Abstract
Coenzyme Q10 (CoQ10) or ubiquinone is one of the two electron carriers in the mitochondrial respiratory chain which has an essential role in the process of oxidative phosphorylation. Defects in CoQ10 synthesis are usually associated with the impaired function of CoQ10–dependent complexes I, II and III. The recessively transmitted CoQ10 deficiency has been associated with a number of phenotypically and genetically heterogeneous groups of disorders manifesting at variable age of onset. The infantile, multisystemic presentation is usually caused by mutations in genes directly involved in CoQ10 biosynthesis. To date, mutations in COQ1 (PDSS1 and PDSS2), COQ2, COQ4, COQ6, COQ7, COQ8A/ADCK3, COQ8B/ADCK4, and COQ9 genes have been identified in patients with primary form of CoQ10 deficiency. Here we report novel mutations in the COQ4 gene, which were identified in an infant with profound mitochondrial disease presenting with perinatal seizures, hypertrophic cardiomyopathy and severe muscle CoQ10 deficiency.
Keywords: CoQ10 deficiency, CoQ4, Infantile cardiomyopathy, Encephalomyopathy, Whole exome sequencing
1. Introduction
Coenzyme Q10, also known as ubiquinone, is a lipophilic component of all cellular membranes and has multiple metabolic functions. In the mitochondrial electron-transport chain, it transfers electrons from complex I (NADH dehydrogenase-ubiquinone oxidoreductase) and complex II (succinate dehydrogenase) to complex III (ubiquinone-cytochrome c reductase). It also receives electrons from de novo pyrimidine biosynthesis, fatty acid, amino acid, and sulfide oxidation [1]. In addition to its central role in the mitochondrial respiratory chain as an electron carrier, CoQ10 also plays a role as an antioxidant in the cell membrane [2] and may modulate apoptosis [3]. Diseases associated with CoQ10 deficiency are phenotypically heterogeneous and can manifest at any age. Five major clinical phenotypes have been described: (i) an encephalomyopathic form, characterized by mitochondrial myopathy and recurrent myoglobinuria and central nervous signs [4], [5]; (ii) an ataxic form dominated by cerebellar ataxia and cerebellar atrophy [6], [7]; (iii) an isolated myopathic form with lipid storage and respiratory dysfunction [8], [9]; (iv) a multisystemic infantile form with encephalopathy and renal disease [10], [11] and v) nephropathy [12], [13], [14].
The precise biosynthesis pathways of CoQ10 in mammals are yet to be characterized. However, in yeast, and possibly in mammals, at least 12 mitochondrial enzymes are known to participate in CoQ10 biosynthesis [15]. Mutations in PDSS1 (MIM607429) [16], PDSS2 (MIM610564) [10], COQ2 (MIM609825) [16], [17], COQ4 (MIM 616227) [18], [19], COQ6 (MIM614647) [20], COQ7 (MIM616733) [21], COQ8A/ADCK3 (MIM 612016) [22], COQ8B/ADCK4 (MIM615573) [12], and CoQ9 (MIM614654) [23] genes have been described in patients with early-onset multisystemic mitochondrial encephalopathy, cardiomyopathy, and renal failure associated with severe CoQ10 deficiency. To date, primary CoQ10 deficiency due to mutations in COQ4 (CoQ10 D7, MIM616276) has been described in twelve patients from nine unrelated families. All affected patients but one harbored either homozygous or compound heterozygous mutations in the COQ4 gene [18], [19]. A single case of CoQ10 deficiency was reported to be associated with haploinsufficiency of COQ4 [24].
Here we report novel mutations in COQ4 identified in an infant presenting with early onset cardiomyopathy, hypotonia, hearing loss, seizures, and lactic acidosis associated with severe muscle CoQ10 deficiency.
