Abstract
The “integrated diagnosis” for infiltrating gliomas in the 2016 revised World Health Organization (WHO) classification of tumors of the central nervous system requires assessment of the tumor for IDH mutations and 1p/19q codeletion. Since TERT promoter mutations and ATRX alterations have been shown to be associated with prognosis, we analyzed whether these tumor markers provide additional prognostic information within each of the five WHO 2016 categories. We used data for 1206 patients from the UCSF Adult Glioma Study, the Mayo Clinic and The Cancer Genome Atlas (TCGA) with infiltrative glioma, grades II–IV for whom tumor status for IDH, 1p/19q codeletion, ATRX, and TERT had been determined. All cases were assigned to one of 5 groups following the WHO 2016 diagnostic criteria based on their morphologic features, and IDH and 1p/19q codeletion status. These groups are: 1-Oligodendroglioma, IDH-mutant and 1p/19q-codeleted; 2- Astrocytoma, IDH-mutant; 3- Glioblastoma, IDH-mutant; 4- Glioblastoma, IDH-wildtype; and 5-Astrocytoma, IDH-wildtype. Within each group, we used univariate and multivariate Cox proportional hazards models to assess associations of overall survival with patient age at diagnosis, grade, and ATRX alteration status and/or TERT promoter mutation status. Among Group 1 IDH-mutant 1p/19q-codeleted oligodendrogliomas, the TERT-WT group had significantly worse overall survival than the TERT-MUT group (HR: 2.72, 95%CI: 1.05–7.04, p=0.04). In both Group 2, IDH-mutant astrocytomas and Group 3, IDH-mutant glioblastomas, neither TERT mutations nor ATRX alterations were significantly associated with survival. Among Group 4, IDH-wildtype glioblastomas, ATRX alterations were associated with favorable outcomes (HR: 0.36, 95% CI: 0.17–0.81, p=0.01). Among Group 5, IDH-wildtype astrocytomas, the TERT-WT group had significantly better overall survival than the TERT-MUT group (HR: 0.48, 95% CI: 0.27–0.87), p=0.02). Thus, we present evidence that in certain WHO 2016 diagnostic groups, testing for TERT promoter mutations or ATRX alterations may provide additional useful prognostic information.
Keywords: Glioma classification, ATRX alteration, TERT promoter mutation, brain tumor prognosis, telomere maintenance
INTRODUCTION
A large body of literature indicates that certain glioma molecular alterations define subgroups that are prognostic and can be used in the clinical management of infiltrating glioma patients. Two molecular alterations that have well-established associations with prognosis are isocitrate dehydrogenase (IDH) mutations and 1p/19q-codeletions [6]. Because these two markers separated gliomas into more biologically distinct entities than histological classification alone, the World Health Organization (WHO) incorporated IDH mutation and 1p/19q codeletion into an “integrated diagnosis” in the 2016 revised 4th edition of the classification of tumors of the central nervous system [15].
Alterations in two telomere maintenance-related genes, telomerase reverse transcriptase (TERT) and Alpha Thalassemia/Mental Retardation Syndrome X-Linked (ATRX) have also been the subject of many investigations into glioma classification and prognosis. Telomeres, the nucleoprotein complexes at the ends of all eukaryotic chromosomes, are composed of several hundred nucleotide repeats which progressively shorten with each mitosis. Telomerase is a reverse transcriptase that carries its own RNA molecule used as a template to add nucleotides to telomeres. Tumors maintain their telomere length either via re-activation of telomerase or through telomerase-independent mechanisms collectively called alternative lengthening of telomeres (ALT). Hotspot mutations in the TERT promoter lead to increased telomerase activity and are found in gliomas and many other tumors [11]. TERT promoter mutations occur in 70–83% of glioblastomas, 74–78% of oligodendrogliomas, 25–50% of oligoastrocytomas and 10–25% of astrocytomas [1, 6, 11]. Many cancers with ALT harbor mutations in ATRX or death-domain associated protein (DAXX) genes encoding ATRX and DAXX proteins, which are central components of the chromatin remodeling complex required for the incorporation of H3.3 histone proteins into telomeres [8–10]. ATRX mutations occur in nearly 75% of grade II–III astrocytomas and secondary glioblastomas [9, 10, 13]. ATRX mutations are widely distributed across the gene and are mostly truncating (including frameshift and nonsense variants) and less commonly missense mutations [8–10]. Loss of ATRX protein expression on immunohistochemistry (IHC) can be used as a surrogate marker of ATRX mutations with high sensitivity and specificity and is near perfectly correlated with ALT pathway activation [8–10]. In some glioma subgroups ATRX alterations and TERT promoter mutations may be mutually exclusive, likely due to functional redundancy. Over 90% of IDH-mutant diffuse gliomas have mutations in one or both genes [3, 6].
Alterations in TERT and ATRX are not only associated with certain histologic subgroups of gliomas, but also are associated with variable prognosis [3, 6, 12, 16, 21, 26, 28]. Our research team previously showed that molecular classification of gliomas based on IDH, TERT promoter mutations and 1p/19q codeletion status yields five subgroups that are independently associated with prognosis in grade II and III gliomas [6]. A similar classification of grade II and III infiltrating gliomas using only IDH and TERT promoter mutation status was also suggested [28]. Other groups demonstrated a role for ATRX in the classification of gliomas [12, 16]. Data from The Cancer Genome Atlas (TCGA) suggest lower grade gliomas (WHO grades II and III) can be grouped using IDH and p53 mutation and 1p/19q codeletion status; ATRX inactivation differs significantly among these groups [3]. Others showed that IDH, 1p/19q and ATRX can be used in clinical classification [21].
The literature regarding glioma telomere maintenance mechanisms has been rapidly growing. Their prognostic and predictive roles are of great interest and may guide clinical management of glioma patients. As of yet, there are no studies specifically analyzing the distribution and importance of ATRX alterations and TERT promoter mutations among diagnostic entities in the WHO 2016 classification. The purpose of this study is to evaluate the distribution of ATRX alterations and TERT promoter mutations among each of the five major WHO 2016 diagnostic categories of infiltrating glioma, and to analyze whether either or both of these two markers provide additional prognostic information in each category.
MATERIAL AND METHODS
Study population
We included all patients with infiltrative glioma, grades II, III, and IV from the archives of the University of California, San Francisco (UCSF) Adult Glioma Study (AGS), the Mayo Clinic glioma case-control study and The Cancer Genome Atlas (TCGA) for whom tumor status for IDH mutations, 1p/19q codeletion, ATRX alterations, and TERT promoter mutations had been determined. All TCGA cases in this current study were previously included in Ceccarelli et al.[3], and all Mayo Clinic cases and 216 of 347 UCSF cases were previously included in Eckel-Passow et al. [6]. Because cases in this current study were not population based, the percent distributions of the glioma diagnostic groups will not be representative of the general population. In particular, grade II/III patients are over-represented in this study.
Assessment of histology and molecular features
Two pathologists (TT and CG) reviewed histologic sections for UCSF AGS and Mayo Clinic cases as previously described [6, 27]. Pathology data for TCGA cases were obtained from Supplemental Table S1 in Ceccarelli et al. [3]. For UCSF AGS and Mayo Clinic cases, 5-micron sections obtained from routinely processed formalin-fixed paraffin embedded (FFPE) tissue were used for IHC and FISH, and DNA extracted from the FFPE tissue using standard methods was used for sequencing. Status of all molecular alterations for the TCGA cases presented here was obtained from Ceccarelli et al.[3].
