Abstract
Background and Objectives:
Helicobacter pylori (H. pylori) infection is cause of several gastrointestinal diseases in humans. Virulence genes of H. pylori are associated with severity of disease and vary geographically. The aim of present study was to detect H. pylori in formalin-fixed paraffin-embedded (FFPE) tissues and further investigate prevalence of babA2, cagA, iceA1, iceA2, vacA s1/s2 and vacA m1/m2 genotypes in H. pylori from gastric cancer (GC) and gastric ulcer (GU) patients’ biopsy samples.
Methods:
We used FFPE tissues of 35 GC and 10 GU patients’ biopsy samples. Using Polymerase Chain Reaction (PCR), detection of H. pylori strain was performed by using specific primers targeting 16S rRNA and ureC encodes for phosphoglucosamine mutase genes. We have identified different virulence genes of H. pylori by PCR.
Results:
Of all the 45 samples tested, 20 GC and all 10 GU samples were positive for identification of H. pylori using specific genes (16S rRNA and ureC). The prevalence of babA2(100%) was significantly higher in GC as compared to GU (40%) samples. The rate of virulence genes vacAs1 was higher in both GU 8 (80%) and GC (100%).
Conclusions:
Our study finds that vacAs1am1 and babA2 are most prominent genotypes and may play role in development of Gastric cancer.
Keywords: H. pylori, Genotyping, Gastric ulcer, Gastric cancer
INTRODUCTION
Helicobacter pylori (H. pylori) is a pathogenic bacterium that inhabits gastric mucosa of humans and cause several gastrointestinal diseases including gastric cancer (GC). In developing countries prevalence of H. pylori infection may exceed as compared to developed countries where 20-50% are affected with infection.1 Previous studies suggest that H. pylori is genetically variable and certain genotypes are only detected in certain populations.2 Several virulence factors of H. pylori strain, such as babA2, cagA, vacA, and iceA1 have been identified and involved in pathogenesis of infection.3
Recently reported blood group antigen-binding adhesin (BabA), a membrane protein of H. pylori help in binding to gastric epithelium. Three different bab alleles have been reported where only the babA2 is functional for binding activity.4 Previous studies have reported prevalence of babA2 positive strain in peptic ulcer and GC but with conflicting results.5 Therefore, relationship between H. pylori genotype and disease condition may vary from one geographic region to other.
The cytotoxin-associated gene A (cagA) is present in almost 50% of H. pylori strains and is constituent of genomic pathogenecity island (cag-PAI) responsible for type IV secretion system. The cagA-positive strains of H. pylori are responsible for mucosal inflammation and interleukin-8 (IL-8) production and are associated with pathogenesis of gastric cancer.6 In Asian countries, rate of cagA positivity has been reported in almost all strains of H. pylori isolated from infected cases.7
The vacuolating cytotoxin gene (vacA) encodes for vacuolating cytotoxin and found in all strains of H. pylori. It is involved in pathogenesis of peptic ulcer and GC by injuring gastric epithelial cells.8 All strains of H. pylori contain the vacA gene, and have two variable parts i.e signal region (s1/s2) and middle region (m1/m2). Previous studies have reported vacA allelic variations in different geographical regions as well as their toxic activity.9
Recently another virulence gene of H. pylori, iceA (the induced by contact with epithelium) has been reported. The iceA gene has two allelic types, iceA1 and iceA2 where iceA1 is significantly expressed and associated with peptic ulceration.10 The ureC encodes for phosphoglucosamine mutase, and renamed as glmM gene. The ureC (glmM) gene is easily detectable in H. pylori strains and involved in the development of the bacterial cell wall and growth.11
In Saudi Arabia, H. pylori infection rate is high in the Eastern, Central and Western region of the Kingdom. Previous studies have reported some virulence genes such as cagA, iceA1, and iceA2 in peptic ulcer and gastric ulcer in Saudi population.12,13 Detection of virulence genes of H. pylori mentioned above has not been reported yet in GC patients in Saudi Arabia. The genotyping of H. pylori is useful to determine epidemiological importance of H. pylori strain. Therefore, we designed a study to detect the presence of H. pylori using specific glmM and 16S rRNA. Further prevalence of virulence genes babA2, cagA, iceA1, iceA2, vacA s1/s2 and vacA m1/m2 from paraffin-embedded (FFPE) gastric biopsies collected from GC and GU Saudi patients have been studied.
