE4 enhances age-dependent decline in cognitive function by down-regulating an NMDA receptor pathway in EFAD-Tg mice. Mol. Neurodegener. 10 7 10.1186/s13024-015-0002-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti B., Berteotti C., Contestabile A. (2005). Dysregulation of memory-related proteins in the hippocampus of aged rats and their relation with cognitive impairment. Hippocampus 15 1041–1049. 10.1002/hipo.20099 [DOI] [PubMed] [Google Scholar]
- Morris J. C. (1993). The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43 2412–2414. [DOI] [PubMed] [Google Scholar]
- Mouravlev A., Dunning J., Young D., During M. J. (2006). Somatic gene transfer of cAMP response element-binding protein attenuates memory impairment in aging rats. Proc. Natl. Acad. Sci. U.S.A. 103 4705–4710. 10.1073/pnas.0506137103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe F. (1969). Missile Wounds of the Brain: A Study of Psychological Deficits. London: Oxford University Press. [Google Scholar]
- Ohkubo N., Mitsuda N., Tamatani M., Yamaguchi A., Lee Y. D., Ogihara T., et al. (2001). Apolipoprotein E4 stimulates cAMP response element-binding protein transcriptional activity through the extracellular signal-regulated kinase pathway. J. Biol. Chem. 276 3046–3053. 10.1074/jbc.M005070200 [DOI] [PubMed] [Google Scholar]
- Packard C. J., Westendorp R. G., Stott D. J., Caslake M. J., Murray H. M., Shepherd J., et al. (2007). Association between apolipoprotein E4 and cognitive decline in elderly adults. J. Am. Geriatr. Soc. 55 1777–1785. 10.1111/j.1532-5415.2007.01415.x [DOI] [PubMed] [Google Scholar]
- Paula J. J., Miranda D. M., Moraes E. N., Malloy-Diniz L. F. (2013). Mapping the clockworks: what does the clock drawing Test assess in normal and pathological aging? Arq. Neuropsiquiatr. 71 763–768. 10.1590/0004-282X20130118 [DOI] [PubMed] [Google Scholar]
- Payton A., Sindrewicz P., Pessoa V., Platt H., Horan M., Ollier W., et al. (2016). A TOMM40 poly-T variant modulates gene expression and is associated with vocabulary ability and decline in nonpathologic aging. Neurobiol. Aging 39 217e1–7. 10.1016/j.neurobiolaging.2015.11.017 [DOI] [PubMed] [Google Scholar]
- Pozzi L., Sacchetti G., Agnoli L., Mainolfi P., Invernizzi R. W., Carli M. (2011). Distinct changes in CREB phosphorylation in frontal cortex and striatum during contingent and non-contingent performance of a visual attention task. Front. Behav. Neurosci. 5:65 10.3389/fnbeh.2011.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Qiao F., Gao X. P., Yuan L., Cai H. Y., Qi J. S. (2014). Apolipoprotein E4 impairs in vivo hippocampal long-term synaptic plasticity by reducing the phosphorylation of CaMKIIalpha and CREB. J. Alzheimers Dis. 41 1165–1176. 10.3233/JAD-140375 [DOI] [PubMed] [Google Scholar]
- Radloff L. S. (1977). The CES-D sclae: a self-report depressive scale for research in the general population. J. Appl. Psychol. Meas. 1 385–401. 10.1177/014662167700100306 [DOI] [Google Scholar]
- Rapanelli M., Lew S. E., Frick L. R., Zanutto B. S. (2010). Plasticity in the rat prefrontal cortex: linking gene expression and an operant learning with a computational theory. PLoS ONE 5:e8656 10.1371/journal.pone.0008656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Tornos F. M., San Aniceto I., Cubelos B., Nieto M. (2013). Enrichment of conserved synaptic activity-responsive element in neuronal genes predicts a coordinated response of MEF2, CREB and SRF. PLoS ONE 8:e53848 10.1371/journal.pone.0053848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnlund M., Nyberg L., Backman L., Nilsson L. G. (2005). Stability, growth, and decline in adult life span development of declarative memory: cross-sectional and longitudinal data from a population-based study. Psychol. Aging 20 3–18. 10.1037/0882-7974.20.1.3 [DOI] [PubMed] [Google Scholar]
- Rouleau I., Salmon D. P., Butters N., Kennedy C., McGuire K. (1992). Quantitative and qualitative analyses of clock drawings in Alzheimer’s and Huntington’s disease. Brain Cogn. 18 70–87. 10.1016/0278-2626(92)90112-Y [DOI] [PubMed] [Google Scholar]
- Sambamurti K., Kinsey R., Maloney B., Ge Y. W., Lahiri D. K. (2004). Gene structure and organization of the human beta-secretase (BACE) promoter. FASEB J. 18 1034–1036. 10.1096/fj.03-1378fje [DOI] [PubMed] [Google Scholar]