IDH1 and IDH2 mutation
IDH mutation status for UCSF AGS cases was evaluated by Sanger sequencing of IDH1 and IDH2 genes or by IHC (IDH1R132H, H09, Dianova GmbH, Hamburg, Germany) using standard techniques [6]. Data solely from IHC results were used only when the IHC staining was positive for the presence of mutation; negative IHC results were validated by sequencing. IDH mutation status for all Mayo Clinic and TCGA cases was assessed via Sanger sequencing. IDH mutation denotes either an IDH1 or IDH2 mutation in the tumor [3, 6].
ATRX mutation or loss of expression
ATRX alterations for UCSF AGS cases were assessed by IHC (HPA001906, Sigma Aldrich, St. Louis, MO) performed at the UCSF Brain Tumor Research Center using previously-published methods [8]. Briefly, loss of nuclear staining in the majority of the tumor cells in the presence of an internal positive control was interpreted as loss of ATRX expression [6]. IHC for Mayo Clinic cases was performed at the Memorial Sloan-Kettering Cancer Center or at the Mayo Clinic using the same methods and interpreted by the same criteria [6]. ATRX data for TCGA cases reflected sequencing-based somatic mutations as described in Ceccarelli et al. [3]. ATRX-mutant (ATRX-MUT) denotes loss of ATRX expression for UCSF AGS and Mayo Clinic cases, and presence of any exonic mutation for TCGA cases. ATRX-wildtype (ATRX-WT) denotes retained nuclear expression of ATRX for UCSF AGS and Mayo Clinic cases and wildtype ATRX for TCGA cases.
1p/19q codeletion
1p/19q codeletion for UCSF AGS cases was assessed using clinical FISH assays. Tumors with 1p/19q codeletion almost invariably have IDH and TERT promoter mutations and 1p/19q codeletion is almost mutually exclusive with ATRX mutations. Therefore, IDH-wild type gliomas and IDH-mutant gliomas with ATRX alterations were not tested for 1p/19q codeletion unless it was previously performed for clinical reasons. These tumors were classified as 1p/19q-intact for the purposes of this study. 1p/19q codeletion status was assessed in all Mayo Clinic cases either by FISH as a clinical test or by array comparative genomic hybridization (aCGH) using an Agilent custom 8×60K array. 1p/19q codeletion status for the TCGA cases was obtained from Supplemental Table S1 in Ceccarelli et al. [3].
TERT promoter mutation
TERT promoter mutation status for UCSF AGS and the Mayo Clinic cases was assessed by a previously published method [6] using a 2-step PCR and Sanger sequencing of a 244 base pair segment of TERT promoter region spanning the C228T and C250T mutations to assign cases as TERT-mutant (TERT-MUT) or TERT-wildtype (TERT-WT). For UCSF AGS cases, mutation calls were independently made by two reviewers (MP, HMH), and a consensus call was used for discrepant results. At the Mayo Clinic, cases were re-reviewed if the results from two pathologists were ambiguous, and a consensus call was used for discrepant results. For the TCGA cases, TERT promoter mutation status was obtained from sequencing data if available and expression data if sequencing data was not available [3].
Study groups
All cases were assigned to one of five study groups following the WHO 2016 diagnostic criteria based on their morphologic features and IDH and 1p/19q codeletion status [14]. These groups are: 1-Oligodendroglioma, IDH-mutant and 1p/19q-codeleted, including WHO grades II and III; 2- Astrocytoma, IDH-mutant, including WHO grades II and III; 3- Glioblastoma, IDH-mutant; 4- Glioblastoma, IDH-wildtype; and 5- Astrocytoma, IDH-wildtype, including both morphologic oligodendroglioma and astrocytoma and both WHO grades II and III. A subset of cases, originally diagnosed as anaplastic oligodendroglioma or anaplastic oligoastrocytoma, showing IDH mutations but not 1p/19q codeletion or lacking IDH mutations, were re-reviewed and appropriately reclassified according to the WHO 2016 diagnostic criteria as either astrocytoma or glioblastoma based on the absence/presence of microvascular proliferation and/or necrosis.
Statistical Methods
Differences in age at diagnosis by histologic grade and molecular alterations in each WHO 2016 group were evaluated and compared using the Kruskal-Wallis test. Within each group, separate Cox proportional hazards univariate models were used to determine whether ATRX alterations and TERT promoter status were associated with overall survival. When appropriate, based on the fit of the model, multivariate models within each group included adjustment or stratification for age at diagnosis and histologic grade. In order to assess the independent effects of TERT and ATRX status within each group, we present separate multivariate models for each group that includes both TERT and ATRX status whether or not each was statistically significant in the corresponding univariate model. All models were tested for assumptions and stratification was used if proportionality assumptions were violated. For the three groups in which stratification by age was necessary (Groups 1, 4 and 5), we determined the best age groupings by examining hazard ratios for each decile of age at diagnosis and grouping deciles with similar hazard ratios. The resulting age strata cut points are listed below each table in which results from models with age stratification are presented. ATRX alteration status and TERT promoter mutation status were included in each model along with an interaction term whenever applicable; since none of the interactions were statistically significant, we do not present the interaction results. Hazard ratios and confidence intervals were calculated for all models; the referent group used for each categorical variable (grade, TERT or ATRX status) was that with the largest frequency. A p value less than 0.05 was considered statistically significant. Survival curves stratified by either TERT or ATRX status are generated from Cox proportional hazards models with adjustment for age and/or grade as appropriate. All analyses were conducted in R version 3.3.0 [20]. Among relevant clinical and treatment variables (i.e. extent of resection, performance status, chemotherapy type, and radiation), the only variable that was consistently collected in all three data sources was radiation treatment (given versus not given). We included radiation history in secondary analyses of each of the multivariate models presented below and found that radiation treatment did not confound any of the multivariate results with respect to statistical significance of TERT or ATRX alterations (results not shown). Other clinically relevant molecular markers such as MGMT methylation and TP53 mutation were also not available for a sufficient number of cases to assess their impact on the findings.
RESULTS
Of 1208 glioma cases identified for these analyses, two cases with 1p/19q codeletion but without IDH mutations were not included in further analyses. Of the remaining 1206 cases, 347 were from the UCSF AGS, 296 were from the Mayo Clinic glioma study and 563 were from TCGA (Table 1). Data for the 643 UCSF and Mayo cases are provided as supplementary material (Supplementary Table 1). These 1206 study cases were assigned to the WHO 2016 groups as follows: Group 1-Oligodendroglioma, IDH-mutant and 1p/19q- codeleted (n=291), Group 2- Astrocytoma, IDH-mutant (n=401), Group 3- Glioblastoma, IDH-mutant (n=51), Group 4- Glioblastoma, IDH-wildtype (n=309), and Group 5- Astrocytoma, IDH-wildtype (n=154). Age, median survival, TERT and ATRX status and other characteristics of each group are also shown in Table 1. Univariate survival statistics assessing the entire study population showed distinct survival curves for groups 1 through 4 corresponding to most favorable through least favorable survival outcomes (Figure 1); Group 5 astrocytoma, IDH-wildtype patients had intermediate survival between Group 3 IDH-mutant glioblastoma and Group 4 IDH-wildtype glioblastoma for approximately six years after diagnosis, after which, survival is more similar to Group 3 patients.
Table 1.
