Abstract
Members of the genus Aeromonas that commonly occur in various aquatic ecosystems are taken into account as vectors spreading antibiotic resistance genes (ARGs) in the environment. In our study strains of Aeromonas spp. (n = 104) not susceptible to ampicillin were isolated from municipal sewage of different levels of purification – raw sewage, activated sludge and treated wastewater. The crucial step of the study was the identification of β-lactamase resistance genes. The identified genes encode β-lactamases from 14 families – blaTEM, blaOXA, blaSHV, blaCTX-M, blaMOX, blaACC, blaFOX, blaGES, blaPER, blaV EB, blaKPC, cphA, imiH, and cepH. There were no significant differences in number of identified ARGs between isolation points. BlaOXA, blaFOX variants and, characteristic for Aeromonas genus, metallo-β-lactamase cphA-related genes were the most commonly identified types of β-lactam resistance determinants. Moreover, we found four extended-spectrum β-lactamases (blaSHV -11, blaCTX-M-27, blaCTX-M-98, and blaPER-4) – and seven AmpC (blaACC, blaFOX-2-like, blaFOX-3, blaFOX-4-like, blaFOX-9, blaFOX-10-like, and blaFOX-13-like) types and variants of genes that had never been found among Aeromonas spp. before. Five of the β-lactamases families (blaTEM, blaOXA, blaFOX, blaV EB, and cphA) were identified in all three isolation sites, which supports the hypothesis that wastewater treatment plants (WWTPs) are hot spots of ARGs dissemination. The obtained ARGs sequences share high identity with previously described β-lactamases, but new variants of those genes have to be considered as well. Characterization of antibiotic susceptibility was performed using disk the diffusion method with 12 different antibiotics according to CLSI guidelines. Over 60% of the strains are unsusceptible to cefepime and chloramphenicol and the majority of the strains have a multidrug resistance phenotype (68%). Finally, analysis of plasmid profiles among the resistant strains showed that 62% of the isolates from all three points of the WWTP carry plasmids of different sizes. Among some of the isolated plasmids blaFOX-4-like and blaGES genes have been found. To sum up, the results strongly suggest that Aeromonas spp. can be considered as agents of antibiotic resistance dissemination from wastewater to the natural environment.
Keywords: Aeromonas, β-lactamases, plasmid, integron, antibiotic resistance gene, horizontal gene transfer, wastewater treatment plant
Introduction
The World Health Organization (WHO) identified the development of antibiotic resistance as one of the major global threats to the society and recommended intensive monitoring for the identification and surveillance of critical hot spots, aimed at reducing resistance dissemination (World Health Organization [WHO], 2013). The results of many studies show that wastewater treatment plants (WWTPs) are one of the key reservoirs of antibiotic resistance (Bouki et al., 2013). Therefore, WWTPs are now considered as one of the main hot spots of the potential emergence and spread of antibiotic resistance bacteria (ARB) and antibiotic resistance genes (ARGs) in the environment (Rizzo et al., 2013). ARB and ARGs are massively discharged into the municipal sewage system with wastewater of different origins (Kümmerer, 2009). As a reason many ARB species have been already found in WWTPs and among the most common are bacteria from the genus Aeromonas. Recent publications place Aeromonas spp. among dominant genera in wastewater communities (Chen C.-Y. et al., 2016; Hu et al., 2016). Aeromonas spp. are ubiquitous especially in all kind of aquatic environments, such as lakes, rivers, sea water, estuaries, pristine water, aquacultures, drinking water, or wastewater (Piotrowska and Popowska, 2014). Members of this genus are opportunistic food and waterborne pathogens of humans and animals, especially fish. From a public health point of view Aeromonas spp. infections are not very important, but these bacteria are taken into consideration as important vectors of ARGs in the environment (Berendonk et al., 2015).
Nowadays, there is a growing number of literature data on the dissemination of ARGs among Aeromonas spp. (Piotrowska and Popowska, 2014). However, the research on WWTP samples is still insufficient (Zhang et al., 2015; Varela et al., 2016). The ARGs that have been found in wastewater encode resistance to quinolones, β-lactams, aminoglycosides and tetracyclines, with the first two groups being predominant. Current research pays more attention to searching for quinolone and β-lactam resistance genes, which are potentially plasmid-mediated (Marti et al., 2014; Varela et al., 2016). However, all these studies concentrate on the most prevalent variants of these genes. In the case of β-lactam resistance genes the variety of types and variants of genes is very wide, which prompts researchers to narrow the spectrum of research and focus on a very small group of β-lactamases. So far in wastewater-derived Aeromonas spp. blaTEM, blaOXA, blaCTX-M, blaSHV, blaKPC, blaPSE1/CARB1, and cphA genes have been found (Figueira et al., 2011; Igbinosa and Okoh, 2012; Picão et al., 2013). However, the occurrence of different types of β-lactamases has been shown in studies with isolates from different environments, such as aquacultures, rivers, lakes, or sea (Henriques et al., 2006; Girlich et al., 2010, 2011). Among these strains, also the extended-spectrum β-lactamase AmpC and extended-spectrum β-lactamases (ESBL) genes blaFOX, blaV EB, blaPER, blaGES have been found in addition to those listed above. In this case searching for a wider range of β-lactamase genes in wastewater seems to be justified and informative regarding the flow of these genes between different water environments.
In this study we focus on the variety of β-lactam resistance genes, which occurs among Aeromonas spp. isolated from wastewater of different purification levels: raw sewage, liquid phase of activated sludge and treated wastewater. The sampling points have been chosen in such a way to show the flow of β-lactam resistance genes through the treatment process. Such a diversity of β-lactamase types and variants has never been studied before in a particular wastewater environment.
Materials and Methods
Characteristics of Study Sites and Sample Collection
Samples of raw sewage, activated sludge and effluent were collected from the urban wastewater treatment plant (UWTP) located in Warsaw, Poland (52.351° N, 20.959° E). The UWTP is located in the north-east of Warsaw, near the Vistula river where the final effluent is discharged. Studied UWTP is a secondary treatment facility – CAS WWTP (Conventional Activated Sludge) – with average daily throughput equal to 435,300 m3/d, at a load of 2,400,000 PE (People Equivalents). This UWTP collects domestic, urban and hospital sewage from Warsaw and suburban area.
The strains were isolated from four time periods (September 2011, October 2011, April 2014, and June 2014) with the final pool comprising four samples of each type: raw sewage (influent), activated sludge and treated effluent (effluent). The samples were collected in 5 L sterile glass bottles, transported refrigerated to the laboratory and subjected to biological analyses within 6 h.
Bacterial Count and Identification
A 100 mL volume of each sample was filtered through cellulose nitrate membranes (pore size 0.45 μm, Merck Millipore, Germany) and the filters were rinsed with saline. The total number of each: heterotrophs (R2Agar medium), ceftazidime resistant bacteria (R2Agar supplemented with ceftazidime) and bacteria belonging to Aeromonas spp. (Ryan Aeromonas Medium Base) were quantified using dilution plating procedure. Diluted (10–1000 times) and undiluted 0.1 mL aliquots were plated on R2 Agar complete medium (Graso Biotech, Poland), supplemented or not supplemented with ceftazidime or meropenem to a final concentration of 16 μg/mL, or Ryan Aeromonas Medium Base (Oxoid, England), supplemented with Ampicillin Selective Supplement (Oxoid, England). The concentrations of antibiotics were selected according to CLSI guidelines (Clinical and Laboratory Standards Institute [CLSI], 2010). R2A Agar is dedicated to the recovery and isolation of aerobic and facultative anaerobic heterotrophic bacteria. Ryan is a selective diagnostic medium for the isolation of Aeromonas hydrophila from clinical and environmental specimens. The plates were incubated for 24–48 h at room temperature. For each assay, plate counts were performed in triplicate with four different dilutions for each sample. Strains were stored at 4°C on agar plates supplemented with antibiotic and in LB medium with 10% glycerol at -70°C.
All isolates were identified to genus level by sequencing partial nucleotide sequence of 16S rRNA gene obtained using PCR. Amplification reactions were performed using the conditions described elsewhere (Devereux and Willis, 1995). PCR products were separated in 0.8% agarose gel by electrophoresis and purified using Clean-up Concentrator Kit (A&A Biotechnology, Poland) or Gel-out Concentrator Kit (A&A Biotechnology, Poland) according to the manufacturer’s instructions. PCR amplicons were sequenced in Genomed (Warsaw, Poland) using BigDye® Terminator v3.1 from Applied Biosystems (Life Technologies) where nucleotide sequences were determined. The resulting 16S rRNA gene sequences were compared with the GenBank database using BLAST software (Altschul et al., 1990). Genus-level identifications were performed using the following criterium: a bacterium was assigned to a particular genus when identity with the genus sequences in the database was more than 95%.