2. Case presentation
The infant male was born at term to a 24-year-old gravida 3 para 1 mother. The pregnancy was uncomplicated with normal serology and ultrasounds. Apgar scores were 1 at 1 min and 7 at 5 min and positive pressure ventilation was required at delivery. On the second day of life, the infant had an apneic event with bradycardia and reduction in oxygen saturation. A chest X-ray was performed which noted cardiomegaly. Biventricular hypertrophy was observed by echocardiogram. Lactate levels were persistently elevated at 4–6 mM (nl < 2.4 mM) during the first several weeks of life but subsequently normalized. Seizures were noted on the fifth day of life and the patient required intubation after the initiation of anti-epileptic medications. The patient was successfully extubated at 11 days of age. Seizure management was initially acceptable, but the patient eventually became refractory to multi-drug therapy (phenobarbital, topiramate, clobazam). Additional problems included gastroesophageal reflux requiring fundoplication, delayed visual maturation without structural abnormality of the eyes, bilateral hearing loss, profound hypotonia and absence of development. There was no evidence of renal dysfunction, transaminitis or liver synthetic dysfunction.
Brain MRI studies at one week of age showed focal regions of cortical increased T1 signal and magnetic resonance spectroscopy identified enlarged lactate peaks. Subsequent MRI at 10 weeks showed microcephaly with volume loss and increasing prominence of lactate peaks.
His initial hospitalization lasted for 4 months. He was discharged on palliative measures but was readmitted six days later with profound acidosis and respiratory failure. Ventilator support was withdrawn with the agreement of his parents and he expired.
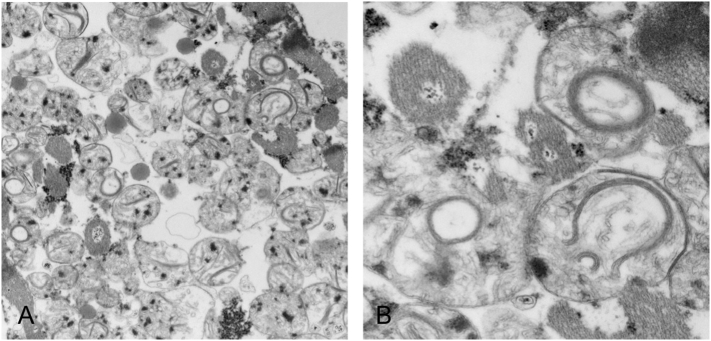
At autopsy, there was marked bi-ventricular cardiac hypertrophy. EM studies of cardiomyocytes mitochondria showed swollen mitochondria with loss of cristae and some mitochondria with semi-circular arrangements of cristae (Fig. 1). Light microscopy and EM studies of mitochondria in the kidney and liver were unremarkable.
Fig. 1.
Electron micrographs of cardiac muscle show strikingly abnormal mitochondrial morphology, with unusual semi-circular arrangements of crista noted in multiple myofiber mitochondria, which are non-artefactual in nature. A & B, transmission electron microscopy, A × 17,200; B × 57,500.
3. Biochemical studies
CoQ10 concentration in muscle extract and in skin fibroblasts were measured by reversed-phase high-performance liquid chromatography and electrochemical detection system methodology as described previously [7]. The levels of CoQ10 in autopsied skeletal muscle (6.9 μg/g tissue; normal range 19.6–46.8) and in skin fibroblasts (19.1 μg/mg protein; normal range 45.4–65.7) were found to be severely reduced. Due to the tissue availability and specimen quality, we were able to determine the activities of the electron transport complexes only in skin fibroblasts using the methods described [25]. As shown in the table, consistent with the low CoQ10 level found in fibroblasts, the combined activities of CoQ10-dependent complexes i.e., rotenone-sensitive NADH cytochrome c reductase (complex I + III) and succinate-cytochrome c reductase (complex II + III) were also found to be significantly reduced, whereas the activities of other complexes were essentially comparable to those found in normal controls.