Demographics, survival, study site and combined TERT/ATRX status by integrated 2016 WHO glioma entities.
| 2016 WHO Group | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| All | Group 1 ligodendroglioma, IDH-mutant and 1p/19q-codeleted |
Group 2 Astrocytoma, IDH-mutant |
Group 3 Glioblastoma, IDH-mutant |
Group 4 Glioblastoma, IDH-wildtype |
Group 5 Astrocytoma, IDH-wildtype |
|
| Total | 1206 | 291 | 401 | 51 | 309 | 154 |
|
| ||||||
| Age mean (median) | 47 (46) | 45 (44) | 38 (36) | 42 (38) | 59 (59) | 50 (52) |
| Male gender n (%) | 722 (60%) | 167 (57%) | 243 (61%) | 36 (71%) | 192 (62%) | 84 (55%) |
| Percent deceased | 45% | 19% | 30% | 67% | 81% | 57% |
| Median survival (years) | 5.3 | 17.5 | 9.3 | 3.6 | 1.2 | 1.9 |
| Study Site | ||||||
| UCSF | 347 (29%) | 86 (30%) | 116 (29%) | 17 (33%) | 76 (25%) | 52 (34%) |
| Mayo | 296 (25%) | 68 (23%) | 87 (22%) | 25 (49%) | 97 (31%) | 19 (12%) |
| TCGA | 563 (47%) | 137 (47%) | 198 (49%) | 9 (18%) | 136 (44%) | 83 (54%) |
| TERT/ATRX | ||||||
| WT/WT | 187 (16%) | 11 (4%) | 63 (16%) | 10 (20%) | 60 (19%) | 43 (28%) |
| WT/MUT | 371 (31%) | 2 (1%) | 311 (78%) | 32 (63%) | 10 (3%) | 16 (10%) |
| MUT/WT | 629 (52%) | 273 (94%) | 20 (5%) | 6 (12%) | 238 (77%) | 92 (60%) |
| MUT/MUT | 19 (2%) | 5 (2%) | 7 (2%) | 3 (6%) | 1 (0%) | 3 (2%) |
| TERT-MUT n (column %) | 648 (54%) | 278 (96%) | 27 (7%) | 9 (18%) | 239 (77%) | 95 (62%) |
| ATRX-MUTa n (column %) | 390 (32%) | 7 (2%) | 318 (79%) | 35 (69%) | 11 (4%) | 19 (12%) |
UCSF and Mayo Clinic cases has ATRX alterations identified by immunohistochemistry while TCGA cases had ATRX alterations identified by sequencing. ATRX alterations were found in 36% of UCSF cases, 35% of Mayo cases and 28% of TCGA cases.
Fig. 1.
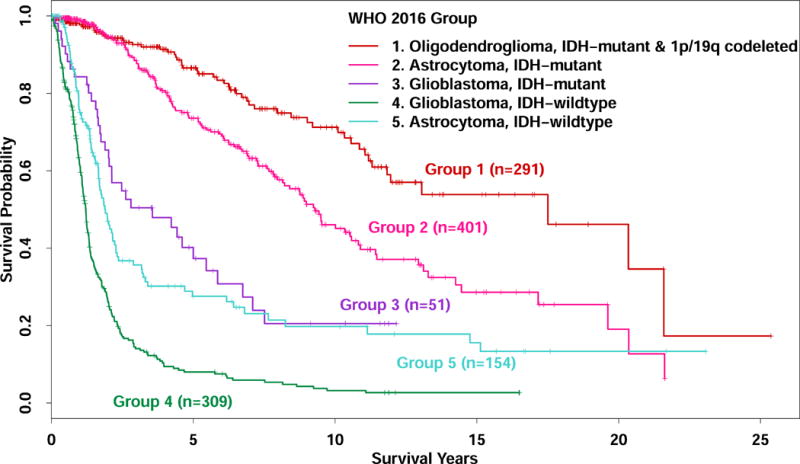
Kaplan-Meier survival curves for glioma cases classified into five WHO 2016 entities. (n=1206)
Group1 - Oligodendroglioma, IDH-mutant and 1p/19q-codeleted
Group 1 included 173 grade II and 118 grade III gliomas per WHO 2016 grading criteria with a median age at diagnosis of 44 (range 20–75) years (Table 2). Patients with grade II tumors were significantly younger than those with grade III tumors (median ages 42 and 48, respectively, Kruskal-Wallis test p=0.006). As shown in Table 1 and Figure 2a, almost all cases (n=273, 94%) had TERT promoter mutations only, five (1.7%) cases demonstrated both TERT and ATRX alterations, eleven (4%) had neither TERT nor ATRX alterations, and two had only ATRX alterations. In univariate analyses (Table 2), higher age at diagnosis, higher grade, and lack of TERT promoter mutation were significantly associated with decreased overall survival. In the final multivariate model which included grade, TERT and ATRX status and was stratified by age at diagnosis, neither grade nor TERT status were significantly associated with overall survival. (HR for grade: 1.61, 95%CI: 0.92–2.82, p=0.10HR for TERT: 2.46, 95%CI: 0.94–6.42, p=0.07). However, in a multivariate model excluding grade, absence of TERT mutation was significantly associated with worse survival (HR: 2.72, 95% CI: 1.05–7.04, p=0.04) (Table 2a and Figure 2a). With respect to age at diagnosis, patients whose tumors that had TERT but did not have ATRX mutations (n=273, median age at diagnosis 44), did not significantly differ in age from those with tumors that had both TERT and ATRX mutations (n=5, median age at diagnosis 38) (Kruskal-Wallis test p=0.99) or from those patients with tumors that had neither TERT nor ATRX mutations (n=11, median age at diagnosis 46) (Kruskal-Wallis p=0.28). In a multivariate model controlling for grade and stratified by age, compared to cases with only TERT mutation, the cases with neither TERT nor ATRX alterations had significantly worse survival (HR: 2.61, 95%CI:1.00–6.85, p=0.05). For nine of 13 TERT-wildtype tumors, we were able to confirm that the 1p/19q codeletions were indeed whole arm deletions (not partial arm deletions) by manual examination of tracings from genomic data, and of these one showed ATRX mutation by sequencing. The remaining four of the 13 TERT-wildtype tumors had 1p/19q codeletion tested by FISH, and none showed evidence of co-polysomy. Of these, three were TP53-wildtype, and one did not have TP53 mutation data, but showed loss of ATRX expression by IHC.
Table 2.
Associations of age at diagnosis, tumor grade and TERT and ATRX status with overall survival in Group 1: IDH-mutant and 1p/19q-codeleted oligodendrogliomas.
| N (column %) | Age Median (range) | Percent deceased | Median survival years (95% CI) | Univariate analysis | Multivariate analysisb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |||||
| All/Age (continuous) | 291 (100%) | 44 (20–75) | 19% | 17.5 (11.8-NA) | 1.08 | (1.05–1.11) | <0.0001 | |||
|
| ||||||||||
| Grade II | 173 (59%) | 42 (20–74)a | 16% | 17.5 (13.1-NA) | Ref | Ref | ||||
| Grade III | 118 (41%) | 48 (22–75)a | 22% | 11.2 (8.5-NA) | 2.00 | (1.17–3.42) | 0.01 | 1.61 | (0.92–2.82) | 0.10 |
|
| ||||||||||
| TERT-WT | 13 (4%) | 46(27–74) | 38% | 6.9 (5.1-NA) | 3.25 | (1.27–8.29) | 0.01 | 2.46 | (0.94–6.42) | 0.07 |
| TERT-MUT | 278 (96%) | 44 (20–75) | 18% | 17.5 (12-NA) | Ref | Ref | ||||
|
| ||||||||||
| ATRX-WT | 284 (98%) | 44(20–75) | 19% | 17.5 (11.8-NA) | Ref | Ref | ||||
| ATRX-MUT | 7 (2%) | 46 (31–74) | 14% | 11.3 (11.1-NA) | 0.69 | (0.1–5.03) | 0.72 | 0.57 | (0.08–4.17) | 0.58 |
Patients with grade III tumors were significantly older at diagnosis than patients with grade II tumors (p=0.006).