Detection of β-Lactamase Genes and Integrons
To identify β-lactamase genes, a molecular investigation by PCR amplification method was performed, mainly by Multiplex PCR using the conditions described in previous works (Perez-Perez and Hanson, 2002; Henriques et al., 2006; Dallenne et al., 2010). For this study five multiplex sets of primers and two simplex pairs of primers (Supplementary Table S1), characteristic for Aeromonas spp. metallo-β-lactamase chromosomal gene cphA and ESBL β-lactamase genes from blaCTX-M8/25 group were selected. As positive controls genomic DNA of clinical strains, obtained from the National Medicines Institute (Warsaw, Poland), carrying selected β-lactams resistance genes (Supplementary Table S2) and well-characterized foodborne Aeromonas hydrophila ATCC 7966 strain (Seshadri et al., 2006) were used. As a negative control Escherichia coli ATCC 25922 strain was used. PCR products were separated in 0.8–2% agarose gel by electrophoresis and purified using the same kits as in bacterial identification protocol. PCR products of the expected sizes were sequenced by Genomed (Warsaw, Poland). Products were compared with GenBank database, using BLAST N and BLAST X tools (Altschul et al., 1990) and with Antibiotic Resistance Database (ARDB) (Liu and Pop, 2009). Sequence analysis and assembly were performed using Clone Manager 8 (Sci-Ed Software, USA) and chromatogram viewer FinchTV (Geospiza, USA). Reference sequences have been taken from Lahey Clinic β-lactamases database1.
Integrase genes intI1, intI2, and intI3 were identified in total DNA from isolated strains by PCR using specific primers as described previously (Henriques et al., 2006). As positive controls genomic DNA of Aeromonas spp. strains with confirmed presence of integrase genes was used (environmental collection of our research group). The obtained products were processed in the same way as ARGs, i.e., they were sequenced and compared with GenBank database using BLAST N.
BlaFOX sequences were aligned using the built-in MUSCLE (default parameters), and a phylogenetic tree was built using the Neighbor joining method with default parameters and 1000 bootstrap replications with the MEGA6 software. Accession numbers for the reference genes are as follows: blaFOX-1 from Klebsiella pneumoniae – NG_049098.1; blaFOX-2 from E. coli – NG_049102.1; blaFOX-3 from Klebsiella oxytoca – NG_049103.1; blaFOX-4 from E. coli – NG_049104.1; blaFOX-9 from K. pneumoniae – NG_049108.1; blaFOX-10 from K. pneumoniae – NG_049099.1; blaFOX-13 from Providencia rettgeri – NG_049101.1.
Antibiotic Susceptibility Testing and Determination of Multiple Antibiotic Resistance (MAR) Index
The susceptibility to 12 antibiotics was determined using the agar diffusion method and Clinical and Laboratory Standards Institute guidelines M45A2E (Clinical and Laboratory Standards Institute [CLSI], 2010). The antibiotics tested were ciprofloxacin (CIP, 5 μg), tetracycline (TET, 30 μg), amikacin (AK, 30 μg), gentamicin (CN, 10 μg), chloramphenicol (C, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), cefepime (FEP, 30 μg), aztreonam (AZT, 30 μg), ertapenem (ERT, 10 μg), imipenem (IMP, 10 μg), and meropenem (MEM, 10 μg). Inhibition zones larger than R (resistant) and smaller than S (susceptible) were classified as intermediate resistance and excluded from the resistance percentage calculations. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were included as quality controls.
The Multiple Antibiotic Resistance (MAR) index was calculated for each isolate based on the results of the disk diffusion method analysis. The MAR index for a single isolate was calculated as the number of antibiotics to which an isolate is resistant (a) divided by the total number of antibiotics against which the isolate was tested (b) (Zhang et al., 2015).
Plasmids Isolation and Conjugation Experiments
In order to determine the location of β-lactamase resistance genes, plasmid DNA was extracted and purified using Plasmid Mini AX Gravity kit (A&A Biotechnology, Poland), according to the manufacturer’s instruction. Samples were separated by electrophoresis in 0.8% agarose gels.
Southern Hybridization Protocol
Plasmid DNA was separated by electrophoresis in 0.8% agarose gels. Gels were then stained in ethidium bromide, and visualized using UV transilluminator. DNA was transferred to a nylon membrane (Roche Diagnostics GmbH, Germany) and hybridized with previously prepared probes. As probes, PCR-amplified and digoxigenin (DIG)-labeled resistance genes identified at the previous stages of this research were used. The process of preparing labeled probes and hybridization was performed using DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Science, Germany), following the manufacturer’s procedure.
Statistical Analyses
Statistical analyses were performed using the R 3.2.5 software (R Core Team, 2016). The prevalence of antibiotic resistance phenotypes and ARGs were compared among strains from different origin of isolations: influent, activated sludge, and effluent. In case of antibiotic resistance phenotypes generalized linear model – GLM with Bernoulli distribution was applied. Significance of the distribution of ARGs was determined using chi-square test. In both tests the approved level of significance was p-value < 0.05.
Results
Bacterial Count and Identification of Aeromonas Isolates
Plate counting was performed from four independent isolations. These data are shown in Table 1. In each replicate the total number of heterotrophic bacteria was the highest in samples from activated sludge and the lowest in the effluent from 1 × 107 to 4.2 × 107 and from 2.2 × 104 to 5.6 × 104, respectively. As the present study was aimed to detect Aeromonas spp. resistant to β-lactams, we applied three different media to obtain as many as possible non-repetitive isolates. The percentage of ceftazidime resistant bacteria was the highest in influent and varied between 39.8 and 0.8% and the lowest in activated sludge – from 4.9 to 1.0%; both were therefore higher than the percentage of meropenem resistant bacteria, which varied between 1.6 and 0.1% in the influent and from 0.4 to 0.001% in activated sludge. The total number of Aeromonas spp. isolates was assessed only twice, but each time the percentage of isolates was the highest in the effluent, ranging from 9.1 to 6.5%; in the influent it varied from 5.3 to 2.2% and in activated sludge – from 0.7 to 0.2%, being the lowest result.
Table 1.
Total cultivable heterotrophs, Aeromonas spp. and ceftazidime and meropenem resistance bacterial counts (expressed in CFU/mL) and percentage of resistant isolates (% resistants) isolated from influent, activated sludge and effluent of UWTP.
| Time | Sampling point | Total heterotrophs | Total ceftazidime resistant | % Resistants | Total meropenem resistant | % Resistant | Total Aeromonas | % Aeromonas |
|---|---|---|---|---|---|---|---|---|
| September 2011 | Influent | 2.40E+06 | 9.56E+05 | 39.8 | 3.90E+04 | 1.6 | nd | – |
| Activated sludge | 1.00E+07 | 4.24E+05 | 4.2 | 4.47E+04 | 0.4 | nd | – | |
| Effluent | 2.30E+04 | 1.14E+03 | 5.0 | 4.90E+01 | 0.2 | nd | – | |
| October 2011 | Influent | 1.43E+06 | 4.80E+05 | 33.6 | 5.00E+03 | 0.3 | nd | – |
| Activated sludge | 4.20E+07 | 8.80E+05 | 2.1 | 7.50E+04 | 0.2 | nd | – | |
| Effluent | 2.20E+04 | 1.20E+03 | 5.5 | 2.10E+02 | 1.0 | nd | – | |
| April 2014 | Influent | 1.00E+07 | 8.00E+04 | 0.8 | 6.00E+03 | 0.1 | 5.30E+05 | 5.3 |
| Activated sludge | 1.02E+07 | 5.00E+05 | 4.9 | 1.00E+04 | 0.1 | 7.05E+04 | 0.7 | |
| Effluent | 5.00E+04 | 2.00E+03 | 4.0 | 5.00E+02 | 1.0 | 3.25E+03 | 6.5 | |
| June 2014 | Influent | 4.30E+06 | 5.60E+05 | 13.0 | 4.52E+03 | 0.1 | 9.63E+04 | 2.2 |
| Activated sludge | 2.23E+07 | 2.13E+05 | 1.0 | 2.54E+02 | 0.0 | 4.50E+04 | 0.2 | |
| Effluent | 8.60E+04 | 2.20E+03 | 2.6 | 2.30E+01 | 0.0 | 7.80E+03 | 9.1 |
Finally, 104 non-repetitive, phenotypically and morphologically different isolates were identified as Aeromonas spp.: 39 from influent, 45 from activated sludge, and 20 from effluent. Species level identification was impossible to obtain, as according to literature data and our own experience the gene encoding 16S rRNA is not enough diverse, which means that among Aeromonas spp. this sequence is highly conserved (Janda and Abbott, 2010). In order to determine the species of identified Aeromonas strains, sequencing of other housekeeping genes, such as gyrB and rpoD, is necessary.