- Samieri C., Proust-Lima C., M Glymour M., Okereke O. I, Amariglio R. E., Sperling R. A., et al. (2014). Subjective cognitive concerns, episodic memory, and the APOE epsilon4 allele. Alzheimers Dement 10 752–759e751. 10.1016/j.jalz.2014.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Huertas C., Rico B. (2011). CREB-Dependent Regulation of GAD65 Transcription by BDNF/TrkB in Cortical Interneurons. Cereb. Cortex 21 777–788. 10.1093/cercor/bhq150 [DOI] [PubMed] [Google Scholar]
- Schiepers O. J., Harris S. E., Gow A. J., Pattie A., Brett C. E., Starr J. M., et al. (2012). APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Mol. Psychiatry 17 315–324. 10.1038/mp.2010.137 [DOI] [PubMed] [Google Scholar]
- Shao Z., Janse E., Visser K., Meyer A. S. (2014). What do verbal fluency tasks measure? Predictors of verbal fluency performance in older adults. Front. Psychol. 5:772 10.3389/fpsyg.2014.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. L., Pozueta J., Gong B., Arancio O., Shelanski M. (2009). Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proc. Natl. Acad. Sci. U.S.A. 106 16877–16882. 10.1073/pnas.0908706106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stek M. L., Gussekloo J., Beekman A. T., van Tilburg W., Westendorp R. G. (2004). Prevalence, correlates and recognition of depression in the oldest old: the Leiden 85-plus study. J. Affect. Disord. 78 193–200. 10.1016/S0165-0327(02)00310-5 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Scheet P., Giusti B., Bandinelli S., Piras M. G., Usala G., et al. (2009). Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet. 84 477–482. 10.1016/j.ajhg.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis K. Q., Duman R. S., Newton S. S. (2008). CREB binding and activity in brain: regional specificity and induction by electroconvulsive seizure. Biol. Psychiatry 63 710–720. 10.1016/j.biopsych.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telese F., Ma Q., Perez P. M., Notani D., Oh S., Li W., et al. (2015). LRP8-reelin-regulated neuronal enhancer signature underlying learning and memory formation. Neuron 86 696–710. 10.1016/j.neuron.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi M. H., Greer T. L. (2014). Cognitive dysfunction in unipolar depression: implications for treatment. J. Affect. Disord. 15 19–27. 10.1016/j.jad.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Trompet S., de Craen A. J., Jukema J. W., Pons D., Slagboom P. E., Kremer D., et al. (2011). Variation in the CBP gene involved in epigenetic control associates with cognitive function. Neurobiol. Aging 32 549.e1–549.e8. 10.1016/j.neurobiolaging.2009.12.019 [DOI] [PubMed] [Google Scholar]
- Vandesquille M., Baudonnat M., Decorte L., Louis C., Lestage P., Beracochea D. (2013). Working memory deficits and related disinhibition of the cAMP/PKA/CREB are alleviated by prefrontal alpha4beta2∗-nAChRs stimulation in aged mice. Neurobiol. Aging 34 1599–1609. 10.1016/j.neurobiolaging.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Weintraub S., Wicklund A. H., Salmon D. P. (2012). The neuropsychological profile of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2:a006171 10.1101/cshperspect.a006171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C., Linden D. E. (2012). Biological pathways to adaptability–interactions between genome, epigenome, nervous system and environment for adaptive behavior. Genes Brain Behav. 11 3–28. 10.1111/j.1601-183X.2011.00752.x [DOI] [PubMed] [Google Scholar]
- Wolf C., Mohr H., Diekhof E. K., Vieker H., Goya-Maldonado R., Trost S., et al. (2015). CREB1 genotype modulates adaptive reward-based decisions in humans. Cereb. Cortex 26 2970–2981. 10.1093/cercor/bhv104 [DOI] [PubMed] [Google Scholar]
- Yaffe K., Cauley J., Sands L., Browner W. (1997). Apolipoprotein E phenotype and cognitive decline in a prospective study of elderly community women. Arch. Neurol. 54 1110–1114. 10.1001/archneur.1997.00550210044011 [DOI] [PubMed] [Google Scholar]
- Yiu A. P., Rashid A. J., Josselyn S. A. (2011). Increasing CREB function in the CA1 region of dorsal hippocampus rescues the spatial memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology 36 2169–2186. 10.1038/npp.2011.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., et al. (2005). Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102 4459–4464. 10.1073/pnas.0501076102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