Multivariate model includes grade, TERT and ATRX status and was stratified by age (grouped as 20–37, 38–52, and 53–75) because age did not meet the Cox model’s proportionality assumptions. In a multivariate model excluding grade, TERT status became statistically significant (HR: 2.72, 95%CI: 1.05–7.04, p=0.04) and ATRX status remained not significantly associated with survival (HR: 0.55, 95%CI: 0.08–4.07, p=0.56)
Bolded values are statistically significant; 95% CI: 95% confidence interval; Ref: reference group.
Fig. 2.
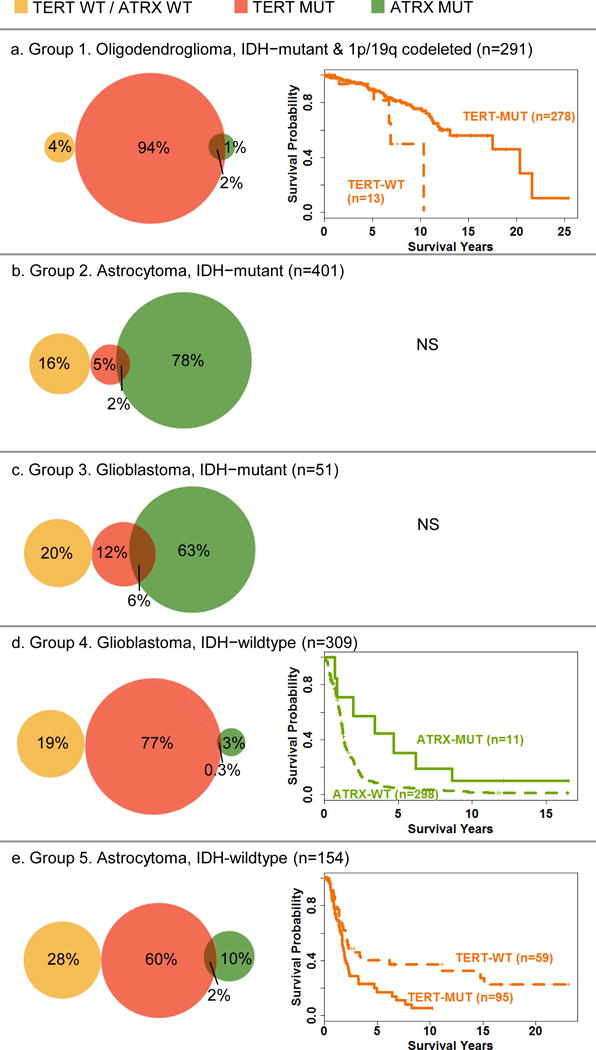
Venn diagrams showing the distribution of TERT/ATRX status by WHO 2016 groups and age (and grade when appropriate) adjusted Cox survival curves for WHO 2016 groups that have a statistically significant association of TERT or ATRX with survival in multivariate models. Vertical lines on survival curves indicated censored observations. NS=No statistically significant associations for survival were found for TERT or ATRX in multivariate models. Note that some percents add up to more than 100 due to rounding errors.
Group 2 - Astrocytoma, IDH-mutant (1p/19q-intact)
Group 2 included 220 grade II and 181 grade III gliomas per WHO 2016 grading criteria with a median age at diagnosis of 36 (range 18–73) years (Table 3). There was no difference between patients with grade II and grade III tumors for age at diagnosis (median age was 37 and 36 years respectively, p=0.60). As shown in Table 1 and Figure 2b, although most cases (n=311, 78%) had ATRX alterations only, seven (2%) cases had both TERT and ATRX alterations, 20 (5%) had TERT mutations only, and 63 (16%) had neither TERT nor ATRX alterations. In univariate analyses (Table 3), older age at diagnosis and higher grade were significantly associated with worse overall survival (HR for age: 1.02, 95%CI: 1.01–1.04, p=0.01; HR for grade: 1.45, 95%CI: 1.01–2.07, p=0.04), but neither TERT nor ATRX status was associated with survival. In a multivariate model including age, TERT and ATRX status, stratified by grade, only older age at diagnosis was significantly associated with worse overall survival (HR: 1.02, 95%CI: 1.01–1.04, p=0.009).
Table 3.
Associations of age at diagnosis, TERT and ATRX status with overall survival in Group 2: IDH-mutant (1p/19q-intact) astrocytomas.
| N (column %) | Age Median (range) | Percent deceased | Median survival years (95% CI) | Univariate analysis | Mulivariate analysisd | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |||||
| All/Age (continuous) | 401 (100%) | 36 (18–73) | 30% | 9.3 (8.2–10.8) | 1.02 | (1.01–1.04) | 0.01 | 1.02 | (1.01–1.04) | 0.009 |
|
| ||||||||||
| Grade II | 220 (55%) | 37 (18–73)a | 29% | 9.4 (8.7–12.9) | Ref | |||||
| Grade III | 181 (45%) | 36 (18–72)a | 32% | 8.9 (6.7–13.1) | 1.45 | (1.01–2.07) | 0.04 | |||
|
| ||||||||||
| TERT-WT | 374 (93%) | 36 (18–73)b | 30% | 9.3 (8.2–10.8) | Ref | Ref | ||||
| TERT-MUT | 27 (7%) | 39 (20–66)b | 30% | 9 (7.6-NA) | 0.75 | (0.36–1.55) | 0.43 | 0.68 | (0.31–1.52) | 0.35 |
|
| ||||||||||
| ATRX-WT | 83 (21%) | 39 (20–69)c | 19% | 8.2 (5.3-NA) | 1.09 | (0.64–1.85) | 0.76 | 1.18 | (0.65–2.12) | 0.59 |
| ATRX-MUT | 318 (79%) | 35 (18–73)c | 33% | 9.4 (8.4–10.9) | Ref | Ref | ||||
Patients with grade II and III tumors had similar age at diagnosis (p= 0.60).
Patients with TERT-mutant tumors were older at diagnosis than those with TERT-wildtype tumors (p=0.007).
Patients with ATRX-mutant tumors were younger at diagnosis than those with ATRX-wildtype tumors (p=0.0004).
Multivariate model includes age, TERT and ATRX status stratified by grade because it did not meet the proportionality assumption.
Bolded values are statistically significant; 95% CI: 95% confidence interval; Ref: reference group.
Group 3 - Glioblastoma, IDH-mutant
Group 3 had 51 cases with median age at diagnosis of 38 (range 21–78) years (Table 4). Patients with tumors with TERT promoter mutations were significantly older than those with TERT-wildtype tumors (median ages 60 and 38, respectively, p=0.006). Patients with tumors that had ATRX alterations were younger at diagnosis than those who had ATRX-wildtype tumors (median ages 35 and 46 years, respectively, p=0.007). As shown in Table 1 and Figure 2c, the majority of the tumors in this group (n=32, 63%) had ATRX alterations only, three (6%) cases had both TERT and ATRX alterations, six (12%) had TERT mutations only, and 10 (20%) had neither TERT nor ATRX alterations. In univariate analyses, older age at diagnosis and presence of TERT promoter mutations were associated with worse overall survival. There was no significant association between ATRX status and survival (p=0.36). In a multivariate analysis including age, TERT status, and ATRX status, only older age remained significantly associated with shorter overall survival.