Identification of β-Lactamases and Integron Genes
The crucial step of this study was to determine the variety of β-lactamase genes among isolated Aeromonas spp. strains. Finally, 13 variants of 14 types of β-lactamase genes were identified (Table 2 and Supplementary Tables S3–S5). According to the PCR products sequencing results, identified ARGs belong to blaTEM, blaOXA, blaSHV, blaCTX-M, blaMOX, blaACC, blaFOX, blaGES, blaPER, blaV EB, blaKPC, cphA, imiH, and cepH types. The most abundant β-lactamase-encoding genes were blaOXA, which were found in 35.58% of strains – 36 isolates (Table 2). The second most prevalent β-lactamase genes were blaFOX and cphA, which were identified in 29.81% (31 isolates) and 27.88% (29 isolates) of strains, respectively.
Table 2.
Number of identified bla, cphA, imiH, cepH and integrase genes and their variants among isolated Aeromonas strains with division into three points of isolation: (a) influent – 39 resistant strains, (b) activated sludge – 45 resistant strains, and (c) effluent – 20 resistant strains.
| Types of ARG | Influent | Activated sludge | Effluent | Summary |
|---|---|---|---|---|
| blaTEM | 3 | 13 | 2 | 18 |
| blaOXA | 15 | 16 | 5 | 36 |
| blaSHV | 2 | 1 | 0 | 3 |
| blaSHV -11 | – | 1 | – | 1 |
| blaSHV -12 | 2 | – | – | 2 |
| blaCTX-M | 1 | 1 | 0 | 2 |
| blaCTX-M-27/98 | 1 | 1 | ||
| blaCTX-M-15 | – | 1 | 1 | |
| blaACC | 1 | 1 | – | 2 |
| blaMOX | 0 | 0 | 2 | 2 |
| blaMOX-10/11 | – | – | 1 | 1 |
| blaMOX-4/8 | – | – | 1 | 1 |
| blaFOX | 15 | 9 | 7 | 31 |
| blaFOX-4-like | 7 | 4 | 4 | 15 |
| blaFOX-10-like | 4 | 1 | 2 | 7 |
| blaFOX-3 | 2 | 2 | – | 4 |
| blaFOX-9 | 2 | – | – | 2 |
| blaFOX-1 | – | 1 | – | 1 |
| blaFOX-2-like | – | – | 1 | 1 |
| blaFOX-13-like | – | 1 | – | 1 |
| blaGES | 7 | 9 | 0 | 16 |
| blaGES | 6 | 9 | – | 15 |
| blaGES-7 | 1 | – | – | 1 |
| blaPER | 6 | 3 | 0 | 9 |
| blaPER-3 | 2 | 1 | – | 3 |
| blaPER-4 | 3 | – | – | 3 |
| blaPER-1/5 | 1 | 2 | – | 3 |
| blaV EB | 3 | 3 | 3 | 9 |
| blaKPC | 0 | 2 | 0 | 2 |
| cphA | 4 | 19 | 6 | 29 |
| imiH | – | 4 | – | 4 |
| cepH | – | 1 | – | 1 |
| intI | 30 | 31 | 14 | 75 |
| intIII | 5 | 2 | 1 | 8 |
BlaTEM genes were observed among 18 Aeromonas spp. isolates: 3 from influent, 13 from activated sludge, and 2 from effluent. The products of primers that were used in this project have 800 bp and cover the blaTEM sequences from 13 to 812 bp (from 5 to 270 aa). Based on the databases blaTEM genes have 861 bp and 286 aa, and variable regions have been found over the entire length of the gene. As the results of comparing our products with reference sequences, using BLAST N tool, our blaTEM genes were 99–100% identical to different variants of blaTEM: blaTEM-1, blaTEM-84 blaTEM-104, blaTEM-122, and blaTEM-163 (J01749, AF427130, AF516719, AY307100, and EU815939). However, using BLAST X all sequences shared 93–100% identity to blaTEM-1. The reason for these differences are changes in amino acids that are outside the PCR product sequences. For instance, between TEM-1 and TEM-84 proteins there is a single mutation at position 272 (D to N). Consequently, it is impossible to determine the variants unambiguously and they all have been classified as blaTEM.
From all identified blaOXA genes only one gene from Aeromonas sp. 217, which was isolated from activated sludge, showed 99% nucleotide and amino acid identity to blaOXA-47 (NG_049751.1) and blaOXA-392 variants (NG_049683.1). The rest of the strains from influent (15 out of 39), activated sludge (16 out of 45) and effluent (5 out of 20) were 100% identical to blaOXA-1 (NG_049392.1) and other variants of these genes (NG_049613.1, GQ924769). Therefore it is also impossible to determine the variant unambiguously, based on this partial sequence.
Genes of blaSHV β-lactamases were identified in two isolates from the influent and one isolate from activated sludge. Both strains in the influent possessed bla genes that showed 100% identity to blaSHV -12 variant (AJ920369). However, bla gene from Aeromonas sp. 368A isolate, which was discovered in activated sludge, belonged to a different variant and was 100% identical to blaSHV -11 (JX268754.1).
As for the blaCTX-M genes, two strains with three different variants were found in the influent and activated sludge. Aeromonas sp. strain 6.41 from influent had a bla gene, which was 100% identical to blaCTX-M-98 (HM755448) and blaCTX-M-27 (AY156923), showing high identity to CTX-M-9 group of β-lactamases of this type. Aeromonas sp. strain T32 from activated sludge had a bla gene with 100% identity to blaCTX-M-15 (KT459668.1), which belongs to CTX-M-1 group of these β-lactamases (Bonnet, 2004).
Among the searched ampC genes only blaMOX, blaACC, cepH, and blaFOX types were found. In one Aeromonas sp. strain 104 from influent and Aeromonas sp. strain 368A from activated sludge blaACC genes were found with 100% identity to blaACC-1 (NG_048588.1) and blaACC-4 (NG_048594.1). Again, it was impossible to determine the variant unambiguously. Two β-lactamase genes that represent blaMOX type were observed only in isolates from the effluent: in Aeromonas sp. strain AWY18 there was 100% nucleotide and amino acid identity to blaMOX-4 (NG_049317.1) and blaMOX-8 (NG_049321.1). In Aeromonas sp. strain 415 there was 100% identity to blaMOX-11 (NG_049313.1) and blaMOX-10 (NG_049312.1). There was no other type of bla gene characteristic only of the effluent. In Aeromonas sp. AKP12 from activated sludge several ARGs were identified, showing 95% identity to cepH gene (NG_047628.1).
The most variable and numerous group of identified ampC genes belongs to blaFOX β-lactamases. Genes from this type observed in this study were classified into seven different variants: blaFOX-1, blaFOX-2, blaFOX-3, blaFOX-4, blaFOX-9, blaFOX-10, and blaFOX-13. Two most prevalent variants that have been found in all three points of isolation are blaFOX-4 and blaFOX-10 genes. In total, blaFOX-4 variant was found in 16 Aeromonas sp. strains. However, the obtained nucleotide sequences were not identical to the deposited reference sequence. Fifteen isolates – seven from influent, four from activated sludge, and four from effluent – showed different nucleotide identity ranging from 95 to 97% to the blaFOX-4 variant of the gene (NG_049104.1). Phylogenetic analysis resulted in five possible diverse phylogenetic branches for genes identified as this variant (Figure 1). In the case of blaFOX-10 identified among seven Aeromonas sp., all of them showed 94–95% identity to blaFOX-10 variant from the database (NG_049099.1). Moreover, phylogenetic analyses divided them into two different branches. In our study only blaFOX-3 was observed in the influent and activated sludge – in each place two isolates carried this type of gene. All of them were 100% identical to blaFOX-3 (NG_049103.1) and on the phylogenetic tree all four strains were located within the same branch. The rest of identified blaFOX variants were unique to the site of isolation and they were: bla from Aeromonas sp. AKP25 100% identical to blaFOX-1 (NG_049098.1), bla from Aeromonas sp. AWY31 97% identical to blaFOX-2 (NG_049102.1), two bla genes with 100% identity to blaFOX-9 (NG_049108.1) and bla from Aeromonas sp. AKP14 with 99% identity to blaFOX-13 (NG_049101.1). All identified blaFOX variants have been confirmed using BLAST X alignment.
FIGURE 1.