METHODS
Biopsy samples Collection and DNA extraction
Gastric FFPE biopsy specimens were collected from 35 GC patients and 10 with GU who had undergone gastric endoscopy in King Abdulaziz University (KAU) hospital in Jeddah, Saudi Arabia, between 2000-2014. Written informed consent was taken from all the patients. This study was approved by the Ethics and Research Committees of the hospital KAU. DNA was extracted from the biopsies using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Extracted DNA was further used for PCR.
Polymerase Chain Reaction (PCR) assays
To detect H. pylori strain in biopsy samples from GC and GU patients specific primers targeting 16S rRNA and ureA genes were used. Further prevalence of virulence factors of H. pylori strain, specific primers targeting babA2, cagA, iceA1, iceA2, vacA s1/s2, vacA m1/m2 were used for PCR (Table-I). Amplification was conducted in a total volume of 25μL. The reaction mixture contained 13 μL, 2X ready PCR mix (Thermo Scientific), 2μL of each forward and reverse primers (Table-I), 1 ug DNA template, and 9μL nuclease free distilled water to a total volume of 25μL. The PCR amplification was performed according to the following program: an initial denaturation at 95°C for five minutes, followed by 35 cycles of denaturation at 95°C for 50s, annealing for 50s (Table-I), and a final extension at 72°C for five minutes. The amplified PCR products were electrophorsed (100 V/35 min) using 1% agarose gel. DNA ladder of 100 bp (Norgen, Canda) was used to determine the size of the amplified bands.
Table-I.
Primers set and conditions used for PCR in this study.
| Genes | Nucleotide sequence (5'-3') | PCR product (bp) | PCR conditions | Reference |
|---|---|---|---|---|
| 16S rRNA | GCGACCTGCTGGAACATTAC CGTTAGCTGCATTACTGGAGA |
138 bP | 95ºC, 50 s; 60ºC, 50s; 72ºC, 50 s (35 cycles) |
28 |
| glmM | AAGCTTTTAGGGGTGTTAGGGGTTT AAGCTTACTTTCTAACACTAACGC |
294 bp | 95ºC, 50 s; 56ºC, 50s; 72ºC, 50 s (35 cycles) |
23 |
| babA2 | CCAAACGAAACAAAAAGCGT GCTTGTGTAAAAGCCGTCGT |
271 bp | 95ºC, 50 s; 44ºC, 50s; 72ºC,50 s (35 cycles) |
29 |
| iceA1 | GTGTTTTTAACCAAAGTATC CTATAGCCATTATCTTTGCA |
247 bp | 95ºC, 50 s; 57ºC, 50s; 72ºC,50 s (35 cycles) |
29 |
| iceA2 | GTTGGGTATATCACAATTTAT TTTCCCTATTTTCTAGTAGGT |
229 bp | 95ºC, 50 s; 57ºC, 50s; 72ºC,50 s (35 cycles) |
28 |
| cagA | TTGACCAACAACCACAAACCGAAG CTTCCCTTAATTGCGAGATTCC |
183 bp | 95ºC, 50 s; 60ºC, 50s; 72ºC,50 s (35 cycles) |
35 |
| vacA s1/s2 | ATGGAAATACAACAAACACAC CTGCTTGAATGCGCCAAAC |
259/286 bp | 95ºC, 50 s; 58ºC, 50s; 72ºC,50 s (35 cycles) |
20 |
| vacA m1/m2 | CAATCTGTCCAATCAAGCGAG GCGTCAAAATAATTCCAAGG |
567/642 bp | 95ºC, 50 s; 54ºC, 50s; 72ºC,50 s (35 cycles) |
14 |
Statistical Analysis
Data of two groups were compared by using chi-square test using SPSS statistical software Version 9 (SPSS Inc., Chicago, IL, USA). A P value less <0.05 was considered as significant.
RESULTS
This study included 45 FFPE gastric biopsy samples from Saudi patients. A total of 45 gastric patients, 13 (28.8%) females and 32 (71.2%) males ranging in age from 18 to 87 years, were included in the study. Among 45 cases, 35 (77.7%) were diagnosed as GC and 10 (22.3%) were GU cases (Table-II).
Table-II.