4. Molecular studies
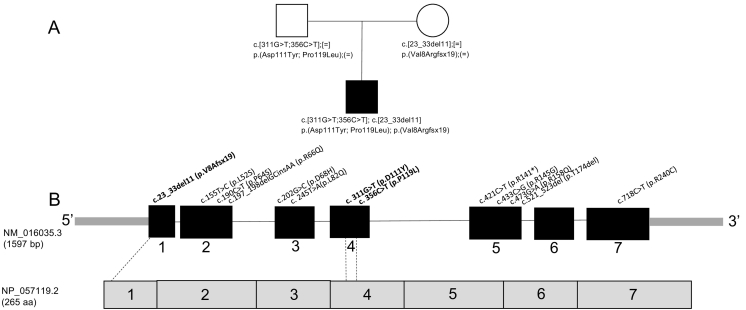
High throughput next generation sequencing of the whole mitochondrial genome as well as 319 mitochondrial nuclear genes (Mito Nuclear Gene Panel, GeneDx® Co., Gaithersburg, MD) identified three previously unreported heterozygous variants in the COQ4 gene: a 11 base pair deletion (c.23_33del11; p.Val8AlafsX19), and two missense mutations: c.331G > T; p.Asp111Tyr and c.356C > T; p.Pro119Leu. Parental testing showed that the frameshift mutation was maternally inherited and the two point mutations were paternally inherited as shown in the family pedigree (Fig. 2A). Sequencing of mitochondrial DNA showed several uncommon variants, but none were pathogenic.
Fig. 2.
Family pedigree and location of CoQ4 mutations.
A. Family pedigree showing inheritance of mutations identified in this study.
B. Location of mutations on CoQ4 gene (novel mutations described here are shown in bold).
The frameshift mutation (c.23_33del11) is not present in the Exome Aggregation Consortium (ExAC) database (Cambridge, Massachusetts, USA; URL: http://exac.broadinstitute.org), dbSNP, or ClinVar. It results in truncation of the COQ4 protein at residue 26 and the mRNA would be subject to nonsense mediated decay. The c.311G > T mutation is present at a frequency of 1/121200 alleles in ExAC, and is also present in dbSNP (rs530213004), but has not been observed in ClinVar. In silico analysis predicts that the resulting coding mutation (p.Asp111Tyr) would be deleterious (Provean, − 7.40) and damaging (SIFT, 0.001) to protein function. The c.356C > T mutation is present in the ExAC database at a frequency of 1/121238 alleles, but is not observed in dbSNP or ClinVar. In silico analysis likewise predicts that the resulting p.Pro119Leu missense mutation would have a deleterious (Provean, − 9.32) and damaging (SIFT, 0.002) effect on protein function.
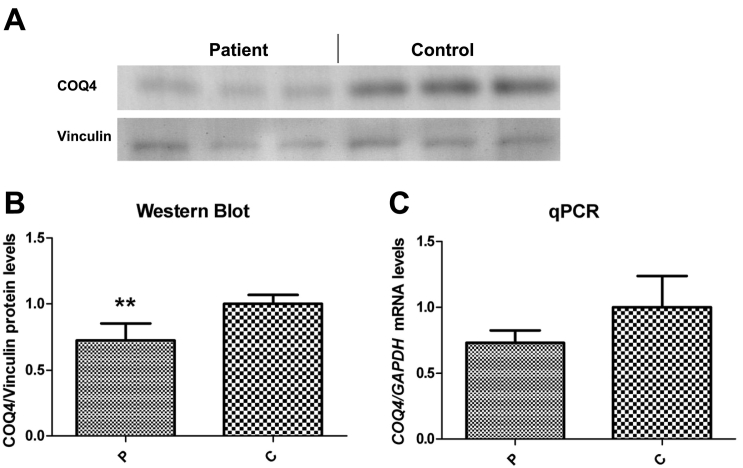
To confirm the expected effects upon the RNA and protein, we performed quantitative real-time PCR analysis (qPCR) for the COQ4 transcript and Western blotting. Western blotting shows a significant loss in the level of the COQ4 protein, consistent with a loss in immunoreactive protein encoded by the frameshift allele (Fig. 3A, B). qPCR showed a reduced level of the COQ4 transcript, which would be anticipated due to nonsense-mediated decay of the frameshift transcript (Fig. 3C).
Fig. 3.
A. Representative Western blot showing the level of CoQ4 protein in skin fibroblasts from patient (left) and control (right). B. Relative level of protein normalized to vinculin. C. Relative level of CoQ4 transcript normalized to GAPDH. Error bars represent standard deviations of six experiments. Mann-Whitney U test. **Indicates a value of p > 0.01.