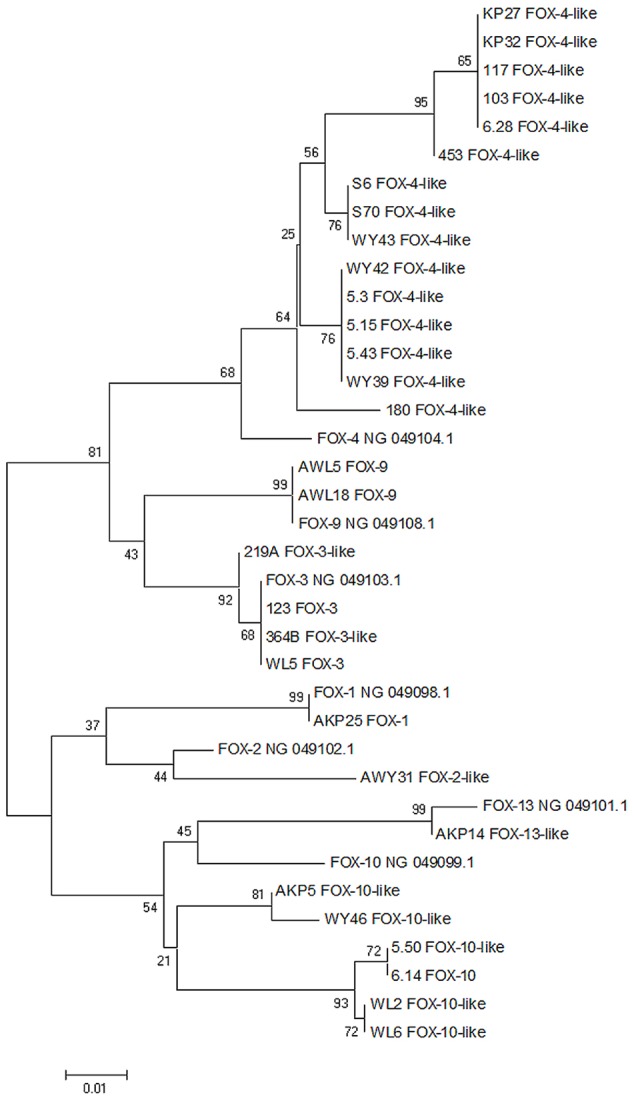
Neighbor joining dendrogram tree based on blaFOX gene sequences of Aeromonas spp. isolated from WWTP. Seven GenBank reference blaFOX sequences also have been attached to the analysis (sequences with NG numbers). The branch numbers refer to the percent confidence as estimated by bootstrap analysis with 1000 replications.
Two variants of blaGES genes were also identified. The first variant isolated from influent was observed only in Aeromonas sp. 6.45 with 99% nucleotide identity to blaGES-7 (NG_049139.1) and to blaGES-17 (NG_049119.1). However, BLAST X analysis resulted in 100% identity to blaGES-7 encoding β-lactamase GES-7 (WP_032490683.1). The second variant was observed in six isolates from influent and in nine cultures from activated sludge. All genes demonstrated 100% nucleotide identity to several blaGES variants and in this case again the variable region was outside the PCR product. As a result it was impossible to conclusively determine the variant of these genes.
Furthermore, genes that encode PER β-lactamases were observed only in the influent and activated sludge and they represented three different variants: blaPER-3, blaPER-4, and blaPER-1/5. In Aeromonas spp. with blaPER-3 all three identified genes showed 100% identity to this variant deposited in the database (NG_049962.1). Three isolates from the influent possessed blaPER-4 genes, however, two of them were 99% identical in nucleotide sequence with gene variant from the database (NG_049963.1) and also with protein sequence of PER-4 (WP_063864593.1). A third Aeromonas sp. 118 strain was 100% identical with the reference gene. The last variant was found in three isolates from the influent (one strain) and activated sludge (two strains); each was 100% identical to blaPER-1 (Z21957) and blaPER-5 (FJ627180). The results of BLAST X analysis also confirmed the identified variants.
Moreover, nine blaV EB genes were identified: in three strains from the influent, in three from the activated sludge and in three strains from the effluent. None of the blaV EB genes that were found in this study can be classified to a particular blaV EB variant based on the obtained part of blaV EB sequence because the variable region was outside the PCR product, and as a result the obtained products have 100% nucleotide identity to the numerous variants of genes deposited in the database.
Finally, in the activated sludge two strains possessed blaKPC genes: Aeromonas sp. 368A and Aeromonas sp. 386. Both genes had 100% nucleotide and protein identity to blaKPC-2 and blaKPC-3 genes (AY034847, AF395881). There was no other type of bla gene that was characteristic only of activated sludge.
Chromosomally encoded metallo-β-lactamase cphA-related genes were identified very frequently in this study. They constituted a very diverse group of genes, which were highly identical to different cphA variants of genes and proteins (Table 3). In total in 29 Aeromonas sp. isolates cphA-related genes were recognized as follows: 4 in influent, 19 in activated sludge, and 6 in effluent. Besides cphA, other metallo-β-lactamases have also been found – in four strains with 97–98% identity to imiH (NG_050414.1). All these genes were identified among strains isolated from activated sludge.
Table 3.
CphA gene variants identified in Aeromonas spp. isolated in this study.
| Aeromonas spp. strain | cphA variant∗ | BLAST N∗∗ | BLAST X∗∗∗ | |
|---|---|---|---|---|
| Influent | 6.28 | cphA1/cphA | 95%, A. salmonicida (NG_047668.1) | 99%, A. salmonicida (WP_059112452.1) |
| 7.47 | cphA/cphA | 96%, A. aquariorum (JF972621.1) | 98%, A. hydrophila (WP_024941978.1) | |
| WL1 | cphA1/cphA | 95%, A. salmonicida (NG_047668.1) | 99%, A. salmonicida (WP_058395305.1) | |
| WL3 | cphA1/cphA | 96%, A. salmonicida (NG_047668.1) | 99%, A. salmonicida (WP_058395305.1) | |
| Activated sludge | T6 | cphA/subclass B2 metallo-β-lactamase | 96%, A. aquariorum (JF972618.1) | 99%, A. hydrophila (WP_050559324.1) |
| S6 | cphA1/cphA | 94%, A. salmonicida (NG_047668.1) | 96%, A. australiensis (WP_040097284.1) | |
| S12 | cphA/cphA2 | 96%, A. aquariorum (JF972621.1) | 98%, A. hydrophila (WP_063865208.1) | |
| E34 | cphA/cphA | 96%, A. veronii (JF972616.1) | 97%, A. australiensis (WP_040097284.1) | |
| 203 | cphA7/cphA7 | 97%, A. jandaei (NG_050400.1) | 93%, A. jandaei (WP_063865212.1) | |
| 206 | cphA7/cphA | 96%, A. jandaei (NG_050400.1) | 96%, A. hydrophila (WP_049047121.1) | |
| 221 | cphA7/cphA | 96%, A. jandaei (NG_050400.1) | 99%, A. hydrophila (WP_060390377.1) | |
| 343A | cphA7/cphA | 96%, A. jandaei (NG_050400.1) | 99%, A. hydrophila (WP_060390377.1) | |
| 357A | cphA7/cphA | 96%, A. jandaei (NG_050400.1) | 99%, A. hydrophila (WP_060390377.1) | |
| 364B | cphA7/cphA | 95%, A. jandaei (NG_050400.1) | 96%, A. hydrophila (WP_049047121.1) | |
| 368A | cphA7/cphA | 96%, A. jandaei (NG_050400.1) | 95%, A. hydrophila (WP_049047121.1) | |
| 280 | cphA/cphA1 | 96% A. aquariorum (JF972625.1) | 97%, A. hydrophila (WP_063844282.1) | |
| 297 | cphA/cphA1 | 96% A. aquariorum (JF972625.1) | 100%, A. hydrophila (WP_060390377.1) | |
| KO26 | cphA/cphA | 94%, A. aquariorum (JF972618.1) | 97%, A. salmonicida (WP_043135519.1) | |
| AKO1 | cphA/cphA | 96%, A. aquariorum (JF972618.1) | 97%, A. veronii (WP_005343384.1) | |
| AKO16 | cphA-like/ cphA-like | 100%, Aeromonas sp. G.I10.28 (DQ447638.1) | 100%, Aeromonas sp. G.I10.28 (ABE01851.1) | |
| AKP15 | cphA/cphA1 | 99%, A. hydrophila (NG_047667.1) | 99%, A. hydrophila (WP_063844282.1) | |
| AKP19 | cphA-type/cphA | 96%, A. dhakensis (AB765398.1) | 99%, A. hydrophila (WP_043161524.1) | |
| AKP23 | cphA2/cphA | 97%, A. hydrophila (NG_050396.1) | 99%, A. hydrophila (WP_043161524.1) | |
| Effluent | 426 | cphA1/cphA | 97%, A. hydrophila (NG_047671.1) | 100%, A. hydrophila (WP_024945765.1) |
| 481 | cphA1/cphA | 96%, A. hydrophila (NG_047671.1) | 100%, A. hydrophila (WP_024945765.1) | |
| 483 | cphA1/cphA | 97%, A. hydrophila (NG_047671.1) | 100%, A. hydrophila (WP_024945765.1) | |
| WY39 | cphA/cphA | 94%, A. aquariorum (JF972621.1) | 96%, A. allosaccharophila (WP_042063527.1) | |
| WY47 | cphA1/cphA | 96%, A. veronii (NG_047669.1) | 97%, A. australiensis (WP_040097284.1) | |
| AWY14 | cphA/cphA | 97%, A. veronii (JQ814285.1) | 98%, A. australiensis (WP_040097284.1) |
∗Gene variant based on BLAST N/BLAST X; ∗∗Identity, bacterial species and accession number of the most identical alignments using BLAST N; ∗∗∗Identity, bacterial species and accession number of the most identical alignments using BLAST X.