Table 4.
Associations of age at diagnosis, and tumor TERT and ATRX status with overall survival in Group 3: IDH-mutant glioblastomas
| N (column %) | Age Median (range) | Percent deceased | Median survival years (95% CI) | Univariate analysis | Multivariate analysisc | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |||||
| All/Age (continuous) | 51 (100%) | 38 (21–78) | 67% | 3.6 (2–5.9) | 1.05 | (1.02–1.08) | 0.002 | 1.04 | (1.01–1.08) | 0.01 |
|
| ||||||||||
| TERT-WT | 42 (82%) | 38 (21–63)a | 62% | 4.4 (2.5–7.5) | Ref | Ref | ||||
| TERT-MUT | 9 (18%) | 60 (33–78)a | 89% | 1.6 (0.8-NA) | 2.58 | (1.14–5.83) | 0.02 | 1.81 | (0.7–4.7) | 0.22 |
|
| ||||||||||
| ATRX-WT | 16 (31%) | 46 (23–78)b | 69% | 2 (1.2-NA) | 1.40 | (0.68–2.92) | 0.36 | 0.78 | (0.33–1.87) | 0.58 |
| ATRX-MUT | 35 (69%) | 35 (21–63)b | 66% | 3.6 (2.1–6.7) | Ref | Ref | ||||
Patients with TERT-mutant tumors were older at diagnosis than patients with TERT-wildtype tumors (p=0.006).
Patients with ATRX-mutant tumors were younger at diagnosis than patients with ATRX-wildtype tumors (p=0.007);
Multivariate model includes age, TERT and ATRX status. There was no significant interaction of TERT and ATRX status in this group. (p=0.36)
Bolded values are statistically significant; 95% CI: 95% confidence interval; Ref: reference group.
Group 4 - Glioblastoma, IDH-wildtype
Group 4 had 309 cases and a median age at diagnosis of 59 (range 24–89) years (Table 5). Patients with tumors that had TERT promoter mutations were older at diagnosis than those with TERT-wildtype tumors (median ages 62 and 52 years, respectively, p<0.001). Patients who had tumors with ATRX alterations were younger than those who had tumors without ATRX alterations (median ages 38 and 60 years, respectively, p<0.001). As shown in Table 1 and Figure 2d, although a large majority of cases (n=238, 77%) had only TERT promoter mutation, 60 (19%) cases had neither TERT promoter nor ATRX alterations, 10 (3%) cases had only ATRX mutations and one case had both TERT and ATRX mutations. In univariate analyses (Table 5), older age at diagnosis, presence of TERT mutation and absence of ATRX alteration were associated with worse overall survival (p<0.0001, p=0.01 and p=0.001, respectively, Table 5). In the multivariate model including TERT and ATRX status, and stratified for age, presence of ATRX alterations (but not TERT status) remained independently significantly associated with better survival (p=0.01, Table 5 and Figure 2d).
Table 5.
Associations of age at diagnosis, and tumor TERT and ATRX status with overall survival in Group 4: IDH-wildtype glioblastomas.
| N (column %) | Age Median (range) | Percent deceased | Median survival years (95% CI) | Univariate analysis | Multivariate analysisc | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |||||
| All/Age (continuous) | 309 (100%) | 59 (24–89) | 81% | 1.2 (1.1–1.3) | 1.04 | (1.02–1.05) | <0.0001 | |||
|
| ||||||||||
| TERT-WT | 70 (23%) | 52 (24–85)a | 83% | 1.8 (1.2–2.2) | 0.67 | (0.5–0.91) | 0.01 | 0.90 | (0.65–1.25) | 0.53 |
| TERT-MUT | 239 (77%) | 62 (30–89)a | 80% | 1.1 (1.1–1.2) | Ref | Ref | ||||
|
| ||||||||||
| ATRX-WT | 298 (96%) | 60 (24–89)b | 81% | 1.2 (1.1–1.3) | Ref | Ref | ||||
| ATRX-MUT | 11 (4%) | 38 (33–61)b | 64% | 4.7 (2-NA) | 0.29 | (0.14–0.62) | 0.001 | 0.36 | (0.17–0.81) | 0.01 |
Patients with TERT-mutant tumors were older at diagnosis than patients with TERT-wildtype tumors (p<0.001).
Patients with ATRX-mutant tumors were younger at diagnosis than patients with ATRX-wildtype tumors (p<0.001).
Multivariate model includes TERT and ATRX status and was stratified by age (grouped as 24–53, 54–70, and 71–89) because age did not meet the Cox model’s proportionality assumptions.
In a model with ATRX alone, stratified by age at diagnosis, ATRX was significantly associated with survival better (hazard ratio: 0.35, 95% CI: 0.16–0.75, p value: 0.007)
Bolded values are statistically significant; 95% CI: 95% confidence interval; Ref: reference group.
Group 5- Astrocytoma, IDH-wildtype
Group 5 included 42 (27%) grade II and 112 (73%) grade III gliomas and had a median age at diagnosis of 52 (range 18–87) years (Table 6). Patients with grade III tumors were significantly older at diagnosis than patients with grade II tumors (median ages 54 and 46 years, respectively, p=0.006). As shown in Table 1 and Figure 2e, most cases in this group had only TERT promoter mutation (n=92, 60%), but 43 (28%) were wildtype for both TERT and ATRX, 16 (10%) had only ATRX alterations and 3 (2%) had both TERT and ATRX alterations. Patients with TERT-mutant tumors were significantly older at diagnosis than patients with TERT-wildtype tumors (median ages 56 and 37 years, respectively, p<0.001) (Table 6). Patients who had tumors with ATRX alterations were significantly younger than patients who had tumors without ATRX alterations (median ages 38 and 53 years, respectively, p=0.006). In univariate analyses (Table 6), older age at diagnosis, higher grade and presence of TERT promoter mutations were significantly associated with worse overall survival (p=0.0008, 0.03, and p=0.0003, respectively), while ATRX status was not associated with survival (p=0.41). In the multivariate model including grade, TERT and ATRX status, and stratified for age, higher grade and presence of TERT mutations (but not ATRX status) continued to be independently associated with significantly worse survival (Table 6 and Figure 2e).
Table 6.
Associations of age at diagnosis, tumor grade, TERT, and ATRX status with survival in Group 5: IDH-wildtype astrocytomas.
| N (column %) | Age Median (range) | Percent deceased | Median survival years (95% CI) | Univariate analysis | Multivariate analysisd,e | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |||||
| All/Age (continuous) | 154 (100%) | 52 (18–87) | 57% | 1.9 (1.7–2.3) | 1.03 | (1.01–1.04) | 0.0008 | |||
|
| ||||||||||
| Grade II | 42 (27%) | 46 (18–87)a | 52% | 3.4 (1.9–14.8) | Ref | Ref | ||||
| Grade III | 112 (73%) | 54 (23–75)a | 59% | 1.7 (1.4–2.2) | 1.69 | (1.04–2.76) | 0.03 | 1.72 | (1.03–2.88) | 0.04 |
|
| ||||||||||
| TERT-WT | 59 (38%) | 37 (18–75)b | 47% | 3.3 (2.1-NA) | 0.42 | (0.26–0.67) | 0.0003 | 0.48 | (0.27–0.87) | 0.02 |
| TERT-MUT | 95 (62%) | 56 (24–87)b | 63% | 1.7 (1.4–2) | Ref | Ref | ||||
|
| ||||||||||
| ATRX-WT | 135 (88%) | 53 (18–87)c | 56% | 1.9 (1.7–2.3) | Ref | Ref | ||||
| ATRX-MUT | 19 (12%) | 38 (23–70)c | 68% | 2.1 (1.4-NA) | 0.78 | (0.43–1.41) | 0.41 | 0.96 | (0.47–1.96) | 0.91 |
Patients with grade III tumors were older at diagnosis than patients with grade II tumors (p=0.006).