5. Discussion
In the inner membrane of human mitochondria, CoQ10 functions primarily as a mobile electron carrier which is necessary for ATP production through the oxidative phosphorylation system. In addition, CoQ10 also participates in other important cellular functions such as pyrimidine biosynthesis, modulating apoptosis, and activation of mitochondrial uncoupling proteins [15]. Thus, because of its multiple cellular functions, CoQ10 deficiency would be expected to cause the impairment of several different biochemical pathways and produce different clinical diseases [3]. Given that CoQ10 is synthesized endogenously and dietary contribution to the cellular content is negligible, the reduction of CoQ10 observed in CoQ10 defects directly results from defects in the biosynthesis pathway. To date, mutations in nine genes participating in CoQ10 biosynthesis have been associated with primary CoQ10 deficiency. It is intriguing that although all of these gene products function in the same pathway to synthesize CoQ10, their clinical presentations are quite distinct. For example, mutations in PDSS2, encoding for one of the two subunits of COQ1 (the first committed enzyme in the CoQ10 biosynthetic pathway) have been reported in an infant with severe Leigh's syndrome, nephrotic syndrome and CoQ10 deficiency [10], whereas autosomal recessive mutations in COQ2, encoding for the para-hydroxybenzoate-polyprenyl transferase (the second committed enzyme in the pathway) was reported in patients with variable clinical presentation of the multisystem infantile form of CoQ10 deficiency or isolated nephropathy [13], [14], [26], [27], [28]. It is tempting to speculate that this clinical heterogeneity may reflect specific problem associated with defects at different steps in the CoQ10 biosynthetic pathway. However, the fact that different molecular defects in the same gene (i.e. COQ2) cause extremely variable clinical manifestations suggests that the impact may be only a product of the overall loss of CoQ10 synthesis. A correlation between the CoQ10 content of yeast expressing diverse CoQ2 mutations and phenotypes in S. cerevisiae has recently been reported [29].
Although the complex biosynthesis of CoQ10 in yeast has been extensively studied, in humans, its synthetic pathway is less well characterized. In particular, the exact function of the COQ4 protein in this pathway still remains unknown. However, based on studies in yeast and human cells, it may function as the core component in the multisubunit complex required for CoQ10 biosynthesis [30], [31]. To date, eleven recessive mutations in COQ4 have been reported in patients presenting neonatal encephalopathy and severe CoQ10 deficiency (Fig. 2B). Chung et al. [19]reported five recessive missense COQ4 mutations (Arg66Glu, Asp68His, Leu82Gln, Arg158Gln, Arg240Cys) in five patients with severe, early-onset mitochondrial disease presenting encephalopathy and/or cardio-myopathy and lactic acidosis. In this study, biochemical (low muscle CoQ10 content and impaired respiratory chain enzyme activities) data were provided for only one patient who harbored compound heterozygous variants of Leu82Gln/Arg158Gln. Therefore, although the authors provide strong molecular evidence in support of the pathogenicity of these variants, further biochemical and functional data are required to ascertain, with high certainty, the status of Arg66Glu, Asp68His, and Arg240Cys variants (Table 1).
Table 1.
Citrate synthase-normalized activities of mitochondrial electron transport chain enzyme (ETC) in the patient's skin fibroblasts.