The last group of genes whose presence was determined were integrase genes – intI1 and intI3 (Table 2). Type I integrase genes were observed in 72% of all isolates (75 out of 104): in 30 strains from the influent, in 31 strains from the activated sludge and in 14 strains from the effluent. Moreover, intI3 genes were identified in 8% of isolates (8 out of 104): in 5, 2, and 1 strain, respectively. In none of the isolated Aeromonas spp. strains intI2 integrase gene was confirmed.
Antibiotic Susceptibility Phenotypes
Among all 104 Aeromonas spp. isolates a high percentage of bacteria were unsusceptible to cefepime (77%) and chloramphenicol (68%) (Table 4). The lowest percentage of bacteria were resistant to ciprofloxacin (6%), ertapenem (8%), imipenem (10%), cefotaxime (10%), meropenem (12%), and aztreonam (20%). All identified strains were unsusceptible to amikacin. Among the majority of the isolates (68%) MDR (multidrug resistance) phenotypes were observed, which means that every MDR strain was resistant to at least three different antibiotics from three different groups. Moreover, the MAR index calculations also confirmed the high percentage of multiresistant strains. The MAR index values ranged from 0.08 (resistance to one antibiotic) to 0.75 (resistance to nine antibiotics), with the most prevalent variants of 0.33 (resistance to four antibiotics) and 0.42 (resistance to five antibiotics) – each value was represented by 19% of Aeromonas strains (Supplementary Table S6). The analysis between three points of isolation shows that among all isolates there were fewer cefepime resistant strains in the activated sludge and effluent than in the influent (p-value < 0.05). Also the number of meropenem resistant strains was lower in activated sludge than in the influent (p-value < 0.05).
Table 4.
Percentage of phenotypically unsusceptible strains of Aeromonas spp. among ceftazidime, meropenem, and ampicillin resistant Aeromonas spp. isolated from influent, activated sludge, and effluent of UWTP.
| Sampling point | CTX | CAZ | FEP | ATM | ERT | IMP | MEM | CIP | TE | C | CN | AK | MDR > 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Influent | 5% | 56% | 95% | 13% | 3% | 3% | 3% | 3% | 21% | 67% | 38% | 100% | 74% |
| Activated sludge | 13% | 44% | 71% | 27% | 11% | 18% | 22% | 11% | 29% | 69% | 36% | 100% | 69% |
| Effluent | 10% | 45% | 55% | 20% | 10% | 5% | 5% | 0% | 15% | 70% | 30% | 100% | 60% |
| Summary | 10% | 49% | 77% | 20% | 8% | 10% | 12% | 6% | 23% | 68% | 36% | 100% | 68% |
CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; AZT, aztreonam; ERT, ertapenem; IMP, imipenem; MEM, meropenem; CIP, ciprofloxacin; TET, tetracycline; C, chloramphenicol; CN, gentamicin; AK, amikacin; MDR, multidrug resistance strains.
Plasmid DNA Analysis and Determination of bla Genes Localization
Plasmid DNA was isolated from 62% of all Aeromonas spp. (64 out of 104): from 28 isolates from the influent, 25 isolates from the activated sludge and 11 isolates from the effluent. Conventional gel electrophoresis revealed that all isolates from all three places of isolation contained at last one plasmid, but most of them were considered to have more than one extrachromosomal replicon. There number of different plasmid profiles identified among the influent, activated sludge and effluent samples was 26, 19, and 11, respectively, which means that most of the strains have unique plasmid profiles. In the influent there were two profiles that were found in two different strains: the first profile in Aeromonas sp. 111 and Aeromonas sp. 115 and the second in Aeromonas sp. WL2 and Aeromonas sp. WL3. In the activated sludge two profiles were observed in three different strains each: the first profile in Aeromonas sp. S6, Aeromonas sp. S70, and Aeromonas sp. S50 and the second in Aeromonas sp. 343A, Aeromonas sp. 297, and Aeromonas sp. 357A.
According to the results of Southern blot hybridization, most of the identified bla genes were located on chromosomes. However, based on the same method several blaGES and blaFOX genes had an extrachromosomal localization. In the case of blaGES genes, four blaGES genes identified among influent strains – Aeromonas sp. 5.4, Aeromonas sp. 6.45, Aeromonas sp. 111, and Aeromonas sp. 115 – and one blaGES-7 variant from Aeromonas sp. 280 from activated sludge were located on plasmids (Supplementary Tables S3, S4). Only one blaFOX-4-like gene from Aeromonas sp. S6 from activated sludge was mapped to a plasmid.
Discussion
According to the literature data, different types of ARGs have repeatedly been identified among Aeromonas spp. (Piotrowska and Popowska, 2014). The most frequently found are the ARGs determining resistance to quinolones and β-lactams. We based this study on the hypothesis that bacteria from the genus Aeromonas are an important reservoir of acquired β-lactamase genes (Maravić et al., 2013). We chose WWTP as a study object because of the poor knowledge about the prevalence of β-lactamases in this environment and in the context of the hot spot hypothesis that has already been mentioned.
The main aim of this study was the determination of β-lactamase genes diversity in isolated Aeromonas spp. strains. In the literature data three principal classes of β-lactamases are observed in Aeromonas species, according to Ambler classification: class B metallo-β-lactamase (MBL), class C cephalosporinase, and class D penicillinase (Janda and Abbott, 2010). Recent studies discovered a growing number of class A ESBLs among Aeromonas spp. strains as well (Lu et al., 2010; Girlich et al., 2011). In our study we also found numerous genes from classes A, C, B and D, with the most diverse and frequent class A.
Extended-spectrum β-lactamases confer resistance to all penicillins, narrow- and extended-spectrum cephalosporins and monobactams. Carbapenemases from KPC type also belong to this group. Bush and Fisher (2011) reported that almost 600 β-lactamases has been assigned to this class so far. In our study seven types of bla genes from class A have been found: blaTEM, blaSHV, blaCTX-M, blaGES, blaPER, blaV EB, and blaKPC. Some of the identified variants of these genes have been observed for the first time in Aeromonas spp. isolates from WWTP or even among Aeromonas genus in general. To our knowledge blaSHV -11, blaCTX-M-27, blaCTX-M-98, and blaPER had never been observed in Aeromonas spp. before but have been identified among different genera. BlaSHV -11 have been found in many clinical and environmental K. pneumoniae strains (Alibi et al., 2015; Davies et al., 2016; Shahraki-Zahedani et al., 2016), blaCTX-M-27 in many Enterobacteriaceae strains (Matsumura et al., 2016; Rodrigues et al., 2016), blaCTX-M-98 in E. coli strains from China (Liu et al., 2015) and blaPER-4 in Proteus vulgaris clinical strain (NG_049963). The rest of the identified ESBL bla genes have been observed in Aeromonas spp. before and these are: blaTEM, blaSHV -12, blaCTX-M-15, blaPER-3, blaGES, and blaV EB. However, only blaCTX-M-15 had been observed previously in Aeromonas spp. from wastewater (Amos et al., 2014). What is worth emphasizing is that blaKPC gene, which encodes clinically emerging carbapenemase, has also been identified among strains from activated sludge. The KPC (K. pneumoniae carbapenemase) enzyme has been reported to spread worldwide and among several bacterial species, such as Enterobacteriaceae strains, Pseudomonas aeruginosa and Acinetobacter baumannii (Bush and Fisher, 2011). Literature data describes blaKPC-2 genes among Aeromonas sp. isolates from a hospital effluent as well (Picão et al., 2013).