Demographic characteristics and prevalence of virulence genes in different groups.
| Group I (GU) (n=10) | Group II (GC) (n=35) | |
|---|---|---|
| Age: mean | 31.3±7.5 | 58.2±14.6 |
| Gender: | ||
| Male | 3 (30%) | 29 (82.9%) |
| Female | 7 (70%) | 6 (17.1%) |
| babA2 | 4 (4%) | 20 (100%) |
| iceA1 | 0 (0) | 3 (15%) |
| iceA2 | 0 (0) | 3 (15%) |
| cagA | 3 (30%) | 8 (40%) |
| vacA s1 | 5 (50%) | 12 (60%) |
| vacA s2 | 0 (0) | 0 (0) |
| vacA s1 s2 | 3 (30%) | 8 (40%) |
| vacA m1 | 0 (0) | 3 (15%) |
| vacA m2 | 2 (20%) | 3 (15%) |
| vacA m1/m2 | 0 (0) | 0 (0) |
The detection of H. pylori was investigated using PCR. Among 45 FFPE gastric tissues, 20 (57.2%) GC and 10 (100%) GU biopsy samples were positive for H. pylori using 16S rRNA (Fig. 1a and b) and glmM genes. We examined six different H. pylori virulence genes in gastric biopsy samples. Among H. pylori positive gastric tissues prevalence of babA2 gene was higher (100%) in GC tissues as compare to GU (4%). The presence of cagA gene yield a fragment of 183 bp using PCR. Amplification of iceA1 and iceA2 gene was performed using specific primers (Table-II). Prevalence of both iceA1 and iceA2 was detected only in three GC samples (15%) and was negative for GU samples. Only eight samples (40%) from GC and three (30%) from GU was positive for cagA. There was no significant difference for different genotypes among two different goups of patients (P > 0.05).
Fig.1.
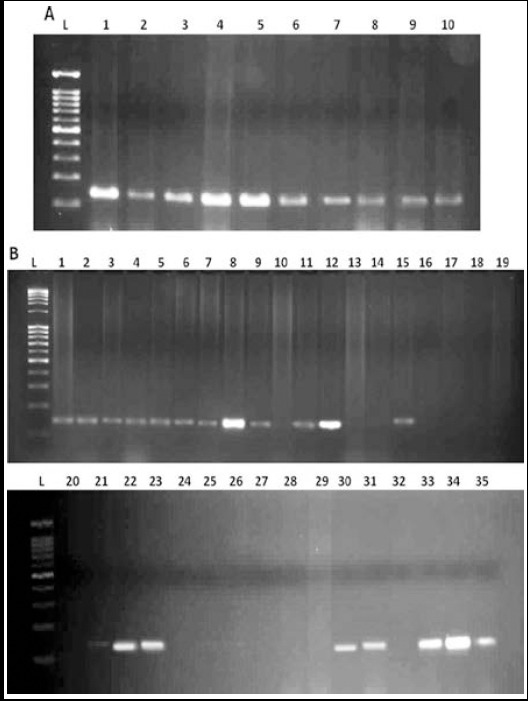
Detection of 16s rRNA gene. (A) Lane L, marker; 1-10, GU samples (B) Lane L, marker; 1-35, GC samples.
The detection of vacA s1/s2 allele showed presence in 20 (100%) samples from GC and 8 (80%) from GU. The most virulent vacA s1 was detected in most of the samples as it was detected in 12 (60%) samples from GC and 5 (50%) samples from GU. Whereas, vacA s2 was not detected in any sample. Combined vacA s1/s2 was detected in 8 (40%) GC and three (30%) GU samples. Detection of vacA m1/m2 yielded fragment of 567/642 bp product. Only two (20%) samples were positive for vacA m2 from GU samples. While for GC samples, allele vacA m1 was detected in 3 (15%) samples and vacA m2 was also detected in 3 (15%) samples. Combined vacA m1/m2 was not detected in any sample (P > 0.05). In our study vacA s1 was common allele in GC while vacA s1/s2 was predominant genotype in GU samples (Table-II).
DISCUSSION
Gastric cancer is an important cause of the death not only in Saudi Arabia but all over the world. Several previous studies have reported the importance of the H. pylori virulence genotypes. However, very few studies are available regarding H. pylori genotypes in Saudi population. Our study has confirmed high prevalence of H. pylori infection in GC and GU biopsy samples. In this study we have investigated the prevalence of various virulence factors (babA2, cagA, iceA1, iceA2, vacA s1/s2, vacA m1/m2) from H. pylori positive GC and GU biopsy samples using PCR. Prevalence of H. pylori and Gastric Cancer is high in Asian countries such as Japan and Korea as compared to other countries.14 In this study we have seen high prevalence of H. pylori virulence factors in GC and GU samples. Different genes contribute in colonization of H. pylori to gastric epithelium of humans.15 The glmM and 16S rRNA gene has more sensitivity than other genes for detection in gastric biopsies samples.16 Therefore, we have used glmM and 16S rRNA for detection of H. pylori in biopsy samples.