| MRC enzyme | Patient | Control (range) |
|---|---|---|
| Complex I/CS | 0.63 | 0.86–1.70 |
| Complex II/CS | 1.86 | 1.53–1.87 |
| Complex I + III/CS | 0.11 | 0.55–1.30 |
| Complex II + III/CS | 0.08 | 0.20–0.79 |
| Complex IV/CS | 0.54 | 0.37–1.74 |
| Complex V/CS | 8.02 | 5.07–11.35 |
| Citrate synthase (nmol/min/mg protein) | 48.15 | 49.60–56.18 |
In the study reported by Brea-Calvo et al. [18], six pathogenic, autosomal recessive, variants were reported in five patients with early-onset mitochondrial disease and CoQ10 deficiency (Fig. 2B). The authors validated the pathogenicity of these variants by conducting extensive functional studies in a recombinant yeast model. Although the clinical presentations of these patients were heterogeneous, all patients had early-onset of the disease and four out of five had early fatal outcome. The only surviving member of this cohort in the study who reached adulthood is an 18-year-old male proband in who harbored a homozygous missense (c.190C > T; p. Pro64Ser) variant in exon 2 of COQ4 gene. Interestingly, although this variant was accompanied by a profound reduction in ETC complex I, a 70% reduction in complex II + III, and a 50% reduction in complex III, there was minimal effect on the level of CoQ10 in the muscle when compared to that found in controls [18]. Therefore, from these data it becomes tempting to hypothesize that perhaps, at least in human, the c.190C > T transition in the second exon of CoQ4 gene may have minimal functional impact on the CoQ4 protein and consequently on the CoQ10 biosynthesis. The underlying cause of the profound reduction in the complex I activity detected in this patient in the cohort, was not explained. However, it is noteworthy that deficiency of complex I activity was found also in fibroblasts carrying PDSS2 mutations [10]. Moreover, analysis by blue native gel electrophoresis in cerebellum of Coq9x/x mice showed reduced level of CI, supporting the hypothesis that CoQ10 deficiency causes instability of super complex I + III [32]. In Chung et al. [19], five out of five patients had hypotonia and five out of six patients presented with cardiomyopathy, yet the prevalence of hypotonia and cardiomyopathy in the cohort reported by Brea-Calvo et al. [18]was one in five and two in five respectively. Nevertheless, amongst all reported patients (including ours) harboring COQ4 mutations, cardiomyopathy is the most prominent features in 8/12 patients.
Cardiomyopathy is a frequent manifestation of the multisystemic form of CoQ10 deficiency, and has been described in association with mutations in PDSS1 [16], PDSS2 [33], COQ2 [28], and COQ9 [11], [34]. Intriguingly, whereas nephropathy is a prevalent manifestation in patients harboring mutations in COQ1 (PDSS2 subunit) [10], COQ2 [17], COQ6 [20], and COQ9 [23] genes, to date, none of the mutations of COQ4 gene identified in patients with CoQ10 deficiency were associated with renal dysfunction. This could be due to the different level of COQ4 expression in different tissues.
6. Conclusion
In this study, we report three novel variants in the COQ4 gene that are associated with a severe early-onset mitochondrial disease and profound CoQ10 deficiency. Identification of disease-causing mutations in genes involved in CoQ10 biosynthesis is important as it may lead not only into a new insight in the pathogenesis of CoQ10 deficiency, but also to early diagnosis and treatment of these disorders.
Declaration of conflict of interest
None.
Ethical approval
This retrospective case report did not require ethics committee approval at our institution.
Acknowledgements
Part of this research was supported by National Institutes of Health grants 5P01 HD080642-02 NIH/NICHD (to ABN).
References
- 1.Vafai S.B., Mootha V.K. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491(7424):374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 2.Bentinger M., Brismar K., Dallner G. The antioxidant role of coenzyme Q. Mitochondrion. 2007;7(Suppl):S41–S50. doi: 10.1016/j.mito.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Quinzii C.M., DiMauro S., Hirano M. Human coenzyme Q10 deficiency. Neurochem. Res. 2007;32(4–5):723–727. doi: 10.1007/s11064-006-9190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Giovanni S. Coenzyme Q10 reverses pathological phenotype and reduces apoptosis in familial CoQ10 deficiency. Neurology. 2001;57(3):515–518. doi: 10.1212/wnl.57.3.515. [DOI] [PubMed] [Google Scholar]
- 5.Ogasahara S. Improvement of abnormal pyruvate metabolism and cardiac conduction defect with coenzyme Q10 in Kearns-Sayre syndrome. Neurology. 1985;35(3):372–377. doi: 10.1212/wnl.35.3.372. [DOI] [PubMed] [Google Scholar]
- 6.Lamperti C. Cerebellar ataxia and coenzyme Q10 deficiency. Neurology. 2003;60(7):1206–1208. doi: 10.1212/01.wnl.0000055089.39373.fc. [DOI] [PubMed] [Google Scholar]
- 7.Musumeci O. Familial cerebellar ataxia with muscle coenzyme Q10 deficiency. Neurology. 2001;56(7):849–855. doi: 10.1212/wnl.56.7.849. [DOI] [PubMed] [Google Scholar]