Class C β-lactamases (AmpC) are located both on chromosomes and plasmids and are represented by many types that have been found worldwide in numerous sources (Jacoby, 2009). Like the chromosomal AmpC β-lactamases, plasmid-mediated enzymes confer resistance to a broad spectrum of β-lactams, including penicillins, oxyimino-β-cephalosporins, cephamycins, and aztreonam. In this study, four types of β-lactamases encoding genes from this class have been identified: blaMOX, blaACC, blaFOX, and cepH. The most prevalent and diverse was blaFOX group with seven different variants identified. Nevertheless, only three variants of these genes had 100% identical sequences to bla genes deposited in the NCBI database: blaFOX-1, blaFOX-3, and blaFOX-9. The rest of the variants demonstrated lower sequence identity and in this case should be proposed as blaFOX-2-like, blaFOX-4-like, blaFOX-10-like, and blaFOX-13-like. All selected reference blaFOX genes originate from clinical pathogens and on phylogenetic trees they are located on different but closely related branches with blaFOX-like genes that were isolated in this study. The wide range of possible new variants of blaFOX-like genes among environmental strains seems to be unexplained, although it was previously observed among water samples of Aeromonas spp. (Voolaid et al., 2013). According to our knowledge, all identified variants have never been observed among Aeromonas spp. wastewater isolates before and, besides blaFOX-1, neither among Aeromonas genus.
Among the identified ampC genes also blaMOX and blaACC genes were found. BlaMOX variants encoding β-lactamases are derived from K. pneumoniae blaMOX-1 and most of the variants – from blaMOX-3 to blaMOX-12 have been found among environmental Aeromonas spp. so far (GenBank numbers: NG_049316.1, NG_049317.1, NG_049318.1, NG_049319.1, NG_049320.1, NG_049321.1, NG_049312.1, NG_049313.1, NG_049314.1). In this study blaMOX-4/8 and blaMOX-10/11 have been identified and according to our knowledge this is the first observation of these variants in wastewater isolates. Furthermore, the identified blaACC genes have also been found for the first time in Aeromonas spp. Previously, blaACC-1 genes were described to reside within plasmids isolated from K. pneumoniae and Salmonella enterica subsp. enterica (Hasman, 2005; Markovska et al., 2012).
Only one type of bla genes from class D was found in this study – blaOXA. Genes from blaOXA type are widely spread in many environments, also among Aeromonas spp., and their name derived from their oxacillin-hydrolyzing abilities. They have been found in natural waters (Henriques et al., 2006; Picão et al., 2008), aquacultures (Jacobs and Chenia, 2007), fishes (Verner-Jeffreys et al., 2009) and wastewater as well (Moura et al., 2007; Varela et al., 2016). In our study we also found many blaOXA genes, which was an expected result.
Class B MBLs, unique carbapenemases which require zinc ion at the active site, are widely distributed in clinical and environmental species. Among Aeromonas spp. few MBL β-lactamases have been found: AsbM1, IMP-19, VIM, ImiS, ImiH, and most prevalent CphA (Janda and Abbott, 2010). CphA enzymes have a very specific substrate range with high hydrolytic activity against penems and carbapenems (Segatore et al., 1993). In this study cphA-related genes were commonly identified among isolated Aeromonas spp. and they were the most divergent group of genes. All recognized cphA genes showed high level of identity ranging from 94 to 99% to deposited variants. Wu et al. (2012) have found 51 cphA-positive Aeromonas spp. isolates with 94% identity with cphA gene from database (AE036). The occurrence of cphA-related genes is also species-related and assignment to species level of our Aeromonas spp. isolates should be performed. Moreover, the rules concerning nomenclature and depositing the cphA genes in the databases are still unclear and unspecified, which makes the determination of new variants even more complicated.
Generally, there were no significant differences between the number of β-lactamase resistance genes among all three points of isolation. This could be explained by inefficiency of the applied technology of wastewater treatment in reducing the number of ARGs or ARB during treatment process. The dissemination of ARGs and ARB from WWTP to environment is poorly understood and depending on type of treatment and studied bacteria the results differ (Rizzo et al., 2013). However, according to the literature, three factors contribute to this constant level of ARGs: (i) the optimal nutrition conditions for Aeromonas spp. prevailing in the biological reactor, (ii) high constant inflow of diverse clinical and environmental Aeromonas spp. strains brought to the reactor along with the wastewater of different origin and (III) the accumulation of the antibiotics considered a selective pressure (Novo and Manaia, 2010; Bouki et al., 2013). Our findings also support the hypothesis that Aeromonas spp. play an important role as vectors in dissemination of β-lactamases into the natural environment. Besides the reduced number of Aeromonas spp. isolates in the final effluent, the number of ARGs was not significantly decreased.
Integrons are non-replicative genetic elements, which are able to capture and incorporate gene cassettes by site-specific recombination. Most of these elements belong to the 1st, 2nd, or 3rd class of integrons and contain intI1, intI2, or intI3 integrase genes, respectively. 1st and 2nd class of integrons have been found in the WWTP with predominance of the first one (Igbinosa and Okoh, 2012). Class 1 has been described at all stages of the WWTP process among Aeromonas spp. isolates with different frequency. However, their existence in effluents indicates the inefficiency of the treatment process in removing them (Moura et al., 2007). A large diversity of ARGs among gene cassettes (GCs) of integrons have been found in WWTPs, including β-lactamase genes: blaOXA, blaV IM-2, blaIMP, blaGES-5, and blaGES-7 (Zhang et al., 2009; Stalder et al., 2012). In our study, as expected, many intI1 genes have been identified in all three points of isolation. There were no significant differences between points of isolation. On the other hand, the occurrence of intI3 genes was unexpected, which is of particular importance. Previously, class 3 integrases were observed among Aeromonas allosaccharopila LIM82 originated from sludge in France and it has been the only described case of intI3 among Aeromonas spp. (Simo Tchuinte et al., 2016). Furthermore, most of the known class 3 integrons possess GCs encoding resistance to β-lactams, which makes this class particularly interesting for our future studies (Arakawa et al., 1995; Papagiannitsis et al., 2015).
The percentage of Aeromonas spp. in WWTP is low and equals up to 9.1% of all bacteria. The percentage of β-lactam unsusceptible bacteria was also low and the highest levels ranged from 39.8 to 1.6%, depending on antibiotic – ceftazidime or meropenem, respectively. However, the results of disk-diffusion phenotypical resistance tests showed a high number of MDR strains among all isolated Aeromonas spp. strains. The majority of the isolates were resistant to at least three different antibiotics (68%), but what is even more concerning, 25% of the strains were resistant to at least six antibiotics. This data stands in accordance with the recent study of Varela et al. (2016), in which MDR Aeromonas sp. strains were also predominant (92.9%). A comparison of MDR isolates between points of isolation did not yield any significant differences. This was also confirmed by means of MAR index values, which were very similar regardless of the origin of the strains. This result suggests that the studied WWTP does not reduce the level of antibiotic resistance in Aeromonas spp. during the treatment process, in contrast to the outcome of other studies (Zhang et al., 2015).
The great majority of studied Aeromonas spp. strains (77%) were unexpectedly unsusceptible to cefepime which is fourth generation cephalosporin. In comparison to the literature, the percentage of cefepime unsusceptible strains in our study was more similar to the results of Picão et al. (2013) from hospital effluent than to WWTP, which were 41.7 and 7% respectively. This is an important issue related to the literature reports which show that clinical strains of Aeromonas spp. are susceptible to cefepime (Wang et al., 2009; Chen P.-L. et al., 2016; Soltan Dallal et al., 2016).
The low number of carbapenem resistance among Aeromonas spp. isolates demonstrated in this study is similar to previous observations. In all recent works the percentage of imipenem, ertapenem, or meropenem resistance in Aeromonas sp. strains among WWTP isolates was at a similar level – here it reached the value of about 10% for each antibiotic. In all studies, including both clinical and environmental strains, the percentage of carbapenem resistant isolates was less than 20% (Janda and Abbott, 2010; Figueira et al., 2011; Picão et al., 2013; Varela et al., 2016).
In this project the percentage of ciprofloxacin unsusceptible Aeromonas spp. strains (6%) was generally lower in comparison to previous works. In a Brazilian study by Picão et al. (2013) 33.3 and 11.3% of total Aeromonas spp. strains isolated from hospital effluent and WWTP, respectively, were resistant to ciprofloxacin. In a Portuguese study by Figueira et al. (2011) and Varela et al. (2016) over 40 and 66% of WWTP Aeromonas spp. strains revealed this phenotype respectively. In Portugal, outpatient usage of quinolones was three times higher than in Poland, which could be an explanation for these differences (Ferech et al., 2006). However, the genetic background of this phenomenon should be studied in the future, especially considering the use of fluoroquinolones as one of the first choice drugs in Aeromonas spp. infections (Parker and Shaw, 2011).