Patients with TERT-mutant tumors were older at diagnosis than patients with TERT-wildtype tumors (p<0.001).
Patients with ATRX-mutant tumors were younger at diagnosis than patients with ATRX-wildtype tumors (p=0.006).
Multivariate model includes grade, TERT and ATRX status and was stratified by age (grouped as 18–36, 37–42, 43–56, 57–66, and 67–87) because age did not meet the Cox model’s proportionality assumptions.
In a model with grade and TERT (without ATRX) and stratified by age at diagnosis, TERT status was still significantly associated with survival (hazard ratio: 0.48, 95% CI: 0.27–0.85, p value: 0.01)
There was no statistically significant interaction between TERT and ATRX in this group (p=0.31)
Bolded values are statistically significant; 95% CI: 95% confidence interval; Ref: reference group.
Summary
Figure 2 and Table 7 summarize the predominant telomere maintenance mechanism and which telomere maintenance mechanism alterations are associated with overall survival within WHO 2016 entities. TERT promoter mutation predominates in Group 1, IDH-mutant 1p/19q-codeleted tumors (96%), Group 4, glioblastoma, IDH-wildtype (77%), and Group 5, astrocytoma, IDH-wildtype (62%), while ATRX alterations predominate in Group 2, astrocytoma, IDH-mutant (79%) and Group 3, glioblastoma, IDH-mutant (69%). When TERT and ATRX alterations were modeled together, TERT promoter mutation was significantly associated with worse survival in IDH-wildtype astrocytomas (Group 5), while TERT mutation was significantly associated with better survival in IDH-mutant, 1p/19q-codeleted oligodendrogliomas (Group 1). Among IDH-wildtype glioblastoma patients (Group 4), ATRX mutation was significantly associated with better survival. Neither TERT nor ATRX status was significantly associated with survival among IDH-mutant astrocytomas (Group 2) or IDH-mutant glioblastoma (Group 3).
Table 7.
Summary of predominant telomere maintenance mechanism and which telomere maintenance mechanisms are associated with overall survival within WHO 2016 glioma entities.
| WHO 2016 Group | |||||
|---|---|---|---|---|---|
| Group 1 Oligodendroglioma, IDH-mutant and 1p/19q-codeleted |
Group 2 Astrocytoma, IDH-mutant |
Group 3 Glioblastoma, IDH-mutant |
Group 4 Glioblastoma, IDH-wildtype |
Group 5 Astrocytoma, IDH-wildtype |
|
| Predominant telomere maintenance (TERT promoter or ATRX alteration) | TERT (96%) | ATRX (79%) | ATRX (69%) | TERT (77%) | TERT (62%) |
|
| |||||
| Tumor telomere maintenance alteration most relevant to prognosis | TERT-WT ↓ survival | Neither significant | Neither significant | ATRX-WT ↓ survival | TERT-MUT ↓ survival |
|
| |||||
| Corresponding groups from Eckel-Passow et al. [6] based on presence or absence of IDH mutation, TERT promoter mutation, and 1p/19q codeletion. | Triple Positive | IDH-Mutant Only AND IDH +TERT-mutant | IDH-Mutant Only AND IDH +TERT-mutant | TERT-Mutant Only AND Triple Negative | TERT-Mutant Only AND Triple Negative |
DISCUSSION
The revised 4th edition of WHO 2016 Classification of tumors of the central nervous system incorporated molecular classification with histology to provide an integrated diagnosis [14]. For diffuse gliomas other than midline gliomas with H3K27M mutations, IDH mutations and 1p/19q codeletion constitute the main components of the integrated WHO 2016 diagnosis. However, because our current knowledge about genetic and epigenetic alterations in gliomas extends beyond these two markers, the prognostic roles of additional markers within gliomas diagnosed according to the WHO 2016 criteria is warranted. Proposed molecular classifications in the literature commonly include some combination of IDH, 1p/19q codeletion and the tumor’s telomere maintenance mechanism, defined by alterations in either TERT or ATRX (reviewed in [25]). To determine whether TERT or ATRX alteration status provide additional prognostic information, we analyzed their associations with overall survival in glioma patients classified according to the WHO 2016 criteria.
Upon review of the molecular and histologic features, we were able to classify nearly all study patients according to the new WHO 2016 criteria [14]. These groups demonstrated different survival outcomes with IDH-mutant, 1p/19q-codeleted, oligodendrogliomas (Group 1) having the most favorable results, followed by IDH-mutant astrocytomas (Group 2), IDH-mutant glioblastomas (Group 3), and IDH-wildtype glioblastomas (Group 4). The survival experience of IDH-wildtype astrocytomas (Group 5) was in between that of Groups 3 and 4 for about the first six years, and afterwards, was more similar to Group 3.
Group 1: Oligodendroglioma, IDH-mutant and 1p/19q-codeleted
Consistent with the literature [1, 24], the predominant telomere maintenance mechanism in this group was TERT promoter mutation (96%). A subset (n=5) showed concurrent TERT and ATRX alteration, but was too small for further statistical evaluation. This group may represent either misclassified tumors due to imperfect specificity of ATRX for assessing ALT status or tumors that acquired ATRX mutations as a later event. Patients with tumors lacking TERT promoter mutation had worse survival. A possible explanation could be that these tumors have false positive 1p/19q codeletion results, which would have led to misclassification of these cases as prognostically favorable oligodendrogliomas. It is known that small deletions in 1p and 19q may result in false positive 1p/19q codeletion results as tested by FISH in the absence of whole arm deletions [5]. In addition, 1p/19q codeletion in this study included those with relative 1p/19q codeletion (deletions in 1p and 19q in the presence of polysomy), which was reported to be associated with worse overall prognosis [4]. However, these possibilities are low since the whole arm deletions were confirmed through manual examination of tracings in nine out of these 13 cases. Among the remaining four cases, three were TP53-wildtype, which argues against a false positive 1p/19q codeletion. One case with unknown TP53 status had ATRX loss, which may be a concern regarding misclassification of an astrocytoma. While a single case is not sufficient to make a generalized conclusion, it suggests that tumors with discordant ATRX and 1p19q deletion results may warrant further testing to establish a definitive diagnosis.
Group 2- Astrocytoma, IDH-mutant (1p/19q-intact)
Among Group 2 IDH-mutant astrocytomas, significant prognostic indicators were age at diagnosis and grade in univariate models. Consistent with the literature [9, 10, 21, 24], the predominant telomere mechanism in Group 2 was ATRX alteration (79%). Given that rates of ATRX alterations are high in Group 2 IDH-mutant (1p/19q-intact) astrocytoma patients but rare in Group 1 IDH-mutant, 1p/19q-codeleted oligodendroglioma patients, ATRX status may be a useful diagnostic tool among IDH-mutant lower grade gliomas. However, 83 (21%) of all Group 2 IDH-mutant grade II and III gliomas lack both ATRX alterations and 1p/19q codeletion. Thus, the absence of ATRX alterations cannot be used as evidence for an IDH-mutant, 1p/19q-codeleted oligodendroglioma diagnosis without confirmation of 1p/19q codeletion. Previous studies reported frequent TP53 mutations in IDH-mutant, 1p/19q-intact gliomas, which correlated with significant p53 staining on IHC [3]. As noted in the methods, we did not have sufficient data about TP53 mutation status or p53 staining to examine this marker in the statistical analyses.