Virulence genes of H. pylori cagA and vacA are important adherence factors involved in gastrointestinal diseases.17-19 In our study we found low frequency of cagA in both groups. cagA is associated with the development of gastric carcinoma and it is an important marker for the most virulent strains associated with gastric severe infection.20 Previous studies have shown relationship of gastric cancer and cagA positive H. pylori21 while other show contradictory results like we have in our study. We have used VacAs1/s2 and VacAm1/m2 type virulence factor for detection and according to our results prevalence of VacAs1 is high in both GU and GC. VacAs1 genotype is associated with high toxin activity and severe diseases. Previous study from China has also reported similar results.22
The babA2 gene has been shown to be associated with high risk of gastric cancer and also has strong relation with VacAs1.8 In GC samples we have found high prevalence of babA2 and VacAs1. A previous study has reported similar results where these genotypes were associated with increased risk of developing cancer.23
H. pylori iceA1 positive strains are associated with peptic ulcers due to the production of IL-8 produce by these strains.24 In our study prevalence of iceA1 and iceA2 is lower in both GC and GU samples. Our study is consistent with some previous studies25 while contradictory results are found in other.26 Therefore, the both alleles only detected in GC samples but not in GU. The babA2 gene of H. pylori is an important contributor for higher risk of GU and GC development. Our results are similar to previous study where H. pylori strains carrying babA2, cagA, and vacAs1 genotypes were associated with the risk of intestinal cancer.8 Several virulence factors contribute in the pathogenesis of H. pylori have been studied here.
CONCLUSIONS
Our study report first time high incidence of H. pylori virulence genes in Saudi patients with GC and GU. Prevalence of cagA genotype is not associated with severity of disease and iceA1 allele was detected in GC patients only. We have seen in our study high frequency of genes babA2 and VacAs1 in GC patients.
Authors’ Contributions
FB made substantial contributions to design this study.
SA, MY, and EIA were involved in PCR and data interpretation.
AJF, SAS, and AS were responsible for sample collection and clinical databases.
FB drafted the manuscript.
All authors have read and approved the final manuscript.
Footnotes
Declaration of interest: The authors declare that they have no conflict of interests.
Funding: This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH)–King Abdulaziz City for Science and Technology the Kingdom of Saudi Arabia award number (12BIO2725-03). The authors also, acknowledge with thanks Science and Technology Unit, King Abdulaziz University for technical support.
REFERENCES
- 1.Miwa H, Go MF, Sato N. H. pylori and gastric cancer:the Asian enigma. Am J Gastroenterol. 2002;97(5):1106–1112. doi: 10.1111/j.1572-0241.2002.05663.x. doi:10.1111/j.1572-0241.2002.05663.x. [DOI] [PubMed] [Google Scholar]
- 2.Sicinschi LA, Correa P, Peek RM, Camargo MC, Delgado A, Piazuelo MB, et al. Helicobacter pylori genotyping and sequencing using paraffin-embedded biopsies from residents of Colombian areas with contrasting gastric cancer risks. Helicobacter. 2008;13(2):135–145. doi: 10.1111/j.1523-5378.2008.00554.x. doi:10.1111/j.1523-5378.2008.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erzin Y, Koksal V, Altun S, Dobrucali A, Aslan M, Erdamar S, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter. 2006;11(6):574–580. doi: 10.1111/j.1523-5378.2006.00461.x. doi:10.1111/j.1523-5378.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 4.Pride DT, Meinersmann RJ, Blaser MJ. Allelic variation with in Helicobacter pylori babA and babBInfect. Immun. 2001;69(2):1160–1171. doi: 10.1128/IAI.69.2.1160-1171.2001. doi:10.1128/IAI.69.2.1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattar R, dos Santos AF, Eisig JN, Rodrigues TN, Silva FM, Lupinacci RM, et al. No correlation of babA2 with vacA and cagA genotypes of Helicobacter pylori and grading of gastritis from peptic ulcer disease patients in Brazil. Helicobacter. 2005;10(6):601–608. doi: 10.1111/j.1523-5378.2005.00360.x. doi:10.1111/j.1523-5378.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 6.Van der Ende A, Pan ZJ, Bart A, et al. cagA-positive Helicobacter pylori populations in China and the Netherlands are distinct. Infect Immun. 1998;66(5):1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito Y, Azuma T, Ito S, et al. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35(7):1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Angchan A, Ripa B, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12(1):30–36. doi: 10.1016/j.ijid.2007.03.012. doi:10.1016/j.ijid.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Atherton JC, Cao P, Peek RM. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori or Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 10.Peek RM, Jr, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene iceA that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110(6):531–544. [PubMed] [Google Scholar]