- 8.Horvath R. Characterization of human SCO1 and COX17 genes in mitochondrial cytochrome-c-oxidase deficiency. Biochem. Biophys. Res. Commun. 2000;276(2):530–533. doi: 10.1006/bbrc.2000.3495. [DOI] [PubMed] [Google Scholar]
- 9.Lalani S.R. Isolated mitochondrial myopathy associated with muscle coenzyme Q10 deficiency. Arch. Neurol. 2005;62(2):317–320. doi: 10.1001/archneur.62.2.317. [DOI] [PubMed] [Google Scholar]
- 10.López L.C. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 2006;79(12):1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman S. Neonatal presentation of coenzyme Q10 deficiency. J. Pediatr. 2001;139(3):456–458. doi: 10.1067/mpd.2001.117575. [DOI] [PubMed] [Google Scholar]
- 12.Ashraf S. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J. Clin. Invest. 2013;123(12):5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diomedi-Camassei F. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J. Am. Soc. Nephrol. 2007;18(10):2773–2780. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- 14.Salviati L. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65(4):606–608. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- 15.Turunen M., Olsson J., Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660(1–2):171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Mollet J. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J. Clin. Invest. 2007;117(3):765–772. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinzii C.M. A mutation in para-hydroxybenozoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am. J. Hum. Genet. 2006;78(2):345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brea-Calvo G. COQ4 mutations cause a broad spectrum of mitochondrial disorders associated with CoQ10 deficiency. Am. J. Hum. Genet. 2015;96(2):309–317. doi: 10.1016/j.ajhg.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung W.K. Mutations in COQ4, an essential component of coenzyme Q biosynthesis, cause lethal neonatal mitochondrial encephalomyopathy. J. Med. Genet. 2015;52(9):627–635. doi: 10.1136/jmedgenet-2015-103140. [DOI] [PubMed] [Google Scholar]
- 20.Heeringa S.F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 2011;121(5):2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freyer C. Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J. Med. Genet. 2015;52(11):779–783. doi: 10.1136/jmedgenet-2015-102986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagier-Tourenne C. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am. J. Hum. Genet. 2008;82(3):661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan A.J. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am. J. Hum. Genet. 2009;84(5):558–566. doi: 10.1016/j.ajhg.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salviati L. Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J. Med. Genet. 2012;49(3):187–191. doi: 10.1136/jmedgenet-2011-100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spinazzi M. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 2012;7(6):1235–1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- 26.Jakobs B.S. A novel mutation in COQ2 leading to fatal infantile multisystem disease. J. Neurol. Sci. 2013;326(1–2):24–28. doi: 10.1016/j.jns.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy H.J. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin. J. Am. Soc. Nephrol. 2013;8(4):637–648. doi: 10.2215/CJN.07200712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scalais E. Early myoclonic epilepsy, hypertrophic cardiomyopathy and subsequently a nephrotic syndrome in a patient with CoQ10 deficiency caused by mutations in para-hydroxybenzoate-polyprenyl transferase (COQ2) Eur. J. Paediatr. Neurol. 2013;17(6):625–630. doi: 10.1016/j.ejpn.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Desbats M.A. The COQ2 genotype predicts the severity of coenzyme Q10 deficiency. Hum. Mol. Genet. 2016;25(19):4256–4265. doi: 10.1093/hmg/ddw257. [DOI] [PubMed] [Google Scholar]
- 30.Belogrudov G.I. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 2001;392(1):48–58. doi: 10.1006/abbi.2001.2448. [DOI] [PubMed] [Google Scholar]
- 31.Casarin A. Functional characterization of human COQ4, a gene required for coenzyme Q10 biosynthesis. Biochem. Biophys. Res. Commun. 2008;372(1):35–39. doi: 10.1016/j.bbrc.2008.04.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Corzo L. Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum. Mol. Genet. 2013;22(6):1233–1248. doi: 10.1093/hmg/dds530. [DOI] [PubMed] [Google Scholar]
- 33.Rötig A. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356:391–395. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- 34.Duncan A.J. Decreased ubiquinone availability and impaired mitochondrial cytochrome oxidase activity associated with statin treatment. Toxicol. Mech. Methods. 2009;19(1):44–50. doi: 10.1080/15376510802305047. [DOI] [PubMed] [Google Scholar]