Whether ATRX status has a prognostic role among Group 2 IDH-mutant gliomas has been previously investigated. A prior study, which includes data that overlap with this study, suggested that among patients with IDH-mutant 1p/19q-intact lower grade gliomas, loss of ATRX expression may be associated with longer survival as compared to those with retained ATRX expression; however, the latter group was small (n=9) and there was poor statistical power [12]. Another study classifying gliomas into four subgroups as “IDH-mutant and 1p/19q-codeleted,” “IDH-mutant/ATRX-lost,” “IDH-mutant only,” and “IDH-wildtype” reported similar results; however, they did not provide a pairwise comparison between “IDH-mutant/ATRX-lost” and “IDH-mutant only” groups [16]. In addition, glioblastomas were included in the study and the final multivariate analysis with proposed groups did not include grade as a variable. In our study, neither TERT promoter mutation nor ATRX alterations were associated with overall survival in this group of astrocytoma, IDH-mutant patients. This suggests ATRX status may not have prognostic significance among IDH-mutant lower grade gliomas, beyond its almost mutually exclusive distribution with 1p/19q codeletion.
Group 3- Glioblastoma, IDH-mutant
IDH-mutant glioblastomas represented a small group, and frequently carried ATRX alterations, which were not associated with survival. Presence of TERT promoter mutation was associated with unfavorable outcomes in a univariate model, but not after adjusting for age. The small subset of patients with TERT-mutant tumors was significantly older than those with TERT-wildtype tumors, which may partially explain the unfavorable outcome associated with TERT promoter mutations. Nevertheless, the number of TERT-mutant IDH-mutant glioblastomas was too small to make a generalized statement. Given that Sanger sequencing is accepted as the gold standard for assessment of IDH status with high specificity, it is unlikely that these cases represent misclassified IDH-wildtype primary glioblastomas. Whether TERT promoter mutations represent additional molecular alterations during the progression of tumors in this group should be further studied. It should also be noted that 20% of IDH-mutant glioblastomas had neither TERT nor ATRX alterations and 6% had both, therefore; ATRX status cannot be used to infer TERT status in this group.
Group 4- Glioblastoma, IDH-wildtype
Consistent with the literature [1], most IDH-wildtype glioblastomas had TERT promoter mutations (77%). Only 4% of cases had ATRX alterations; these patients were younger at diagnosis and had significantly better overall survival, even in multivariate models stratified by age at diagnosis and controlling for radiation treatment. Assessment of ATRX status among IDH-wildtype glioblastomas is necessary to identify this group. Overall survival did not differ among those with or without TERT mutation. Furthermore, TERT promoter mutation status was not independently associated with overall survival in the multivariate models. Thus, clinical utility of routine TERT promoter mutation analysis among IDH-wildtype glioblastomas is unclear.
Previous studies demonstrated an interaction between the predictive role of MGMT methylation and TERT status in this group [2, 17]. One study showed that the prognostic influence of MGMT promoter methylation depended on the presence of TERT promoter mutations among IDH-mutant glioblastoma patients who were treated with standard chemoradiation [17]. Another study showed a significant interaction between TERT status and MGMT methylation status among IDH-wildtype glioblastoma patients treated with radiation and temozolamide, and proposed four subgroups [2]. However, they have only provided the hazard ratios without confidence intervals or p values to assess the significance of any of the pairwise associations. Unfortunately, our data regarding the MGMT methylation is limited to the TCGA cases only, which does not have sufficient power to perform a subgroup analysis. In addition, some of our cases were treated before the temozolamide era, and chemotherapy regimens in this current study are not well-defined precluding further assessment of “predictive” role of TERT mutations.
Group 5- Astrocytoma, IDH-wildtype
Among the astrocytoma, IDH-wildtype group, TERT promoter mutation was common, occurring in (62%) of patients. These patients were significantly older than those without TERT promoter mutation and had shorter overall survival, similar to our previous report using a dataset with significant overlap to that used here [6]. These gliomas most likely represent the difficulty in pathologic classification of under-sampled IDH-wildtype glioblastomas displaying intra-tumoral heterogeneity. Another possibility is that the tumor is in an early phase of evolution and not all histological features that are typically associated with high grade are present.
A subset of IDH-wildtype astrocytomas without TERT or ATRX alterations may correspond to those that share genetic and epigenetic features with pilocytic astrocytomas [3]. All patients in this dataset are “adults”, however, there is the possibility that there are “pediatric type” tumors, which we have not accounted for. For example, we cannot exclude the possibility that some of the IDH-wildtype astrocytomas indeed represent pediatric-type oligodendrogliomas without IDH mutations or 1p/19q codeletion. Similarly, there may be an unaccounted for group of pediatric-type high grade astrocytomas among IDH-wildtype gliomas in groups 4 and 5. We do not have sufficient data regarding recurrent mutations in pediatric high grade tumors including those in the histone coding genes. H3 G34R (or less likely G34V) mutations exclusive to hemispheric gliomas, which are frequently seen in teenagers and young adults, and associated with favorable prognosis may correspond to a subset of IDH-wildtype astrocytomas with ATRX mutations in this study [23]. We have also not included one of the new diagnostic entities in 2016 WHO, “diffuse midline glioma, H3 K27M-mutant,” in our classification. These tumors may have been mixed among IDH-wildtype glioblastomas and astrocytomas. These tumors are expected to be TERT-wildtype and a subset may show ATRX mutations [5]. Future studies analyzing additional molecular alterations are necessary for further classification of IDH-wildtype infiltrating gliomas.
Role of histologic grade
The criteria for histologic grading of diffuse gliomas are from the pre-IDH era, and do not account for the prognostic effects of the molecular classification. We found that among patients with IDH-mutant and 1p/19q-codeleted oligodendrogliomas, those with grade III tumors were older and had reduced overall survival. While several studies in the literature reported grade to be a significant predictor of outcome among oligodendrogliomas, they had not considered molecular features in their diagnosis [7, 18]. While Scheie et al. reported grade and 1p/19q status to be independent prognostic factors among cases with histologic oligodendroglial component, it was unclear whether grade was significant within the 1p/19q-codeleted group or if its significance was driven by cases with higher grade tumors including the no-longer recognized group of “glioblastoma with oligodendroglioma, grade IV” [22]. More recent studies reported that grade was not significantly associated with survival among IDH-mutant 1p/19q-codeleted oligodendrogliomas [19, 24]. Our results indicate that among molecularly defined oligodendrogliomas, grade was not significantly associated with overall survival in multivariate models. A caveat is that we did not include treatment data beyond radiation in our model. Furthermore, there is an inherent bias in almost all retrospective studies regarding the treatment given as part of standard of care, since chemotherapy with or without radiation treatment is usually reserved for patients with grade III tumors. Similar to our study, most studies in the literature are retrospective in nature and do not account for treatment variation. Additional studies including prospective randomized treatment data are warranted to further assess this issue.