- 11.Espinoza MGC, Vazquez RG, Mendez IM, Vargas CR, Cerezo SG. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J Clin Microbiol. 2011;49(4):1650–1652. doi: 10.1128/JCM.00461-10. doi:10.1128/JCM.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.BinSaeed AA. Glimpse of the epidemiological research on Helicobacter pylori in Saudi Arabia. Saudi J Gastroenterol. 2009;15(2):85. doi: 10.4103/1319-3767.48963. doi:10.4103/1319-3767.48963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadi RH, Halawani EM, Abdelkader HS. Prevalence of H.pylori strains harbouring cagA and iceA virulence genes in Saudi patients with gastritis and peptic ulcer disease. Microbiology Discovery. 2014;2(1):1. doi:10.1155/2015/753710. [Google Scholar]
- 14.World Cancer Report 2014. World Health Organization. 2014;(Chapter 5. 4) ISBN 9283204298. [Google Scholar]
- 15.Wang G, Maier RJ. A RecB-like helicase in Helicobacter pylori is important for DNA repair and host colonization. Infect Immunity. 2009;75(1):286–291. doi: 10.1128/IAI.00970-08. doi:10.1128/IAI.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol. 1997;179(11):3488–3493. doi: 10.1128/jb.179.11.3488-3493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanih NF, McMillan M, Naidoo N, Ndip LM, Weaver LT, Ndip RN. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in South African patients with upper gastrointestinal diseases. Acta Tropica. 2010;116(1):68–73. doi: 10.1016/j.actatropica.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Miehlike S, Schuppler M, Frings C, et al. Helicobacter pylori vacA, iceA and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease. Am J Gastroenterol. 2001;96(4):1008–1030. doi: 10.1111/j.1572-0241.2001.03685.x. doi:10.1111/j.1572-0241.2001.03685.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA cagA and vacA status and clinical outcome:studies in four different countries. J Clin Microbiol. 1999;37(7):2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chih-Ho L, Ju-Chun H, Chiang-Ni C, Ju-Pi L, Lii-Tzu W, Hua-Shan W, et al. Mixed Infections of Helicobacter pylori Isolated from Patients with Gastrointestinal Diseases in Taiwan. Gastroenterol Res Pract. 2014;2014 doi: 10.1155/2016/7521913. Article ID 7521913, 7 pages. doi:10.1155/2016/7521913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiwari SK, Manoj G, Kumar GV, Sivaram G, Hassan SI, Prabhakar B, et al. Prognostic significance of genotyping Helicobacter pylori infection in patients in younger age groups with gastric cancer. Postgrad Med J. 2008;84(990):19. doi: 10.1136/pgmj.2007.065060. doi:10.1136/pgmj.2007.065060. [DOI] [PubMed] [Google Scholar]
- 22.Aziz F, Chen X, Yang X, Yan Q. Prevalence and correlation with clinical diseases of Helicobacter pylori cagA and vacA genotype among gastric patients from Northeast China. BioMed Research international. 2014;2014 doi: 10.1155/2014/142980. Article ID 142980, 7 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höcker M, Hohenberger P. Helicobacter pylori virulence factors—one part of a big picture. Lancet. 2005;362(9391):1231–1233. doi: 10.1016/S0140-6736(03)14547-3. [DOI] [PubMed] [Google Scholar]
- 24.Al Qabandi A, Mustsfa AS, Siddique I, Khajah AK, Madda JP, Junaid TA. Distribution of vacA and cagA genotypes of Helicobacter pylori in Kuwait. Acta Trop . 2005;93(3):283–288. doi: 10.1016/j.actatropica.2005.01.004. doi:10.1016/j.actatropica.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Lin HJ, Perng CL, Lo WC, Wu CW, Tseng GY, Li AFY, et al. Helicobacter pylori cagA, iceA and vacA genotypes in patients with gastric cancer in Taiwan. World J Gastroenterol. 2004;10(17):2493–2497. doi: 10.3748/wjg.v10.i17.2493. doi:10.3748/wjg.v10.i17.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SI, Kirsch C, Oyedeji KS, Arigbabu AO, Coker AO, Bayerdöffer E, et al. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in Nigerian patients with duodenal ulcer disease. J Med Microbiol. 2002;51(10):851–854. doi: 10.1099/0022-1317-51-10-851. doi:10.1099/0022-1317-51-10-851. [DOI] [PubMed] [Google Scholar]