Plasmids are ubiquitous among all prokaryotes, including those inhabiting the WWTP environment, as they play a significant role in horizontal gene transfer and interspecies dissemination of virulence and resistance determinants. In our study, as a result of determining the genomic localization of identified bla genes, blaFOX-4-like, blaGES and blaGES-7 have been found within the genomes of isolated plasmids. Based on literature data, blaGES-7 gene was identified on 60 kb plasmid of A. veronii isolated from Seine River in France (Girlich et al., 2011). Plasmid location of blaFOX genes has never been confirmed among Aeromonas spp. before. Besides that, also blaOXA-1, blaOXA-10, blaCMY -2, and blaCTX-M-15 have already been confirmed as plasmid-mediated among Aeromonas spp. of different environmental origin (Piotrowska and Popowska, 2015). In our study, according to our methodology, besides blaGES and blaFOX, the rest of identified bla genes were located on bacterial chromosomes.
Conclusion
Our major findings in this study were: (i) the identification of ESBL (blaSHV -11, blaCTX-M-27, blaCTX-M-98, and blaPER-4) and AmpC (blaACC, blaFOX-2-like, blaFOX-3, blaFOX-4-like, blaFOX-9, blaFOX-10-like, and blaFOX-13-like) variants of genes, which have never been found among Aeromonas spp. before; (ii) the identification of plasmid-mediated blaGES and blaFOX-4-like genes with the special emphasis on the second type, which have never been observed within plasmid DNA among Aeromonas spp. before; (iii) the lack of significant differences in number of ARGs between points of isolation and (iv) unexpectedly high number of the strains resistant to cefepime. These findings make Aeromonas spp. strains an important research object and in the light of these results the study of the spread of β-lactamase genes and searching for hot spots of their dissemination among Aeromonas spp. seems to be justified. However, our knowledge about Aeromonas spp. resistance in WWTPs is still not complete and requires more comprehensive and in-depth studies, as well as regular monitoring. This seems to be of particular importance especially given the increasing number of Aeromonas spp. infections and MDR strains that are spreading around the world (Esteve et al., 2015; Batra et al., 2016; Qamar et al., 2016).
Author Contributions
MPi: contributed to the establishment and coordination of the collaborations, manuscript design, data collection, data analysis, and drafting and writing of the manuscript. DP and KM: contributed equally to the data collection. MPo: contributed to manuscript design, writing and editing the manuscript, coordination of research and coordination of the collaborations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financed by a grant from the National Centre of Science, Poland (UMO-2014/15/N/NZ9/00439). The authors would like to thank the National Institute of Medicines for giving us DNA of the positive control strains for PCR assay. The authors would like to thank the Czajka Warsaw Wastewater Treatment Plant (Warsaw, Poland) for providing samples of wastewater for our study. The authors would like to also acknowledge the COST Action ES1403 NEREUS “New and emerging challenges and opportunities in wastewater reuse,” supported by COST (European Cooperation in Science and Technology, www.cost.eu) for stimulating the work presented in this paper. We also thank Marcin Studnicki (Department of Experimental Design and Bioinformatics, Warsaw University of Life Science – SGGW, Poland) for statistical analysis, Aleksandra Pawłot (Department of Bacterial Genetics, University of Warsaw, Poland) for linguistic revision and Rafał Ostrowski (Department of Applied Microbiology, Faculty of Biology, University of Warsaw, Poland) for technical support in phylogenetic analysis.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00863/full#supplementary-material
References
- Alibi S., Ferjani A., Boukadida J. (2015). Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella pneumoniae clinical strains from a Tunisian Hospital. Méd. Mal. Infect. 45 139–143. 10.1016/j.medmal.2015.01.010 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amos G. C. A., Hawkey P. M., Gaze W. H., Wellington E. M. (2014). Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 69 1785–1791. 10.1093/jac/dku079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa Y., Murakami M., Suzuki K., Ito H., Wacharotayankun R., Ohsuka S., et al. (1995). A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39 1612–1615. 10.1128/AAC.39.7.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra P., Mathur P., Misra M. C. (2016). Aeromonas spp.: an Emerging Nosocomial Pathogen. J. Lab. Physicians 8 1–4. 10.4103/0974-2727.176234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendonk T. U., Manaia C. M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 310–317. 10.1038/nrmicro3439 [DOI] [PubMed] [Google Scholar]
- Bonnet R. (2004). Growing group of extended-spectrum β-lactamases: the CTX-M Enzymes. Antimicrob. Agents Chemother. 48 1–14. 10.1128/AAC.48.1.1-14.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouki C., Venieri D., Diamadopoulos E. (2013). Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol. Environ. Saf. 91 1–9. 10.1016/j.ecoenv.2013.01.016 [DOI] [PubMed] [Google Scholar]
- Bush K., Fisher J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65 455–478. 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- Chen C.-Y., Wang G.-H., Tseng I.-H., Chung Y.-C. (2016). Analysis of bacterial diversity and efficiency of continuous removal of Victoria Blue R from wastewater by using packed-bed bioreactor. Chemosphere 145 17–24. 10.1016/j.chemosphere.2015.11.061 [DOI] [PubMed] [Google Scholar]
- Chen P.-L., Lamy B., Ko W.-C. (2016). Aeromonas dhakensis, an increasingly recognized human pathogen. Front. Microbiol. 7:793 10.3389/fmicb.2016.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2010). M45-A2 Methods for Antimicrobial Dilution and Disc Susceptibility Testing of Infrequently Isolated of Fastidious Bacteria; Approved Guideline 2nd Edn. Wayne, PA: Clinical and Laboratory Standards Institute [CLSI]. [Google Scholar]
- Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Davies Y. M., Cunha M. P. V., Oliveira M. G. X., Oliveira M. C. V., Philadelpho N., Romero D. C., et al. (2016). Virulence and antimicrobial resistance of Klebsiella pneumoniae isolated from passerine and psittacine birds. Avian. Pathol. 45 194–201. 10.1080/03079457.2016.1142066 [DOI] [PubMed] [Google Scholar]
- Devereux R., Willis S. G. (1995). “Amplification of ribosomal RNA sequences,” in Molecular Microbial Ecology Manual eds Akkermans A. D. L., Van Elsas J. D., De Bruijn F. J. (Dordrecht: Springer; ) 277–287. 10.1007/978-94-011-0351-0_19 [DOI] [Google Scholar]
- Esteve C., Alcaide E., Giménez M. J. (2015). Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 34 137–145. 10.1007/s10096-014-2210-z [DOI] [PubMed] [Google Scholar]
- Ferech M., Coenen S., Malhotra-Kumar S., Dvorakova K., Hendrickx E., Suetens C., et al. (2006). European Surveillance of Antimicrobial Consumption (ESAC): outpatient quinolone use in Europe. J. Antimicrob. Chemother. 58 423–427. 10.1093/jac/dkl183 [DOI] [PubMed] [Google Scholar]
- Figueira V., Vaz-Moreira I., Silva M., Manaia C. M. (2011). Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 45 5599–5611. 10.1016/j.watres.2011.08.021 [DOI] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2010). PER-6 an extended-spectrum β-lactamase from Aeromonas allosaccharophila. Antimicrob. Agents Chemother. 54 1619–1622. 10.1128/AAC.01585-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2011). Diversity of clavulanic acid-inhibited extended-spectrum β-lactamases in Aeromonas spp. from the Seine River, Paris, France. Antimicrob. Agents Chemother. 55 1256–1261. 10.1128/AAC.00921-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H. (2005). β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56 115–121. 10.1093/jac/dki190 [DOI] [PubMed] [Google Scholar]
- Henriques I. S., Fonseca F., Alves A., Saavedra M. J., Correia A. (2006). Occurrence and diversity of integrons and β-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 157 938–947. 10.1016/j.resmic.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Hu Q., Zhang X.-X., Jia S., Huang K., Tang J., Shi P., et al. (2016). Metagenomic insights into ultraviolet disinfection effects on antibiotic resistome in biologically treated wastewater. Water Res. 101 309–317. 10.1016/j.watres.2016.05.092 [DOI] [PubMed] [Google Scholar]