Among patients with IDH-mutant astrocytomas, those with grade III tumors had worse outcomes than patients with grade II tumors. A similar association with grade and survival was also seen in patients with IDH-wildtype astrocytomas. Previous studies in the literature reported a significant survival difference between patients with grade II and grade III astrocytomas; however, the majority of these studies was done in the pre-IDH era and therefore may lack generalizability to modern patient cohorts [18]. More recently, among patients with tumors diagnosed with strict integrated molecular criteria, grade II and grade III astrocytomas did not show significant age or survival differences [21, 24]. Generalization of our results requires caution given our models do not control for concurrent clinical features such as extent of surgical resection, and lack consistent treatment (chemotherapy and/or radiation) across both grades. Furthermore, it is unclear whether the current grading criteria will similarly hold up among IDH-mutant cases versus IDH-wildtype cases. Additional studies specifically examining the role of grading among astrocytomas are needed.
One weakness in this study is the lack of availability for consistently obtained treatment data within each diagnostic group. Owing to the use of three different data sources, the availability of these variables was limited. For example, extent of resection, one of the clinical parameters with known survival effect, was only available in the TCGA subset of lower grade tumors, while 95% of all glioblastoma cases were classified as resection, extent unknown. Similarly, Karnofsky performance status was only available for TCGA cases. Since our analyses are conducted within WHO 2016 diagnostic entities, further reducing the sample sizes by including only the TCGA subset led to insufficient power for an analysis controlling for any of these clinical parameters. Radiation treatment (as given versus not given) was the only treatment variable that was similarly categorized across various data sources. Secondary analyses including radiation in the final multivariate models showed that radiation did not confound any of the associations with TERT or ATRX status in any of the groups. Unfortunately, most retrospective studies, even those with treatment protocol data from a single institution suffer from this bias due to the changes in diagnostic criteria and treatment modalities over time. New prospective clinical trials with standard treatment protocols for diagnostic groups based on integrated diagnoses are essential and these results should be validated in an external dataset.
Our data for ATRX alterations came from either loss of protein expression tested by immunohistochemistry (UCSF AGS and Mayo Clinic), or from sequencing data (TCGA). While the frequency of ATRX alterations in TCGA data was somewhat lower, this may have been due to the different technique. It is generally accepted that protein expression and mutation data are highly correlated. But some missense mutations may not be associated with protein loss, and genetic and epigenetic alterations other than ATRX mutations may lead to loss of protein expression [8–10]. However, earlier studies suggested that loss of protein expression has better correlation with ALT status than ATRX mutations [8]. It is unclear whether the loss of ATRX expression or the presence of ATRX gene mutations is the better prognostic indicator among glioma subtypes, and this topic needs further evaluation.
In summary, this study combines three large cohorts of adult glioma patients that have been successfully re-classified according to the new integrated diagnosis criteria in the revised 4th edition of the WHO 2016. We demonstrated that presence of TERT promoter mutation was associated with favorable outcomes among Group 1 IDH-mutant 1p/19q-codeleted oligodendrogliomas, and with unfavorable outcomes among Group 5 IDH-wildtype astrocytomas. Presence of ATRX alterations was independently associated with favorable outcomes among Group 4 IDH-wildtype glioblastomas. Thus, we present evidence that in certain subgroups, testing for TERT promoter mutation or ATRX alterations may have utility in the clinical management of glioma patients.
Supplementary Material
Acknowledgments
Work at University of California, San Francisco was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, R01CA139020 and R25CA112355), as well as the loglio Collective, the National Brain Tumor Foundation, The Sontag Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen.
Work at the Mayo Clinic was supported by the National Institutes of Health (grant numbers P50CA108961 and P30CA15083), National Institute of Neurological Disorders and Stroke (grant number RC1NS068222Z), the Bernie and Edith Waterman Foundation, and the Ting Tsung and Wei Fong Chao Family Foundation.
This publication was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement # U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred.
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov
The authors wish to acknowledge study participants, the clinicians and research staff at the participating medical centers, the UCSF Helen Diller Family Comprehensive Cancer Center Genome Analysis Core which is supported by a National Cancer Institute Cancer Center Support Grant (5P30CA082103), the UCSF Cancer Registry, the UCSF Neurosurgery Tissue Bank, Katherine Cornelius, the late Dr. Bernd Scheithauer, the Mayo Clinic Center for Individualized Medicine, and the Mayo Clinic Comprehensive Cancer Center Biospecimens and Processing and Genotyping Shared Resources.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ETHICAL APPROVAL
All procedures performed were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
INFORMED CONSENT
Informed consent was obtained from all individual participants included in this study.
References
- 1.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 2.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M, Shimizu S, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4:79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlain MC, Born D. Prognostic significance of relative 1p/19q codeletion in oligodendroglial tumors. J Neurooncol. 2015;125:249–251. doi: 10.1007/s11060-015-1906-y. [DOI] [PubMed] [Google Scholar]
- 5.Clark KH, Villano JL, Nikiforova MN, Hamilton RL, Horbinski C. 1p/19q testing has no significance in the workup of glioblastomas. Neuropathol Appl Neurobiol. 2013;39:706–717. doi: 10.1111/nan.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giannini C, Scheithauer BW, Weaver AL, Burger PC, Kros JM, Mork S, Graeber MB, Bauserman S, Buckner JC, Burton J, et al. Oligodendrogliomas: reproducibility and prognostic value of histologic diagnosis and grading. J Neuropathol Exp Neurol. 2001;60:248–262. doi: 10.1093/jnen/60.3.248. [DOI] [PubMed] [Google Scholar]
- 8.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, Heguy A, Petrini JH, Chan TA, Huse JT. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leeper HE, Caron AA, Decker PA, Jenkins RB, Lachance DH, Giannini C. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015;6:30295–30305. doi: 10.18632/oncotarget.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 14.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer, City; 2016. [Google Scholar]
- 15.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 16.Mur P, Mollejo M, Hernandez-Iglesias T, de Lope AR, Castresana JS, Garcia JF, Fiano C, Ribalta T, Rey JA, Melendez B. Molecular classification defines 4 prognostically distinct glioma groups irrespective of diagnosis and grade. J Neuropathol Exp Neurol. 2015;74:241–249. doi: 10.1097/nen.0000000000000167. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen HN, Lie A, Li T, Chowdhury R, Liu F, Ozer B, Wei B, Green RM, Ellingson BM, Wang HJ, et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2016 doi: 10.1093/neuonc/now189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 19.Olar A, Sulman EP. Molecular Markers in Low-Grade Glioma-Toward Tumor Reclassification. Semin Radiat Oncol. 2015;25:155–163. doi: 10.1016/j.semradonc.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, City; 2016. [Google Scholar]
- 21.Reuss DE, Mamatjan Y, Schrimpf D, Capper D, Hovestadt V, Kratz A, Sahm F, Koelsche C, Korshunov A, Olar A, et al. IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol. 2015;129:867–873. doi: 10.1007/s00401-015-1438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheie D, Meling TR, Cvancarova M, Skullerud K, Mork S, Lote K, Eide TJ, Helseth E, Beiske K. Prognostic variables in oligodendroglial tumors: a single-institution study of 95 cases. Neuro Oncol. 2011;13:1225–1233. doi: 10.1093/neuonc/nor114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 25.Walsh KM, Wiencke JK, Lachance DH, Wiemels JL, Molinaro AM, Eckel-Passow JE, Jenkins RB, Wrensch MR. Telomere maintenance and the etiology of adult glioma. Neuro Oncol. 2015;17:1445–1452. doi: 10.1093/neuonc/nov082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 27.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang P, Cai J, Yan W, Zhang W, Wang Y, Chen B, Li G, Li S, Wu C, Yao K, et al. Classification based on mutations of TERT promoter and IDH characterizes subtypes in grade II/III gliomas. Neuro Oncol. 2016;18:1099–1108. doi: 10.1093/neuonc/now021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