- Igbinosa I. H., Okoh A. I. (2012). Antibiotic susceptibility profile of Aeromonas species isolated from wastewater treatment plant. ScientificWorldJournal 2012:764563 10.1100/2012/764563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs L., Chenia H. Y. (2007). Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int. J. Food Microbiol. 114 295–306. 10.1016/j.ijfoodmicro.2006.09.030 [DOI] [PubMed] [Google Scholar]
- Jacoby G. A. (2009). AmpC β-Lactamases. Clin. Microbiol. Rev. 22 161–182. 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23 35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer K. (2009). Antibiotics in the aquatic environment–a review–part II. Chemosphere 75 435–441. 10.1016/j.chemosphere.2008.12.006 [DOI] [PubMed] [Google Scholar]
- Liu B., Pop M. (2009). ARDB—antibiotic resistance genes database. Nucleic Acids Res. 37 D443–D447. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang Y., Wang G., Xing Q., Shao L., Dong X., et al. (2015). The prevalence of Escherichia coli strains with extended spectrum beta-lactamases isolated in China. Front. Microbiol. 6:335 10.3389/fmicb.2015.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.-Y., Zhang Y.-L., Geng S.-N., Li T.-Y., Ye Z.-M., Zhang D.-S., et al. (2010). High diversity of extended-spectrum beta-lactamase-producing bacteria in an urban river sediment habitat. Appl. Environ. Microbiol. 76 5972–5976. 10.1128/AEM.00711-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravić A., Skočibušić M., Šamanić I., Fredotović Ž, Cvjetan S., Jutronić M., et al. (2013). Aeromonas spp. simultaneously harbouring blaCTX-M-15, blaSHV-12, blaPER-1 and blaFOX-2, in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol. 166 301–308. 10.1016/j.ijfoodmicro.2013.07.010 [DOI] [PubMed] [Google Scholar]
- Markovska R., Schneider I., Marteva-Proevsk Y., Mitov I., Bauernfeind A., Markova B. (2012). First detection of the AmpC beta-lactamase ACC-1 in a Klebsiella pneumoniae isolate in Bulgaria. J. Chemother. 24 307–308. 10.1179/1973947812Y.0000000025 [DOI] [PubMed] [Google Scholar]
- Marti E., Huerta B., Rodríguez-Mozaz S., Barceló D., Jofre J., Balcázar J. L. (2014). Characterization of ciprofloxacin-resistant isolates from a wastewater treatment plant and its receiving river. Water Res. 61 67–76. 10.1016/j.watres.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Pitout J. D. D., Gomi R., Matsuda T., Noguchi T., Yamamoto M., et al. (2016). Global Escherichia coli Sequence Type 131 Clade with bla CTX-M-27 Gene. Emerg. Infect. Dis. 22 1900–1907. 10.3201/eid2211.160519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A., Henriques I., Ribeiro R., Correia A. (2007). Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 60 1243–1250. 10.1093/jac/dkm340 [DOI] [PubMed] [Google Scholar]
- Novo A., Manaia C. M. (2010). Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 87 1157–1166. 10.1007/s00253-010-2583-6 [DOI] [PubMed] [Google Scholar]
- Papagiannitsis C. C., Dolejska M., Izdebski R., Dobiasova H., Studentova V., Esteves F. J., et al. (2015). Characterization of pKP-M1144, a Novel ColE1-Like Plasmid Encoding IMP-8, GES-5, and BEL-1 β-Lactamases, from a Klebsiella pneumoniae Sequence Type 252 Isolate. Antimicrob. Agents Chemother. 59 5065–5068. 10.1128/AAC.00937-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. L., Shaw J. G. (2011). Aeromonas spp. clinical microbiology and disease. J. Infect. 62 109–118. 10.1016/j.jinf.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Perez-Perez F. J., Hanson N. D. (2002). Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40 2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picão R. C., Cardoso J. P., Campana E. H., Nicoletti A. G., Petrolini F. V. B., Assis D. M., et al. (2013). The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 76 80–85. 10.1016/j.diagmicrobio.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Picão R. C., Poirel L., Demarta A., Silva C. S. F., Corvaglia A. R., Petrini O., et al. (2008). Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62 948–950. 10.1093/jac/dkn341 [DOI] [PubMed] [Google Scholar]
- Piotrowska M., Popowska M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64 921–934. 10.1007/s13213-014-0911-2 [DOI] [Google Scholar]
- Piotrowska M., Popowska M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494 10.3389/fmicb.2015.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar F. N., Nisar M. I., Quadri F., Shakoor S., Sow S. O., Nasrin D., et al. (2016). Aeromonas-associated diarrhea in children under 5 years: the GEMS experience. Am. J. Trop. Med. Hyg. 95 774–780. 10.4269/ajtmh.16-0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; Available at: https://www.R-project.org [Google Scholar]
- Rizzo L., Manaia C., Merlin C., Schwartz T., Dagot C., Ploy M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447 345–360. 10.1016/j.scitotenv.2013.01.032 [DOI] [PubMed] [Google Scholar]
- Rodrigues C., Machado E., Fernandes S., Peixe L., Novais Â. (2016). An update on faecal carriage of ESBL-producing Enterobacteriaceae by Portuguese healthy humans: detection of the H 30 subclone of B2-ST131 Escherichia coli producing CTX-M-27: Table 1. J. Antimicrob. Chemother. 71 1120–1122. 10.1093/jac/dkv443 [DOI] [PubMed] [Google Scholar]
- Segatore B., Massidda O., Satta G., Setacci D., Amicosante G. (1993). High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance. Antimicrob. Agents Chemother. 37 1324–1328. 10.1128/AAC.37.6.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R., Joseph S. W., Chopra A. K., Sha J., Shaw J., Graf J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188 8272–8282. 10.1128/JB.00621-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahraki-Zahedani S., Rigi S., Bokaeian M., Ansari-Moghaddam A., Moghadampour M. (2016). First report of TEM-104-, SHV-99-, SHV-108-, and SHV-110-producing Klebsiella pneumoniae from Iran. Rev. Soc. Bras. Med. Trop. 49 441–445. 10.1590/0037-8682-0114-2016 [DOI] [PubMed] [Google Scholar]
- Simo Tchuinte P. L., Stalder T., Venditti S., Ngandjio A., Dagot C., Ploy M.-C., et al. (2016). Characterisation of class 3 integrons with oxacillinase gene cassettes in hospital sewage and sludge samples from France and Luxembourg. Int. J. Antimicrob. Agents 48 431–434. 10.1016/j.ijantimicag.2016.06.018 [DOI] [PubMed] [Google Scholar]
- Soltan Dallal M. M., Mazaheri Nezhad Fard R., Kavan Talkhabi M., Aghaiyan L., Salehipour Z. (2016). Prevalence, virulence and antimicrobial resistance patterns of Aeromonas spp. isolated from children with diarrhea. Germs 6 91–96. 10.11599/germs.2016.1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder T., Barraud O., Casellas M., Dagot C., Ploy M.-C. (2012). Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119 10.3389/fmicb.2012.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela A. R., Nunes O. C., Manaia C. M. (2016). Quinolone resistant Aeromonas spp. as carriers and potential tracers of acquired antibiotic resistance in hospital and municipal wastewater. Sci. Total Environ. 542 665–671. 10.1016/j.scitotenv.2015.10.124 [DOI] [PubMed] [Google Scholar]
- Verner-Jeffreys D. W., Welch T. J., Schwarz T., Pond M. J., Woodward M. J., Haig S. J., et al. (2009). High prevalence of multidrug-tolerant bacteria and associated antimicrobial resistance genes isolated from ornamental fish and their carriage water. PLoS ONE 4:e8388 10.1371/journal.pone.0008388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolaid V., Tenson T., Kisand V. (2013). Aeromonas and Pseudomonas species carriers of ampC FOX genes in aquatic environments. Appl. Environ. Microbiol. 79 1055–1057. 10.1128/AEM.03171-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H., Wang C. Y., Chi C. Y., Ho M. W., Ho C. M., Lin P. C. (2009). Clinical presentations, prognostic factors, and mortality in patients with Aeromonas sobria complex bacteremia in a teaching hospital: a 5-year experience. J. Microbiol. Immunol. Infect. 42 510–515. [PubMed] [Google Scholar]
- World Health Organization [WHO] (2013). Available at: http://www.who.int/drugresistance/activities/wha66_side_event/en/ [Google Scholar]
- Wu C.-J., Chen P.-L., Wu J.-J., Yan J.-J., Lee C.-C., Lee H.-C., et al. (2012). Distribution and phenotypic and genotypic detection of a metallo-β-lactamase, CphA, among bacteraemic Aeromonas isolates. J. Med. Microbiol. 61 712–719. 10.1099/jmm.0.038323-0 [DOI] [PubMed] [Google Scholar]
- Zhang S., Han B., Gu J., Wang C., Wang P., Ma Y., et al. (2015). Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere 135 138–145. 10.1016/j.chemosphere.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Zhang X.-X., Zhang T., Fang H. H. (2009). Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 82 397–414. 10.1007/s00253-008-1829-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


